Abstract
Contraction of skeletal and cardiac muscle is regulated by Ca2+-dependent structural changes in troponin that control the interaction between myosin and actin. We measured the orientations of troponin domains in skeletal muscle fibers using polarized fluorescence from bifunctional rhodamine probes on the C and E helices of troponin C. The C helix, in the regulatory head domain, tilts by ≈30° when muscle is activated in physiological conditions, with a Ca2+-sensitivity similar to that of active force. Complete inhibition of active force did not affect C-helix orientation, and binding of rigor myosin heads did not affect its orientation at saturating [Ca2+]. The E helix, in the IT arm of troponin, tilted by ≈10° on activation, and this was reduced to only 3° when active force was inhibited. Binding of rigor myosin heads produced a larger tilt of the E helix. Thus, in situ, the regulatory head acts as a pure Ca2+-sensor, whereas the IT arm is primarily sensitive to myosin head binding. The polarized fluorescence data from active muscle are consistent with an in vitro structure of the troponin core complex in which the D and E helices of troponin C are collinear. The present data were used to orient this structure in the fiber and suggest that the IT arm is at ≈30° to the filament axis in active muscle. In relaxed muscle, the IT arm tilts to ≈40° but the D/E helix linker melts, allowing the regulatory head to tilt through a larger angle.
Contraction of skeletal and cardiac muscle is triggered by Ca2+ ions binding to troponin in the actin-containing thin filaments (1–4). The regulatory action of troponin is mediated by tropomyosin, which binds to seven actin monomers along each strand of the filament. In resting muscle tropomyosin covers the myosin binding sites on actin. On activation, Ca2+ binding to troponin leads to an azimuthal motion of tropomyosin around the actin filament that uncovers the myosin binding sites, permitting the actin-myosin interaction that drives contraction (5–7).
Recent crystal structures of the troponin core complex (8, 9) have focused attention on this key component of the muscle regulatory machinery (Fig. 1). The “regulatory head” of troponin is the physiological Ca2+-sensor, containing the N-terminal lobe of the troponin C chain (TnC) and the regulatory Ca2+ sites (10, 11). Ca2+ binding to these sites opens the TnC N-lobe and exposes a hydrophobic patch that binds the switch region of troponin I (TnI) (12–15). The “IT arm” is a rigid domain containing a long coiled-coil formed by α-helices from TnI and troponin T (TnT), plus the C-terminal lobe of TnC with its two Ca/Mg binding sites. The relative orientation of the IT arm and the regulatory head differs between the published crystal structures of the core complex (8, 9), suggesting that a flexible junction between the two domains, corresponding to that between the D and E helices of TnC, may allow interdomain motion. In the core complex of skeletal muscle troponin in the Ca2+-saturated state, the D and E helices of TnC are continuous (Fig. 1), but in the Ca2+-free state the D/E linker is melted (9). The junction between the two domains of TnC in the core complex in solution is more mobile in the absence of Ca2+ (16).
Fig. 1.
Structure of the core complex of troponin from skeletal muscle in the Ca2+-saturated form (9), containing troponin C (TnC; red, turquoise, green) and parts of troponin I (TnI; blue) and troponin T (TnT; gold). Bifunctional rhodamine probes cross-linked either cysteines 56 and 63 (yellow spheres) along the C helix (turquoise) or 96 and 103 along the E helix (green) of TnC.
These in vitro studies of the troponin core complex suggest that the relative orientation of the regulatory head and IT arm may play a fundamental role in the regulatory mechanism. However, many components of the regulatory machinery are missing from the core complex, including parts of TnI and TnT, tropomyosin, actin and myosin. Myosin plays an active role in the regulatory mechanism. Thin filaments can be switched on in the absence of Ca2+ at low concentrations of MgATP, when myosin displaces tropomyosin to form a strong “rigor” bond with actin (17), and myosin·ATP complexes can bind to thin filaments in the absence of Ca2+ at low ionic strength (18). These observations emphasize the need to complement in vitro studies of isolated protein fragments or partial complexes by studies of the intact regulatory machinery in the cellular environment.
Here we report measurements of the orientation of the regulatory head and IT arm of troponin in muscle fibers. We introduced bifunctional rhodamine probes (19, 20) into the regulatory head, cross-linking cysteines along the C helix of TnC, and into the IT arm, cross-linking cysteines along its E helix (Fig. 1). We used the polarized fluorescence from these probes to measure the orientation change of each helix when Ca2+ binds to the regulatory sites, and when myosin heads bind to actin. This in situ molecular structural approach preserves the native structure, interrelationships and function of all of the components of the regulatory machinery. The results allowed the regulatory head and IT arm to be oriented on the actin filament, and showed that these two troponin domains have distinct sensitivities to the binding of Ca2+ and myosin heads.
Results
In Situ Orientation of the C Helix of Troponin C.
