Abstract
A chemical synthesis platform with broad applications and flexibility was rationally designed to inhibit biogenesis of adhesive pili assembled by the chaperone–usher pathway in Gram-negative pathogens. The activity of a family of bicyclic 2-pyridones, termed pilicides, was evaluated in two different pilus biogenesis systems in uropathogenic Escherichia coli. Hemagglutination mediated by either type 1 or P pili, adherence to bladder cells, and biofilm formation mediated by type 1 pili were all reduced by ≈90% in laboratory and clinical E. coli strains. The structure of the pilicide bound to the P pilus chaperone PapD revealed that the pilicide bound to the surface of the chaperone known to interact with the usher, the outer-membrane assembly platform where pili are assembled. Point mutations in the pilicide-binding site dramatically reduced pilus formation but did not block the ability of PapD to bind subunits and mediate their folding. Surface plasmon resonance experiments confirmed that the pilicide interfered with the binding of chaperone–subunit complexes to the usher. These pilicides thus target key virulence factors in pathogenic bacteria and represent a promising proof of concept for developing drugs that function by targeting virulence factors.
Keywords: antimicrobials, chaperone–usher pathway, pilicide, urinary tract infection
Extracellular fibers are critical to the persistence of many pathogenic microorganisms that invade the human host. The ability to impair pilus biogenesis represents an ideal strategy to combat bacterial pathogenesis while maintaining host defenses and host flora. Targeting organism-specific virulence factors, in general, has become an attractive approach in the development of new antimicrobial therapies and may provide reduced opportunities for the emergence of resistance mechanisms compared with their bactericidal counterparts.
A multitude of Gram-negative pathogens use the so-called chaperone–usher pathway for the assembly of adhesive fibers that participate in host–pathogen interactions critical for infection (1, 2). Among those fibers, P and type 1 pili, produced by uropathogenic strains of Escherichia coli (UPEC), have served as model systems to understand their structure and function. P pili contain an adhesin, PapG, that recognizes a globoside receptor in the human kidney as part of a mechanism important in the disease process (3, 4). Type 1 pili contain the FimH adhesin that mediates E. coli binding, invasion, and formation of intracellular biofilm-like bacterial communities in the bladder epithelium (5, 6).
Cognate periplasmic chaperones are required for folding, stabilization, and transport of pilus subunits across the periplasm (1). PapD is the prototypic periplasmic chaperone and is required for P pilus assembly. PapD-like chaperones act by binding to and catalyzing folding of pilus subunits newly translocated into the periplasm (7, 8). The chaperone–subunit complexes are then targeted to an outer membrane assembly site called the usher. The usher forms a pore that selectively discriminates among the chaperone–subunit complexes, a process that facilitates the ordered assembly of pili (9, 10).
All PapD-like chaperones have two Ig-like domains (2, 11). Pilus subunits consist of a single, incomplete Ig-like fold that lacks its C-terminal seventh (G) β-strand (12, 13). In a process termed donor-strand complementation, the chaperone's G1 β-strand provides in trans the pilin's seventh strand (12–15). The mechanism of action of PapD depends on the formation of an ion pair between the C-terminal carboxyl group of each subunit and the conserved chaperone cleft residues R8 and K112 (7, 8).
During pilus assembly, the incoming chaperone–subunit complexes bind to the N-terminal domain of the usher. A ternary complex is formed that leads to uncapping of the chaperone, followed by incorporation of the subunit into the growing pilus (16, 17). Assembly occurs by a process termed donor-strand exchange, in which the G1 β-strand of the chaperone is replaced by an N-terminal extension that is present on every subunit (14, 15, 18). Upon exchange, the pilus subunit undergoes a topological transition that triggers the closure of its groove, incorporating its neighbor's N-terminal extension as part of its own Ig fold (15, 19).
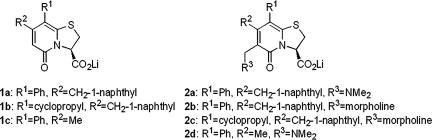
The common structure–function correlates and universal mechanism of action make the periplasmic chaperones prime candidates as broad-range antivirulence targets. The atomic-level structural details of chaperone–subunit interactions and the potential to inhibit pilus biogenesis stimulated the rational design and synthesis of small molecular inhibitors of pilus assembly, termed pilicides. The R8/K112 cleft region of the chaperone was originally targeted (20, 21), followed by compounds 1a and 1b with two substituents (R1 and R2, Fig. 1) (22–24) that bound to PapD and FimC, the P and type 1 pili chaperones, respectively (21, 22, 25). However, the poor water solubility limited their utility. Substitution of the open position in the 2-pyridone scaffold (R3; Fig. 1) (26) resolved this issue, resulting in 2-pyridones, 2a–2d, whereby 2a–2c bound chaperones in the low millimolar range (Fig. 1) (27). 2d, poorly binding to chaperones, was designated as a negative control.