Troponin C with bifunctional rhodamine (BR) cross-linking residues 56 and 63 (TnC-BR56–63) was incorporated into demembranated muscle fibers to measure the orientation of the C helix of TnC in the regulatory head of troponin (Fig. 1). The relationship between the isometric force produced by the fibers and pCa (i.e., −log [Ca2+]) was measured before and after introduction of TnC-BR56–63, and the data were fitted to the Hill equation (see Methods). pCa for half-maximum force, pCa50, was 6.00 ± 0.08 in fibers containing TnC-BR56–63, significantly (P < 0.05) smaller than that in control fibers, 6.40 ± 0.04, but the steepness (nH) of the force–pCa relationship was not significantly affected by TnC-BR56–63 (Table 1). Maximum isometric force was reduced slightly, to 82 ± 6% (n = 5) of that before TnC exchange, but a similar reduction was observed with unlabeled TnC, and is likely to be a nonspecific effect of the TnC exchange protocol (20). Thus, the only functional effect of labeling the C helix of TnC with BR was a modest decrease in Ca2+affinity. The NMR structure of the N-terminal lobe of TnC with bound Ca2+ is not altered by introducing cysteines at positions 56 and 63 and cross-linking them with BR (15).
Table 1.
Ca2+-sensitivity of force and troponin C orientation parameters
| Parameter | C helix; TnC-BR56–63 |
E helix; TnC-BR96–103 |
Native MgATP (n = 11) | ||||
|---|---|---|---|---|---|---|---|
| MgATP (n = 5) | Rigor (n = 4) | MgATP, BTS (n = 4) | MgATP (n = 5) | Rigor (n = 4) | MgATP, BTS (n = 4) | ||
| Force | |||||||
| pCa50 | 6.00 ± 0.08 | 6.30 ± 0.03 | 6.40 ± 0.04 | ||||
| nH | 2.71 ± 0.18 | 2.51 ± 0.18 | 2.62 ± 0.40 | ||||
| 〈P2〉 | |||||||
| pCa50 | 6.02 ± 0.03 | 6.00 ± 0.02 | 6.25 ± 0.03 | 5.98 ± 0.03 | |||
| nH | 1.94 ± 0.15 | 2.01 ± 0.05 | 2.10 ± 0.12 | 2.04 ± 0.15 | |||
| pCa 9 | 0.422 ± 0.021 | 0.207 ± 0.018 | 0.421 ± 0.020 | 0.196 ± 0.007 | −0.005 ± 0.012 | 0.211 ± 0.014 | |
| pCa 4.5 | −0.126 ± 0.006 | −0.142 ± 0.007 | −0.115 ± 0.006 | −0.039 ± 0.007 | −0.123 ± 0.014 | 0.121 ± 0.007 | |
| 〈P4〉 | |||||||
| pCa 9 | 0.128 ± 0.027 | 0.047 ± 0.020 | 0.138 ± 0.032 | −0.135 ± 0.005 | −0.064 ± 0.010 | −0.160 ± 0.006 | |
| pCa 4.5 | −0.012 ± 0.021 | 0.005 ± 0.018 | −0.025 ± 0.015 | −0.045 ± 0.010 | −0.024 ± 0.023 | −0.131 ± 0.004 | |
| θME, ° | |||||||
| pCa 9 | 36.8 ± 1.1 | 47.0 ± 0.9 | 36.8 ± 1.0 | 47.5 ± 0.3 | 57.0 ± 0.6 | 46.8 ± 0.7 | |
| pCa 4.5 | 63.0 ± 0.3 | 63.9 ± 0.4 | 62.5 ± 0.3 | 58.7 ± 0.3 | 62.8 ± 0.8 | 50.9 ± 0.3 | |
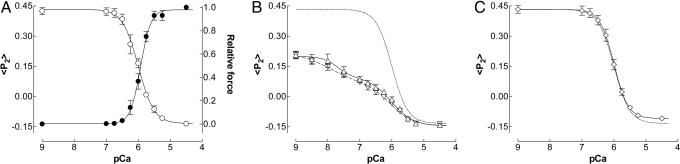
The polarized fluorescence intensities from muscle fibers containing TnC-BR56–63 were used to calculate the order parameters 〈P2〉 and 〈P4〉 that describe the orientation of the BR dipole, and therefore that of the C helix to which it is attached, with respect to the muscle fiber axis (21). 〈P2〉 would be +1 if every BR dipole were parallel to the fiber axis, and −0.5 if they were all perpendicular. 〈P4〉 provides higher resolution angular information, and orientational disorder decreases the absolute values of both 〈P2〉 and 〈P4〉. The average angle between the BR dipole and the fiber axis was described as the mean (θME) of the maximum entropy orientation distribution, the smoothest distribution consistent with the 〈P2〉 and 〈P4〉 data (22).
In relaxing conditions (pCa 9), 〈P2〉 and 〈P4〉 for TnC-BR56–63 were 0.422 ± 0.021 and 0.128 ± 0.027 respectively (Table 1), and θME was 37°. During active contraction (pCa 4.5) 〈P2〉 and 〈P4〉 decreased to −0.126 ± 0.006 and −0.012 ± 0.021 respectively, and θME was 63°. The average angle between the C helix and the fiber axis increases by ≈26° on muscle activation.
The normalized changes in 〈P2〉 and 〈P4〉 had the same Ca2+-dependence in all conditions studied here, and detailed results are presented only for 〈P2〉, which can be measured with greater signal-to-noise ratio. If there are two populations of BR dipoles with distinct orientations, the observed 〈P2〉 is linearly related to the fraction of dipoles in each population. The normalized 〈P2〉–pCa relationship therefore provides a linear measure of the fraction of troponin molecules in the Ca2+-bound conformation. The pCa50 for 〈P2〉 was 6.02 ± 0.03, almost identical to that for force in these fibers (Fig. 2A and Table 1), and nH for 〈P2〉 was 1.94 ± 0.15, close to the value of 2 expected for a structural change that requires binding of two Ca2+ ions. Force had a significantly (P < 0.05) steeper Ca2+-dependence than 〈P2〉, indicating a nonlinear relationship between active force and the fraction of TnC molecules that have bound Ca2+.
Fig. 2.
Ca2+-dependence of 〈P2〉 (open symbols) and force (filled circles) in muscle fibers containing TnC-BR56–63. (A) Activation in the presence of 5 mM MgATP. (B) Rigor with either 1.0 mM (open triangles) or 0.1 mM (inverted open triangles) free Mg2+. (C) Activation with force inhibition by 0.1 mM BTS. Fitted curves from the single (A and C) or double (B) Hill equation (see text for details); the fit to the 〈P2〉 data in A is reproduced as a dotted curve in B and C.
When muscle fibers were transferred from relaxing solution to Ca2+-free rigor (i.e., MgATP-free) solution, conditions in which binding of rigor myosin heads is expected to switch on the thin filament in the absence of Ca2+ (7, 17), 〈P2〉 and 〈P4〉 decreased and θME increased by 10° (Table 1). The effect of rigor myosin heads on the orientation of the C helix at pCa 9 is smaller than that of Ca2+ binding in the presence of MgATP, which increased θME by 26°.
Addition of Ca2+ in rigor produced a further decrease in 〈P2〉 (Table 1). 〈P2〉 and 〈P4〉 at pCa 4.5 were independent of the presence of MgATP, and θME was 63–64° in both conditions. The orientation of the C helix at saturating [Ca2+] is the same in active contraction and rigor. The Ca2+-dependence of the C helix orientation in rigor (Fig. 2B; 1 mM free Mg2+, open triangles) was distinct from that in the presence of MgATP (dotted line). The rigor data were fitted by the sum of two Hill equations, corresponding to Ca2+ binding sites with different affinities. To constrain the fit, the lower affinity sites were assumed to be identical to those observed in the presence of MgATP, with pCa50 6.0, nH 1.94 (Table 1). The double Hill plot (continuous line) gave a good fit to the data (open triangles). The higher affinity sites had pCa50 7.61, nH 1.67, accounting for ≈40% of the total change in 〈P2〉, and may correspond to the Ca/Mg sites in the C-terminal lobe (11). Consistent with this explanation, the 〈P2〉–pCa relation was shifted leftwards when free [Mg2+] was reduced to 0.1 mM (inverted open triangles).
To assess the effect of actively cycling myosin heads on the orientation of the C helix in the presence of MgATP, we used 0.1 mM N-benzyl-p-toluenesulfonamide (BTS; 23) to abolish active force. Strikingly, BTS had no effect on 〈P2〉 and 〈P4〉 for TnC-BR56–63 at any [Ca2+] in the pCa range 9 to 4.5, or on the fitted pCa50 and nH values (Fig. 2C and Table 1). These results show that, in contrast with the effect of rigor myosin heads, actively cycling myosin heads do not influence the orientation of the C helix or the Ca2+ affinity of the regulatory sites. In physiological conditions the in situ orientation of the C helix, and by implication the degree of opening and orientation of the TnC N-lobe, is solely determined by the Ca2+ occupancy of the regulatory sites.
In Situ Orientation of the E Helix of Troponin C.
The orientation of the TnC E helix in the IT arm (Fig. 1) was determined by using a TnC mutant with bifunctional rhodamine cross-linking residues 96 and 103 (TnC-BR96–103). Active isometric force after replacement of native TnC by TnC-BR96–103 was 89 ± 3% (n = 5) of that before TnC exchange. The force–pCa relationship was not significantly affected by introduction of TnC-BR96–103; pCa50 after exchange was 6.30 ± 0.06 and nH was 2.51 (Table 1). Introduction of the BR probe on the E-helix of TnC does not affect its regulatory function.
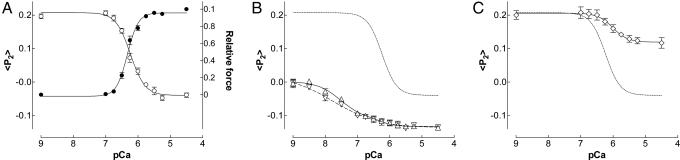
In relaxed fibers (pCa 9), the order parameters 〈P2〉 and 〈P4〉 from TnC-BR96–103 were 0.196 ± 0.007 and −0.135 ± 0.005 respectively, corresponding to θME = 48° (Table 1). During active contraction (pCa 4.5), 〈P2〉 and 〈P4〉 were −0.039 ± 0.007 and −0.045 ± 0.010 respectively, corresponding to θME = 59°. Thus, Ca2+ binding to the N-lobe of TnC produces an 11° change in the mean orientation of the E helix in the C-lobe. During active isometric contraction, the pCa50 of 〈P2〉 for the E helix probe was 6.25 ± 0.03 (Fig. 3A, open circles), not significantly different from pCa50 for force (filled circles) in this group of fibers (Table 1). nH for 〈P2〉 was close to 2, as found for the C helix.