Fig. 1.
Compounds used in the study. A second generation of trisubstituted 2-pyridones 2a–2d were prepared as mimetics of the C terminus of PapG, starting from the disubstituted 2-pyridones 1a–1c.
Here, we report details of the biological activity of these pilicides and show that they interrupt pilus-dependent phenomena crucial to bacterial pathogenesis, including bacterial attachment and biofilm formation. Combining x-ray crystallography and surface-plasmon resonance, we elucidated their mode of action: pilicides selectively disrupted a critical chaperone–usher interaction. Hence, pilicides will not dissociate preformed pili, rather, these small molecules prevent pilus formation. This report presents a class of compounds that selectively disrupts a protein–protein interaction essential for the biogenesis of a bacterial virulence factor.
Results and Discussion
Pilicides Block Bacterial Attachment and Pilus Biogenesis.
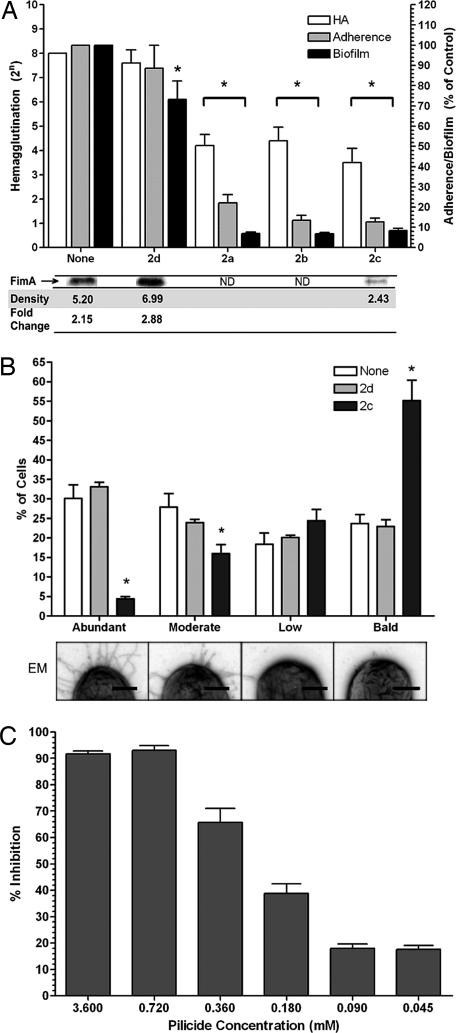
The pilicides were evaluated as inhibitors of type 1 pilus formation by using a mannose-sensitive hemagglutination (MSHA) assay. E. coli strains NU14 and UTI89 were grown in the presence of pilicides 1aand 1b and 2a–2d, and then the MSHA titers were determined. Both strains exhibited reduced MSHA titers with 2a–2c (Fig. 2A) and 1aand 1b (data not shown), whereas the negative control 2d had no effect. 1aand 1b are two of the first generation 2-pyridones that exhibited chaperone-binding properties.
Fig. 2.
Effect of pilicides on pilus biogenesis, hemagglutination, adherence, and biofilm formation. (A) E. coli NU14 was grown in the presence of pilicide (3.5 mM), and HA titers were determined, adherence to 5,637 bladder cells was measured, and the ability to form biofilms was quantified. Adherence and biofilm formation are shown as percentage present relative to each assay when no compound is present. Below the graph are representative FimA immunoblot bands from UTI89 whole-cell 2c pilicide-treated samples. Average summated densitometry measurements (n = 2) are shown. Fold change is densitometry measurements of untreated/2c-treated samples or 2d-treated samples/2c-treated samples. ND, not done. (B) The pili content of 300 control or pilicide (2c or 2b)-treated UTI89 bacteria in triplicate was quantitated by electron microscopy in a blinded study. Bacteria were classified as having one of four degrees of piliation: abundant, moderate, low, or bald (no pili). The percentage of the total number of bacteria counted in each category is displayed. Below the graph are representative images of bacteria from each of the four piliation groups. (Scale bars: 0.25 μm.) (C) Titratable effect of 2c treatment on UTI89 biofilm inhibition. The error bars represent the standard deviation of the mean. ∗ indicates a significant (P < 0.05) difference relative to the included controls.