Fig. 3.
Ca2+-dependence of 〈P2〉 (open symbols) and force (filled circles) in muscle fibers containing TnC-BR96–103. (A) Activation in the presence of 5 mM MgATP. (B) Rigor with either 1.0 mM (open triangles) or 0.1 mM (inverted open triangles) free Mg2+. (C) Activation with force inhibition by 0.1 mM BTS. Fitted curves from the single (A and C) or double (B) Hill equation (see text for details); the fit to the 〈P2〉 data in A is reproduced as a dotted curve in B and C.
Binding of rigor myosin heads to the actin filaments at pCa 9 reduced 〈P2〉 for the E helix to almost zero, and θME increased from 48° to 57° (Table 1). Subsequent addition of Ca2+ in rigor produced a further decrease of 〈P2〉 to −0.123 ± 0.014 at pCa 4.5, and θME increased further to 63°. Thus, Ca2+ binding in rigor increases the angle between the E helix and the filament axis by ≈6°. The orientation of the E helix at pCa 4.5 was significantly different in rigor and during active contraction, in contrast with the result for C helix.
In rigor solution containing 1 mM free Mg2+, pCa50 for 〈P2〉 for the E helix was 7.36 and nH was 0.84 (Fig. 3B, open triangles). When free [Mg2+] was reduced to 0.1 mM (Fig. 3B, inverted open triangles), pCa50 was 7.75 and nH was 0.60. The Mg2+-dependence of this transition suggests that it may be mediated by the Ca/Mg sites in the C-terminal lobe. The observed shift in pCa50 is consistent with a Mg2+ affinity of 2 × 103 M−1, similar to that of isolated TnC (5 × 103 M−1; ref. 11) and reconstituted actin/tropomyosin/troponin (7 × 103 M−1; ref. 24) in solution. Considering the differences in preparation and ionic conditions, the agreement is reasonable. The 〈P2〉 data were also fitted with a double Hill equation in which the parameters for the lower affinity site were fixed to those measured in the presence of MgATP. The fitted line for 1 mM Mg2+ (Fig. 3B, continuous line) has pCa50 = 7.5, nH = 1.19 for the higher affinity site, which accounted for 84% of the amplitude. The higher affinity site is more prominent for the E helix tilt than for that of the C helix (Fig. 2B), although the fitted pCa50 was similar in the two cases.
Inhibition of active force by BTS substantially reduced the change in the orientation of the E-helix reported by 〈P2〉 for the TnC-BR96–103 probe in the presence of MgATP (Fig. 3C). BTS had no effect at pCa 9, but addition of 0.1 mM BTS at pCa4.5 increased 〈P2〉 from −0.039 ± 0.007 to +0.121 ± 0.007, and decreased θME from 59° to 51° (Table 1). BTS significantly (P < 0.05) reduced the Ca2+-sensitivity of the orientation change of the E helix as measured by pCa50, without significant change of nH. Thus, the orientation of the E helix in the IT arm of troponin depends on both [Ca2+] and actively cycling myosin heads.
Discussion
The Regulatory Head of Troponin Is a Two-State Ca2+ Sensor.
Polarized fluorescence from isolated muscle fibers containing bifunctional rhodamine probes attached along the C or E helix of TnC allowed the orientations of those helices to be measured in native regulatory complexes in physiological conditions. The Ca2+-dependence of the C helix orientation during active contraction (Fig. 2A) fitted the Hill equation with nH = 2, as expected for a structural transition that requires binding of two Ca2+ ions to the regulatory sites. The pCa50 was 6.0, consistent with the in vitro Ca2+ affinity of the regulatory sites (11), and equal to pCa50 for active force (Table 1). nH for force was larger, indicating additional cooperativity at a later step in the regulatory pathway.
Complete inhibition of active force had no effect on the C helix orientation or on its Ca2+-dependence (Fig. 2C), providing strong evidence that the Ca2+-affinity of the regulatory sites is not altered by binding of myosin heads to actin during active contraction. Previous fiber studies using monofunctional dansylaziridine probes predominantly attached to Met25 in the N-lobe of TnC (25–27) gave divergent results on this point. This may be related to the unknown basis of the dansylaziridine signals, or to other differences in the experimental protocols. The present conclusion agrees with that based on direct measurements of Ca2+ binding in muscle fibers (28).
Binding of myosin heads in rigor changes the orientation of the C helix at pCa 9, but not at pCa 4.5, when Ca2+ is bound to the regulatory sites. The orientation of the C helix at pCa 4.5 is the same when active force is abolished by BTS, during active contraction, and in rigor (Table 1). The Ca2+-affinity of TnC in the absence of MgATP, as monitored by the change in the orientation of the C helix (Fig. 2B), is greater than that in its presence (Fig. 2A), as observed in previous studies of the effect of myosin head binding in rigor (17, 25, 26). This effect may be mediated by the Ca/Mg sites in the C-lobe of TnC. In physiological conditions however, the orientation of the C helix, and by implication the conformation of the regulatory head of troponin, is solely determined by free [Ca2+].
The IT Arm Is Sensitive to Myosin Head Binding.
In physiological conditions the Ca2+-dependence of the orientation of the TnC E helix, in the IT arm of troponin, (Fig. 3A) is the same as that described above for the C helix in the regulatory head. The Hill coefficient (nH) of 〈P2〉 for the E helix was close to 2, and its pCa50 was almost identical to that of force (Table 1). In contrast with the results for the C helix, however, the orientation change of the E helix was greatly reduced by inhibition of active force (Fig. 3C), showing that the change in its orientation when Ca2+ binds to the regulatory sites during normal active contraction has two components. The larger component, with a θME change of 8°, requires binding of myosin heads, while the smaller, with a change of only ≈3°, is due to Ca2+ binding to the regulatory sites.
Binding of myosin heads in rigor also alters the orientation of the E helix (Fig. 3B), and this effect is still present at high [Ca2+], again in contrast with results for the C helix (Table 1). Thus, at pCa 4.5, θME was 12° larger in rigor than when active force was abolished by BTS, much larger than the ≈3° change related to Ca2+ binding to the regulatory sites. The Ca2+-dependence of the orientation change of the E helix in rigor (Fig. 3B) was dominated by a high-affinity component with pCa50 = 7.5, which may correspond to that of the Ca/Mg sites in the C-lobe, with only a small component, 16% according to the double Hill equation fit, matching the Ca2+-affinity of the N-lobe regulatory sites observed in the presence of MgATP.
The orientation of the E helix in the IT arm of troponin is sensitive to binding of myosin heads to actin, both during active contraction and in rigor, with only a small (≈3°) component that is directly related to Ca2+ binding to the regulatory head. This is in marked contrast to the behavior of the C helix, in the regulatory head, which tilts by 26° in direct response to Ca2+ binding, and is insensitive to myosin head binding in physiological conditions. These distinct tilting motions of the regulatory head and IT arm during activation are consistent with the flexibility between the two domains inferred from in vitro structural studies (8, 9, 16). The multiple conformations of the IT arm observed here may be related to the “blocked,” “closed,” and “open” states of the thin filament described in in vitro biochemical and structural studies (29, 30). The IT arm conformation in relaxed fibers would be expected to correspond most closely to that in the blocked state, and that in Ca2+-rigor to the open state. Both closed and open states may be populated during active contraction in situ.
The Conformation of Troponin in Relaxed and Active Muscle.
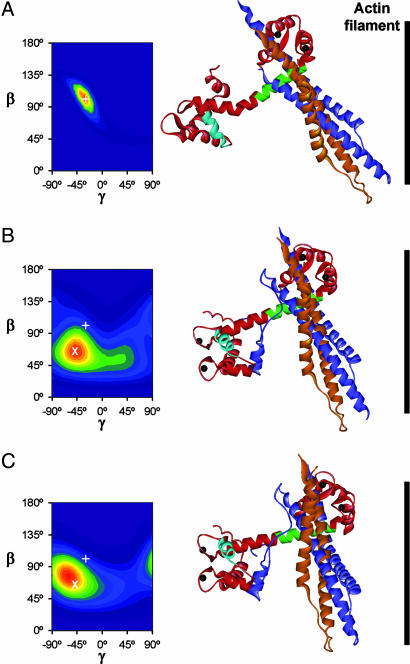
The orientations of the C and E helices of TnC measured here were used to orient in vitro structures of the troponin core complex with respect to the thin filament axis, and to describe the structural changes in the native regulatory complex during activation in situ. Starting from the in vitro structure of the skeletal muscle troponin core complex with bound Ca2+ (Fig. 1) (9), we searched for an in situ orientation that could reproduce the 〈P2〉 and 〈P4〉 values measured during active contraction. To further constrain the orientation, we combined the 〈P2〉 and 〈P4〉 values for the C and E helices (Table 1) with published values for the N and A helices (20). The in situ orientation of the troponin complex was defined by two angles: β, the angle between the TnC D helix and the filament axis, and γ, describing rotation of the molecule around the D helix (20; see Methods). Orientational disorder was incorporated by calculating maximum entropy distributions of β and γ (22); these are the smoothest distributions consistent with the measured order parameters, and can be considered as low resolution approximations to the actual orientation distributions.
The observed 〈P2〉 and 〈P4〉 values for active contraction and Ca2+-rigor were reproduced by well-defined orientations of the in vitro structure of the Ca2+-saturated core complex of skeletal muscle troponin (Fig. 4B and C). The maximum entropy distributions are shown as contour plots (Left), with hotter colors representing a higher probability of that (β,γ) orientation. The peak orientations are represented graphically on the right, with the actin filament (black bar) vertical, and the filament axis and TnC D helix in the plane of the paper. The IT coiled-coil was at 28° and 27° to the filament axis in active contraction (Fig. 4B) and Ca2+-rigor (Fig. 4C) respectively, and the D/E helix at 63° and 71°. These results suggest that the effects of myosin head binding are consistent with a rigid body motion of the troponin core complex, despite the absence of a significant change in the orientation of the C helix.
Fig. 4.