Pilicides 1aand 1b and 2a–2c also blocked P pilus biogenesis in HB101 transformed with pPAP5 (encoding P pili) as determined by a HA assay (see Table 1, which is published as supporting information on the PNAS web site). The pilicides functioned in a dose-dependent manner; hemagglutination (HA) titers decreased with increasing pilicide concentrations (data not shown), and 2d was again inactive. Viable cell counts and growth curves showed that the pilicides did not affect bacterial viability (data not shown).
Having shown that the pilicides reduced both type 1 and P pili biogenesis, we focused our biological assays on the type 1 pili system for which we have more robust, dynamic, and sensitive biological assays. Type 1 pili mediate the binding and invasion of UPEC into bladder epithelial cells, processes critical to infection (5, 28, 29). Deletions of the FimH adhesin or the FimA structural subunit completely abolish the ability of UPEC to cause disease by impairing UPEC colonization and invasion. E. coli strain NU14, grown with pilicides 2a–2c, reduced adherence to cultured bladder 5637 cells by ≈90% compared with both untreated and 2d (negative control)-treated bacteria (Fig. 2A). These results support the argument that treatment with properly substituted 2-pyridone carboxylic acid reduced piliation and, as a consequence, nearly abolished adherence to bladder epithelial cells.
We hypothesized that the biological effects of pilicide treatment should be the result of a reduction in the relative abundance of type 1 pili. Accordingly, immunoblots performed on pilicide-treated UPEC using anti-type 1 pili antibody revealed a 2.88-fold reduction in the FimA major subunit by 2c relative to the 2d control. Also, EM of 2c-treated bacteria revealed a dramatic shift in the proportion of heavily type 1 piliated bacteria to nonpiliated bacteria. In contrast, 2d treatment had no significant effect on levels of piliation (Fig. 2B).
Inhibition of Biofilm Formation.
FimH-mediated invasion into bladder epithelial cells activates a complex genetic cascade resulting in the formation of intracellular bacterial communities (IBCs), highly organized biofilm-like bacterial communities (6). Biofilms and IBC formation depend on type 1 pili (5, 6, 30). We found that the pilicides 2a–2c reduced NU14 biofilm formation in vitro by up to 90%, consistent with the ability of these compounds to inhibit the chaperone–usher-mediated assembly of pili. In contrast, 2d reduced biofilm formation by only 27% (Fig. 2A). The action of 2c on biofilm growth was shown to be titratable in UTI89, such that 50% inhibition occurred between 0.18 and 0.36 mM (Fig. 2C).
Crystal Structure of the PapD–2c Complex.
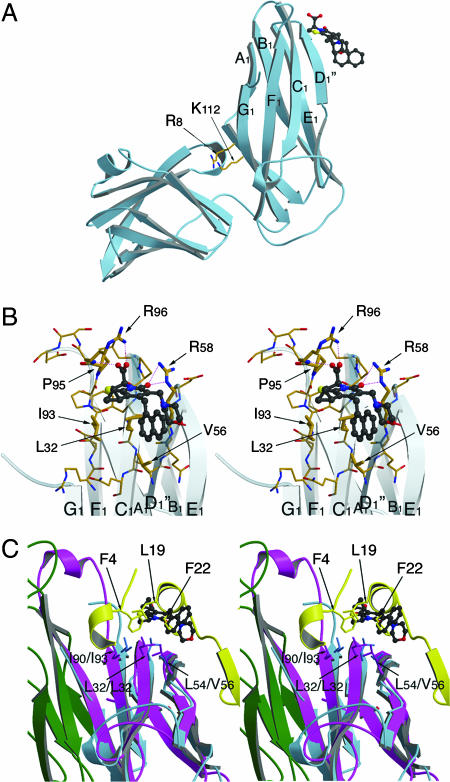
The location of binding and, consequently, the mode of action of the pilicides has been investigated by using chemical shift mapping with 15N-labeled FimC (27). Although two plausible interaction sites were found in this study, one located in the interdomain cleft and one located in the vicinity of the F1–G1 loop, no well defined binding mode could be assigned. In the PapD–PapK complex, the interdomain cleft is sterically hindered, and, thus, access to a molecule that binds the conserved R8 and K112 residues would be blocked. Similarly, mutating R8 and/or K112 would destroy the binding site. However, relaxation-edited NMR spectroscopy revealed that 2c retained most of its affinity for the PapD–PapK complex and for the cleft mutant of PapD, R8A, K112A PapD. To gain a further understanding of pilicide mode of action, we cocrystallized compound 2c with the PapD chaperone and solved the structure by molecular replacement (MR). MR phases were used together with Fo–Fc coefficients to generate an omit electron-density map in which the region of pilicide binding was revealed (Fig. 6, which is published as supporting information on the PNAS web site). Strong, unmistakable electron density was observed at the back of the F1–G1 loop region, and none was observed in the R8/K112 region. Electrostatic and hydrogen bonds are formed between the pilicide carboxylate and the R96 guanidinium group and the R96-P95 backbone amide as well as between the pilicide carbonyl and the R58 guanidinium group (Fig. 3A and B). Furthermore, the plane of the 2-pyridone system and the R1-cyclopropyl, R2-CH2-1-naphthyl and R3-morpholine substituents are in close contact with the hydrophobic patch formed by I93, L32, and V56. Interestingly, this hydrophobic patch running across the back of the F1-C1-D1- β-sheet is highly conserved in all periplasmic chaperones (2) and has been shown by x-ray crystallography to form part of the interaction surface between chaperone–subunit complexes and the usher N-terminal domain (16). This overlap between the pilicide-binding site and the usher-interaction site strongly suggests that this pilicide site is implicated in the observed inhibition of pilus assembly through disruption of the interaction between the chaperone and the usher. Indeed, a superimposition of the PapD–2c structure with that of the type 1 pilus FimC-FimH158–279 chaperone–subunit complex bound to the FimDN N-terminal usher domain (residues 1–125) reveals that binding of the pilicide to the F1–G1 loop-binding site masks the residues implicated in usher binding (Fig. 3C) (16).
Fig. 3.
Structure of the PapD–2c complex. (A) Side view from the PapD chaperone showing the pilicide-binding side at the N-terminal domain, near the F1–G1 loop. The conserved cleft residues R8 and K112 are indicated as a point of reference. In the PapD–2c crystals, no pilicide is seen binding in the cleft; rather, the R8/K112 pair is involved in crystal packing (data not shown). (B) Stereoimage of the pilicide-binding site near the F1–G1 loop. The 2c compound, shown in ball-and-stick representation, forms hydrogen bond and electrostatic interactions with the R96, the P95-R96 backbone amide and R58. Further interactions are formed with the hydrophobic patch made up of the residues I93, L32, and V56. (C) Stereoimage of the PapD–2c complex in overlay with the FimD1–125 N-terminal usher domain in complex with the FimC-FimH158–279 chaperone–adhesin complex. PapD and 2c are shown in light blue ribbon and ball-and-stick representation, respectively. FimC and FimH158–279 are shown in magenta and green, with the FimD1–125 N-terminal domain shown in yellow. The conserved hydrophobic patch across the back of the F1-C1-D1 β-sheet formed by residues I90/I93, L32/L32, and L54/V56 (FimC/PapD) forms part of both the usher-interaction site and the pilicide-binding site. The overlay shows how in the chaperone–pilicide interaction, the plane of the 2-pyridone system and the R1-cyclopropyl and R2-CH2-naphthyl substituents coincide with and mimic the interactions made by the F4, L19, and F22 side chains from the usher N-terminal domain. Finally, the overlay clearly demonstrates the steric clash between the pilicide and the usher N-terminal domain. Figures were generated with Molscript and Raster3D (31, 32).
Pilicide-Interaction Site Probed Through Point Mutations.