In situ conformation of the troponin core complex in relaxation (A), active contraction (B), and rigor at pCa 4.5 (C). Left shows maximum entropy orientation distributions where β denotes the angle between the D helix of TnC and the actin filament axis and γ the twist around this axis. +, peak of distribution in A; X, peak of distribution in B. Molecular graphics show the three peak orientations based on the in vitro structure of the skeletal muscle troponin core complex in the Ca saturated state (Fig. 1), except for the N lobe of TnC in A, which is from ref. 12.
These in situ orientations of the troponin core complex with Ca2+ bound to the regulatory sites are similar but not identical to that proposed recently on the basis of fitting the same core complex structure (9) into electron microscopic reconstructions of isolated thin filaments in the Ca2+-bound state (31). In that case, the IT coiled-coil was estimated to be at 21° to the filament axis, and the D/E helix roughly perpendicular to it. The differences are likely to be due to the limitations of the two techniques rather than the additional effects of myosin head binding in the present experiments. The interpretation of polarized fluorescence data from BR probes on the N, A, C, and E helices of TnC is sensitive to the degree of opening of its N-lobe in the in vitro structure used for the orientation search. In an alternative analysis of these data using a less open NMR structure of the N-lobe (15) combined with a continuous D/E helix, the in situ orientation of the latter at the peak of the maximum entropy distribution was at 82° to the filament axis, and the IT coiled-coil at 49°. The present data cannot distinguish between these two conformations of the TnC N-lobe, and probes at other sites will be required to determine the degree of opening of the N-lobe in situ, and to refine the present estimate of the orientation of the IT arm.
We also attempted to use the in vitro structure of the core complex of skeletal muscle troponin in the Ca2+-free state (9) to determine the in situ orientation of the core complex in relaxed muscle. However, no distribution of orientations of this structure can reproduce the 〈P2〉 and 〈P4〉 data from probes on the N, A, C and E helices in these conditions, suggesting that the in situ conformation of Ca2+-free troponin is significantly different from that seen in crystals of the Ca2+-free core complex.
In the absence of a suitable in vitro structure for the Ca2+-free troponin core complex, we estimated the in situ conformation of the complex in relaxed muscle (Fig. 4A) by separate analysis of the polarized fluorescence data from the regulatory head and IT arm. The IT arm was tilted by 11° from its active conformation to match the tilt of the E helix observed on activation (Table 1). The regulatory head was then added in the orientation calculated from a maximum entropy fit of the 〈P2〉 and 〈P4〉 data for the N, A and C helices, using an NMR structure of the Ca2+-free N-lobe (12), the only in vitro structure of this state consistent with the polarized fluorescence data from relaxed muscle (20). The maximum entropy distribution of the regulatory head (Fig. 4A Left) has a narrow orientation distribution centered on β = 100°, γ = −30°, as described (20).
The resulting in situ conformation of the troponin core complex in relaxed muscle (Fig. 4A Right) has a bend between the D and E helices of TnC. The angle between the IT coiled-coil and the filament axis is 42°, which is larger than the ≈20° estimate derived by fitting the in vitro structure of the Ca2+-free core complex (9) into electron microscopic reconstructions of relaxed thin filaments (31, 32). As noted above, that in vitro conformation is inconsistent with the present polarized fluorescence data from relaxed muscle fibers. On the other hand, our estimate of the in situ conformation of the IT arm in relaxed muscle (Fig. 4A) depends on polarized fluorescence data from a single BR probe on the E helix, and it will be important to refine this estimate, and to test whether the IT arm moves as a rigid unit, using BR probes at other sites.
Methods
Adult New Zealand White rabbits were killed by sodium pentobarbitone injection (200 mg·kg−1). Fiber bundles were dissected from the psoas muscle, demembranated, and stored in relaxing solution containing 50% (vol/vol) glycerol at −20°C (33). Single fiber segments 2.5–3.5 mm long were mounted, via aluminium T-clips, at sarcomere length 2.4 μm between a force transducer and fixed hook in a 60-μl glass trough containing relaxing solution at 10.0 ± 0.5°C.
Experimental solutions contained 25 mM imidazole, 5 mM MgATP (except rigor solutions), 1 mM Mg2+ (except where noted; added as Mg acetate), and 10 mM EGTA (except preactivating solution). Ionic strength was adjusted to 150 mM with potassium propionate (KPr); pH was 7.1 at 10°C. [Ca2+] was set by adjusting K2EGTA:CaEGTA with total [EGTA] 10 mM in all solutions except preactivating solution, which contained 0.2 mM K2EGTA. Solution compositions were calculated as in ref. 34 with apparent stability constants at pH 7.1 and 10°C: CaEGTA, 6.84; MgEGTA, 1.72; CaATP, 3.88; MgATP, 4.22. When required, 0.1 mM N-benzyl-p-toluenesulfonamide (BTS, S949760; Sigma) was added from a 100 mM stock solution in DMSO.
The E56C/E63C and E96C/R103C double-cysteine mutants of chicken skeletal troponin C (TnC) were obtained by site-directed mutagenesis, expressed in Escherichia coli, and purified as described (20). This recombinant TnC has a T130I mutation, which slightly reduces the Ca2+ affinity of the Ca/Mg sites (35). The native cysteine 101 was replaced by alanine. The cysteines were cross-linked with BR (36) to form 1:1 BR:TnC conjugates, which were purified to >95% homogeneity by reverse-phase HPLC (20). Stoichiometry and specificity of BR-labeling were confirmed by electrospray mass spectrometry and tryptic digests.