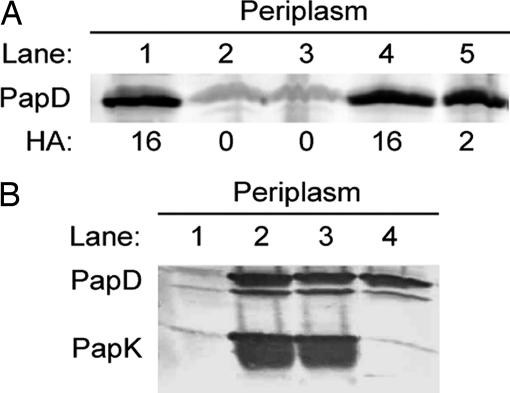
Pilicide binding appears to mask R58 and the residues around it and may result in steric inhibition of the docking or processing of chaperone–subunit complexes at the usher. By using a genetic approach, the R58 pilicide-interactive residue on the PapD chaperone was thus interrogated for its role in pilus assembly. R58 of PapD was mutated to alanine, and R58A PapD was used to complement a papD mutant pap operon in E. coli strain. Immunoblotting demonstrated that the R58A mutation had no effect on PapD stability (Fig. 4A, lane 5). However, the R58A mutation in PapD dramatically reduced P pili, as measured by HA titers (Fig. 4A). The R58A region of the chaperone is not thought to participate in the folding or stabilization of the subunits. Chaperone–subunit interactions have been shown to involve the cleft of the chaperone and critically depend on the anchoring of the C-terminal carboxylate group to the invariant R8 and K112 cleft residues. To investigate whether pilicide binding to PapD had an effect on chaperone–subunit interactions, PapK was coexpressed with wild-type, R58A, or R8A, K112A mutant PapD. Immunoblotting of periplasmic extracts with anti-PapD–PapK antibody revealed that PapK was stable only when expressed with either wild-type PapD (Fig. 4B, lane 2) or R58A PapD (Fig. 4B, lane 3). PapK was unstable when expressed in the absence of PapD (Fig. 4B, lane 1) or when expressed with the R8A, K112A PapD (Fig. 4B, lane 4). Thus, the R58A mutation did not interfere with the ability of PapD to bind, fold, and stabilize subunits. We instead argue that the R58 residue is critical for proper pilus assembly in a step after chaperone–subunit interaction and is likely important for targeting or processing events that take place at the usher (33). Because pilicide binding that masks R58 in PapD and the R58A mutation in PapD both result in decreased pilus assembly, we reasoned that this surface on PapD is critical in chaperone–subunit–usher ternary complex interactions and/or uncapping of the chaperone to allow subunit incorporation. This hypothesis was tested biochemically, as described below.
Fig. 4.
Mutations confirm pilicide binding and function. (A) Western blot developed with anti-PapD antiserum showing periplasm extract of bacterial cells expressing the wild-type pap operon (pPap5; lane 1) or a papD deleted pap operon (pPap37, lane 2) in combination with the vector control (lane 3), wild-type PapD (lane 4), or PapD-R58A (lane 5). The HA titer for each of the constructs is denoted below the blot. (B) Western blot developed with anti-PapDK antiserum showing periplasm extract of bacterial cells expressing PapK alone (lane 1) or in combination with wild-type PapD (lane 2), PapD-R58A (lane 3), or PapD-R8A, K112A (lane 4).
2c Inhibits FimC–FimD Interaction.
The PapD-pilicide crystallographic data suggested that in the type 1 and P pilus systems pilicide 2c inhibits chaperone–subunit targeting to the usher or subsequent processing (16, 17). This conclusion was supported by finding that R58A PapD was able to form stable chaperone–subunit complexes but was unable to assemble pili. To further resolve the mode of 2c action, surface plasmon resonance experiments were conducted to monitor the association between chaperone–adhesin complex FimC-FimH and the N-terminal domain of the usher FimD. A construct FimDN-His consisting of FimD residues 1–125 (16, 34) with a C-terminal (His)6 tag was produced (Fig. 7, which is published as supporting information on the PNAS web site) and immobilized on a Ni-NTA chip in a Biacore 3000 instrument. The association of FimDN-His and FimC-FimH was first monitored in the absence of pilicides (Fig. 8, which is published as supporting information on the PNAS web site). A KD of 12.4 nM was measured, consistent with values observed for full-length FimD (35) but in disagreement with measurements using isothermal titration calorimetry (16).
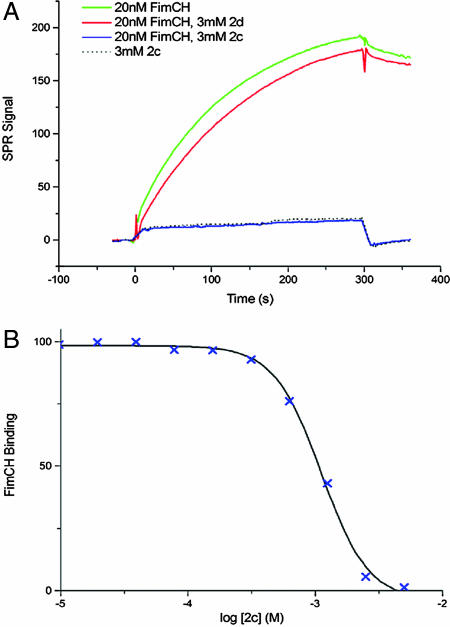
Subsequently, we tested the effect of a 20 nM FimC-FimH solution either alone or containing 3 mM of 2c or 2d (Fig. 5A). Binding of FimC-FimH alone to FimDN-His resulted in a large surface plasmon resonance signal, which was specifically inhibited by the presence of 2c but not 2d. The compound was seen to inhibit the FimDN-His/FimC–FimH interaction in a dose-dependent manner with an IC50 of 1.1 mM. Application of the Cheng–Prusoff relationship (36) gives an inhibition constant (Ki) of 425 μM (Fig. 5B). This experiment confirms the relevance of the binding site elucidated by the x-ray crystal structure of the PapD–2c complex and indicates that 2c specifically inhibits the interaction of the chaperone–subunit with the usher in type 1 pilus biogenesis.