Native TnC was extracted from muscle fiber segments by incubation in a solution containing 0.5 mM trifluoperazine (Fluka 91665), 20 mM 3-(N-morpholino)propanesulfonic acid (Mops), 5 mM EDTA, and 130 mM KPr at pH 7.1 and 10°C (37) for 30 s, followed by 30 s in relaxing solution. After 20 such cycles, no force could be detected in activating solution (pCa 4.5). Fibers were then incubated in relaxing solution containing 0.5–1 mg·ml−1 TnC-BR56–63 or TnC-BR96–103 for 60 min at 10°C.
For polarized fluorescence measurements, a 1.5-mm fiber segment was illuminated vertically from below with 532-nm light from a solid-state laser (GL50s; Bremlas), focused to 0.3-mm width at the fiber. A Pockels cell (Model 340; Conoptics) switched the illumination between polarizations parallel and perpendicular to the fiber axis. Fluorescent light emitted from the fiber propagating vertically (“x-emission”) and horizontally (at 90° to the illuminating beam and fiber axis; “y-emission”) was collected by two objectives (Nikon Plan Fluor ×10, N.A. of 0.30). A 565-nm dichroic mirror in the x-emission path reflected 532-nm light for viewing the fiber via a charge-coupled device camera. Fluorescence emission was selected by 610-nm interference filters with 75-nm bandpass and split into parallel and perpendicular components by Wollaston prisms in the x and y paths. The intensities of the resulting four components were measured by photomultiplier tubes (R4632; Hamamatsu Photonics). The relative transmittances of the excitation and emission optics and photomultiplier sensitivities were determined by using an isotropic film of rhodamine in poly(vinyl alcohol) sandwiched between 45° prisms (38).
The independent order parameters 〈P2d〉, 〈P2〉 and 〈P4〉 were calculated from the fluorescence intensities (21). 〈P2d〉 measures the amplitude of independent rotation of the BR probe with respect to the protein backbone on timescales faster than the fluorescence lifetime, and was always >0.9, indicating that the amplitude of such motion is small. 〈P2〉 and 〈P4〉 are the first two even coefficients in the Legendre polynomial expansion of the orientation distribution of the BR fluorescence dipole with respect to the muscle fiber axis, with the fast local motion factored out. 〈P2〉 and 〈P4〉 were used to calculate the ME distribution for the angle between each probe dipole and the fiber axis (22), and the mean of this distribution (θME) is reported in Results. 〈P2〉 and 〈P4〉 from multiple probes were used to calculate ME distributions of domain orientations (20, 22) in terms of angular coordinates β, the angle between the TnC D-helix axis and the filament axis, and γ, describing rotation around the D helix axis. γ is zero when the A and D helices of TnC are coplanar with the filament axis, and increasing γ denotes counterclockwise twist around the D-helix axis as viewed from Val-83 to Glu-76. The contour plots in Fig. 4 represent ME number densities, i.e., distribution functions multiplied by sin β (22).
Each activation of a muscle fiber segment was preceded by a 1-min incubation in preactivating solution and followed by a 5-min incubation in relaxing solution. Isometric force and fluorescence intensities were measured after steady-state force was established in each activation. Maximum isometric force (pCa 4.5) was recorded before and after each series of activations at submaximal [Ca2+] and was 178 ± 41 mN/mm2 (SD). If isometric force decreased by >10%, the fiber was discarded. The dependence of force, 〈P2〉, and 〈P4〉 on [Ca2+] were fitted to data from individual fibers by using nonlinear least-squares regression to the Hill equation:
where pCa50 is the pCa corresponding to half-maximal change in Y, and nH is the Hill coefficient. All values are given as mean ± SE except where noted, with n representing the number of fibers. Student's t test (two-tailed) was used to estimate statistical significance.
Acknowledgments
Bifunctional rhodamine was generously provided by J. E. T. Corrie and V. R. N. Munasinghe (both of Medical Research Council National Institute for Medical Research). We thank J. E. T. Corrie, B. D. Sykes, and D. R. Trentham for helpful comments on the manuscript. This work was supported by the Medical Research Council.