Fig. 5.
Inhibition of FimDN-His/FimC–FimH interaction by compound 2c. (A) FimC–FimH binding (green) is unaffected by the presence of 3 mM 2d (red). Binding is abolished by the presence of 3 mM 2c (blue); no signal is apparent beyond that produced by 2c alone (dashed line). (B) Measurement of the IC50 of compound 2c. FimDN-His/FimC–FimH binding is reduced by the presence of increasing concentrations of 2c, with IC50 1.1 mM.
Conclusion
In this study, we used rationally designed 2-pyridones to inhibit pilus biogenesis, bacterial adherence, and biofilm formation in uropathogenic bacteria by interfering with chaperones that are key proteins in the highly conserved chaperone–usher pathway. The pilicides target the specific interaction between the chaperone–subunit complexes and the N-terminal domain of the usher, a unique protein–protein interaction site essential to the biogenesis of pili (Fig. 9, which is published as supporting information on the PNAS web site). Given the wealth of protein–protein interaction data currently emerging from concerted mapping efforts, there is considerable interest in the development of small molecule inhibitors capable of disrupting such interactions, but this objective is notoriously difficult to achieve (37). Affinity data for the active pilicide 2c are in the high micromolar range, which in drug discovery terms and considering that the target is a protein–protein interaction, constitutes an excellent starting point.
Pilicides, by blocking chaperone and usher functions, have the potential to inhibit pili formation in a broad spectrum of pathogenic bacteria to prevent critical host–pathogen interactions necessary for many diseases. There are hundreds of diverse cell-surface virulence organelles that are assembled by the chaperone–usher pathway in important bacterial pathogens. Because all chaperones have a conserved structure and mechanism of action, it is reasonable to propose that pilicides likely have broad-spectrum activity. Remarkably, pilicides do not affect bacterial growth, because pilus production is not essential for bacterial survival and replication. These pilicides represent an example of selective, low-molecular-weight, nonpeptidic virulence-determinant inhibitors (38, 39).
Materials and Methods
PapD–2c X-Ray Structure Determination.
PapD was concentrated to 12.3 mg/ml (0.5 mM) in 10 mM Mes (pH 6) and 10 mM NaCl. Compound 2c was added to a final concentration of 1 mM, starting from a 200 mM stock solution in 100% DMSO. PapD–2c crystals were grown by sitting-drop vapor diffusion against a solution containing 16% PEG 4000, 100 mM Tris·HCl (pH 8.5), and 200 mM Li2SO4. Diffraction data were collected to 2.6 Å resolution at the ID23–1 beamline at the European Synchrotron Radiation Facility (Grenoble, France). Crystals belonged to the space group P6 (3)22 with cell parameters of a = b = 94.315 Å, and c = 121.992 Å, and data were integrated and scaled with the programs MOSFLM and SCALA (40). The structure was solved by molecular replacement [program AMORE (41)] by using PapD as a search model, and the resulting model was refined against data ranging from 15–2.6 Å to a final R and free R of 23.24% and 27.26% by using the program CNS (42). The final PapD–2c model contains one molecule of PapD and one 2c molecule per asymmetric unit (see Table 2, which is published as supporting information on the PNAS web site, for data collection and refinement statistics). Initial coordinates for 2c were generated with SYBYL (Tripos, St. Louis, MO), and dictionaries for restrained refinement in CNS were obtained by using XPLO2D (43).
HA, Biofilm, and Adherence Assays.
Bacteria were grown statically for 48 h and passaged for another 48 h of static growth in Luria broth in the presence of 3.6 mM pilicide. This preparation was then used in HA and adherence assays. HA titers were determined as described (44) by using guinea pig red blood cells. HA titers for P pili expressing HB101/pPAP5 were determined according to a published procedure (25). For adherence assays, the bacteria were diluted 1:100 in RPMI medium 1640 plus 10% FCS and applied to tissue culture cell line 5637 (American Type Culture Collection, Manassas, VA) for 1 h at 37°C with 5% CO2. Adherence was determined as reported (45). For the biofilm assays, bacteria were grown in Luria broth in wells of microtiter plates in the presence 3.6 mM pilicide. After 48 h of growth, wells were rinsed and stained with crystal violet, and biofilms were quantified as described (30). Type 1 pili antisera were raised in rabbits by using PAGE gel-purified UTI89 type 1 pili (Sigma–Genosys, St. Louis, MO).
Cloning and Analysis of PapD Mutants.
The papD plasmid (pLS101) is a pMMB91-based vector expressing papD under the control of the Ptac promoter (44). The mutation R58A and dual mutation R8A/K112A were incorporated into pLS101 by using the Quick-Change Mutagenesis kit (Stratagene, La Jolla, CA) and the following primers with their complement: R58A, 5′-ACC CCT CCG GTT CAG GCA CTT GAG CCG GGT GCG-3′; R8A, 5′-CTG GAC AGA ACC GCA GCG GTG TTT GAC GGG-3′; and K112A, 5′-CAG ACC AAA ATA GCA CTT TTT TAT CGC CCG-3′.
Bacterial cells expressing the various PapD constructs were grown in LB broth with appropriate antibiotics to logarithmic phase (OD600 ≈ 0.8) at 37°C. Protein production was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM and continuing growth for 60 min. Periplasmic extracts were prepared by the sucrose/lysozyme method as described (9). Outer membranes were purified by using ultracentrifugation and solubilized in 1% elugent and 20 mM Hepes (pH 7.5) (35). All samples were heated in SDS buffer at 95°C and analyzed by SDS/PAGE, followed by immunoblotting with a polyclonal anti-PapD or anti-PapDK antibody (Sigma) as the primary antibody and alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma) as the secondary antibody.
Cloning, Expression, and Purification of FimC–FimH and pFimDN-His.
Construct pFimDN-His, consisting of residues 1–125 of mature FimD and the C-terminal sequence LVPRGSSARGSHHHHHH (representing a thrombin cleavage site (underlined), linker residues SA, and RGS-His6 tag), was produced by PCR amplification of a FimD gene fragment from E. coli strain J96 by using oligonucleotides 5′-GC TAT CGG TCT CAG GCC GAC CTC TAT TTT AAT CCG CGC TTT TTA GC-3′ and 5′-GCT ATC GGT CTC AGC GCT GGA GCC ACG CGG AAC CAG ACG CGC GCG ATT ACT CAT AAA TGC CTG AGG-3′ (FimD coding sequence underlined) as forward and reverse primers, respectively. The PCR product was digested with BsaI and transferred into vector pASK-IBA32 (Iba-Go, Göttingen, Germany) in frame with the OmpA leader sequence to give plasmid pFimDN-His.
The top10 cells (Invitrogen, Carlsbad, CA) were transformed with pFimDN-His and grown to an OD600 of 1 at 37°C in LB medium containing 100 μg/ml ampicillin. Expression was induced by the addition of anhydrotetracycline at 200 μg per liter of culture, and cells were grown for one further hour before harvesting. Periplasmic extract was diluted 1:4 in 20 mM Tris, 150 mM NaCl, and 20 mM imidazole (pH 7.2) and loaded onto a 1-ml HiTrap HP nickel chelate column (GE Healthcare, Buckinghamshire, U.K.), and eluted with a gradient to 200 mM imidazole. All treatments were performed at 4°C because temperature-dependent degradation was suspected.
The FimC–FimH complex was expressed from E. coli C600 cells transformed with plasmid pNH212 (pBAD18-kan containing J96 FimC and FimH; Nadine Henderson and David Thanassi, unpublished work). Cells were grown to an OD600 of 1 at 37°C in LB medium containing 30 μg/ml kanamycin. Expression was induced by the addition of arabinose at 1g per liter of culture, and cells were grown for two further hours before harvesting. Periplasmic extract was dialyzed into 20 mM Mes (pH 6.0) and loaded onto a HiTrap SP FF column (GE Healthcare) at room temperature, and a 0–1 M NaCl gradient was applied. FimC-FimH eluted at 110 mM NaCl. FimC-FimH affinity for FimDN-His was then exploited for further purification. FimDN-His was covalently attached by its N terminus to the Sepharose matrix of a HiTrap NHS-activated HP column (GE Healthcare) according to manufacturer's recommendations. Concentrated FimC–FimH complex from the SP step was loaded onto the column, and pure FimC–FimH was eluted with 20 mM Tris (pH 7.2) and 100 mM NaCl (see Fig. 7).