Abbreviations
- TnC
troponin C
- BR
bifunctional rhodamine
- TnC-BR56–63
TnC with BR cross-linking residues 56 and 63
- TnC-BR96–103
TnC with BR cross-linking residues 96 and 103
- BTS
N-benzyl-p-toluenesulfonamide
- ME
maximum entropy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Ebashi S, Endo M, Otsuki I. Q Rev Biophys. 1969;4:351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- 2.Greaser ML, Gergely J. J Biol Chem. 1971;246:4226–4233. [PubMed] [Google Scholar]
- 3.Farah CS, Reinach FC. FASEB J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 4.Gordon AM, Homsher E, Regnier M. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 5.Huxley HE. Cold Spring Harbor Symp Quant Biol. 1973;37:361–376. [Google Scholar]
- 6.Parry DAD, Squire JM. J Mol Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 7.Lehman W, Craig R, Vibert P. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 8.Takeda S, Yamashita A, Maeda K, Maeda Y. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 9.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Proc Natl Acad Sci USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzberg O, James MNG. Nature. 1985;313:653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- 11.Potter JD, Gergely J. J Biol Chem. 1975;250:4628–4633. [PubMed] [Google Scholar]
- 12.Gagné SM, Tsuda S, Li MX, Smillie LB, Sykes BD. Nat Struct Biol. 1995;2:784–789. doi: 10.1038/nsb0995-784. [DOI] [PubMed] [Google Scholar]
- 13.Vassylyev DG, Takeda S, Wakatsuki S, Maeda K, Maeda Y. Proc Natl Acad Sci USA. 1998;95:4847–4852. doi: 10.1073/pnas.95.9.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Spyracopoulos L, Sykes BD. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 15.Mercier P, Ferguson RE, Irving M, Corrie JET, Trentham DR, Sykes BD. Biochemistry. 2003;42:4333–4348. doi: 10.1021/bi027041n. [DOI] [PubMed] [Google Scholar]
- 16.Blumenschein TMA, Stone DB, Fletterick RJ, Mendelson RA, Sykes BD. J Biol Chem. 2005;280:21924–21932. doi: 10.1074/jbc.M500574200. [DOI] [PubMed] [Google Scholar]
- 17.Bremel RD, Weber A. Nat New Biol. 1972;238:97–101. doi: 10.1038/newbio238097a0. [DOI] [PubMed] [Google Scholar]
- 18.Chalovich JM, Eisenberg E. J Biol Chem. 1982;257:2432–2437. [PMC free article] [PubMed] [Google Scholar]
- 19.Corrie JET, Brandmeier BD, Ferguson RE, Trentham DR, Kendrick-Jones J, Hopkins SC, van der Heide UA, Goldman YE, Sabido-David C, Dale RE, et al. Nature. 1999;400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson RE, Sun Y-B, Mercier P, Brack AS, Sykes BD, Corrie JET, Trentham DR, Irving M. Mol Cell. 2003;11:865–874. doi: 10.1016/s1097-2765(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 21.Dale RE, Hopkins SC, van der Heide UA, Marszalek T, Irving M, Goldman YE. Biophys J. 1999;76:1606–1618. doi: 10.1016/S0006-3495(99)77320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Heide UA, Hopkins SC, Goldman YE. Biophys J. 2000;78:2138–2150. doi: 10.1016/S0006-3495(00)76760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung A, Dantzig JA, Hollingworth S, Baylor SM, Goldman YE, Mitchison TJ, Straight AF. Nat Cell Biol. 2002;4:83–88. doi: 10.1038/ncb734. [DOI] [PubMed] [Google Scholar]
- 24.Zot HG, Potter JD. J Muscle Res Cell Motil. 1987;8:428–436. doi: 10.1007/BF01578432. [DOI] [PubMed] [Google Scholar]
- 25.Guth K, Potter JD. J Biol Chem. 1987;262:13627–13635. [PubMed] [Google Scholar]
- 26.Allen TS, Yates LD, Gordon AM. Biophys J. 1992;61:399–409. doi: 10.1016/S0006-3495(92)81846-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martyn DA, Freitag CJ, Chase PB, Gordon AM. Biophys J. 1999;76:1480–1493. doi: 10.1016/S0006-3495(99)77308-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs F, Wang Y-P. Am J Physiol. 1991;261:C787–C792. doi: 10.1152/ajpcell.1991.261.5.C787. [DOI] [PubMed] [Google Scholar]
- 29.McKillop DF, Geeves MA. Biophys J. 1983;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vibert P, Craig R, Lehman W. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 31.Pirani A, Vinogradova MV, Curmi PMG, King WA, Fletterick RJ, Craig R, Tobacman LS, Xu C, Hatch V, Lehman W. J Mol Biol. 2005;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 32.Murakami K, Yumoto F, Ohki S-Y, Yasunaga T, Tanokura M, Wakabayashi T. J Mol Biol. 2005;352:178–201. doi: 10.1016/j.jmb.2005.06.067. [DOI] [PubMed] [Google Scholar]
- 33.Sabido-David C, Brandmeier B, Craik JS, Corrie JET, Trentham DR, Irving M. Biophys J. 1998;74:3083–3092. doi: 10.1016/S0006-3495(98)78015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabiato A, Fabiato F. J Physiol (Paris) 1979;75:463–505. [PubMed] [Google Scholar]
- 35.Golosinska K, Pearlstone JR, Borgford T, Oikawa K, Kay CM, Carpenter MR, Smillie LB. J Biol Chem. 1991;266:15797–15809. [PubMed] [Google Scholar]
- 36.Corrie JET, Craik JS, Munasinghe VRN. Bioconjugate Chem. 1998;9:160–167. doi: 10.1021/bc970174e. [DOI] [PubMed] [Google Scholar]
- 37.Metzger JM, Greaser ML, Moss RL. J Gen Physiol. 1989;93:855–883. doi: 10.1085/jgp.93.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell MG, Dale RE, van der Heide UA, Goldman YE. Biophys J. 2002;83:1050–1073. doi: 10.1016/S0006-3495(02)75230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]