Surface Plasmon Resonance Studies.
All samples were prepared in running buffer [10 mM Hepes (pH 7.4)/150 mM NaCl/50 μM EDTA/0.005% Surfactant P20 (GE Healthcare)]. A Biacore NTA sensor chip was used according to the manufacturer's recommendations. For each measurement cycle, 25 μl of a dilute solution of FimDN-His was injected onto a single flow cell at a flow rate of 20 μl/min, resulting in an initial immobilized amount of 420 ± 15 response units; during analysis, data were normalized to this level. A blank flow cell with no protein immobilized provided the internal reference against which SPR signal was measured. Further measurements used a flow rate of 10 μl/min.
At the end of each cycle, bound proteins were removed with 350 mM EDTA regeneration buffer as per Biacore protocol, supplemented by a 10-μl injection of 50 mM NaOH. Nickel ions were replaced with 500 μM NiCl2 solution.
FimDN-His/FimC–FimH affinity was determined by fitting observed rate kobs to the association phase for each assayed concentration (1.56–800 nM, 2-fold dilution at each step). At concentrations >25 nM, a second, slower exponential phase became apparent from the data; for these concentrations, kobs for only the first, faster phase was included. A straight line was fitted to kobs values plotted against concentration (See Fig. 8). The y-intercept at x = 0 (corresponding to koff) was divided by the slope of the line (kon) to give the dissociation constant KD.
For each assayed concentration of 2c, an initial injection of 10 μl of compound alone was made to account for nonspecific response, followed by a 100-μl injection of the same concentration of compound with 20 nM FimC–FimH. Response to a blank (buffer only) injection was subtracted from each data set to offset instrument noise and to account for the dissociation of bound protein from the Ni-NTA surface. For each measurement, the equilibrium response level was derived from an exponential function fitted to the association curve; nonspecific response from compound alone was subtracted to give the final response level.
Synthesis.
Compounds 2a–d were synthesized by basic hydrolysis of methyl ester-substituted 2-pyridones, which were prepared according to a published procedure (26).
Experimental details: 0.1M aqueous LiOH was added drop-wise to a stirred solution of methyl ester-substituted 2-pyridone in MeOH:THF 7:3 (0.3 mol/ml) at 0°C. The solution was then allowed to attain room temperature and was continuously stirred for 12 h and then concentrated and lyophilized from H2O:MeCN 8:2 to give the lithium carboxylate in quantitative yield.
Supplementary Material
Acknowledgments
We gratefully acknowledge the expertise of Wandy Beatty, Imaging Facility, Washington University School of Medicine. This work was supported by the Swedish Natural Science Research Council and the Knut and Alice Wallenberg Foundation. S.J.H. was supported by National Institutes of Health (NIH) Grants R37 AI029549-15, R01 AI48689-05, and U54 AI057160-3. P.S. was supported by NIH Grant K12-HD00850. G.W., F.B., and H.R. were supported by Medical Research Council Grant 58149.
Glossary
Abbreviations
- HA
hemagglutination
- MR
molecular replacement
- MSHA
mannose-sensitive HA
- UPEC
uropathogenic strains of E. coli.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates of the PapD–2c complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2J7L).
References
- 1.Sauer FG, Remaut H, Hultgren SJ, Waksman G. Biochim Biophys Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Hung DL, Knight SD, Woods RM, Pinkner JS, Hultgren SJ. EMBO J. 1996;15:3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts JA, Marklund BI, Ilver D, Haslam D, Kaack MB, Baskin G, Louis M, Mollby R, Winberg J, Normark S. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 5.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 6.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bann JG, Pinkner JS, Frieden C, Hultgren SJ. Proc Natl Acad Sci USA. 2004;101:17389–17393. doi: 10.1073/pnas.0408072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetsch M, Puorger C, Spirig T, Grauschopf U, Weber-Ban EU, Glockshuber R. Nature. 2004;431:329–332. doi: 10.1038/nature02891. [DOI] [PubMed] [Google Scholar]
- 9.Dodson KW, Jacobdubuisson F, Striker RT, Hultgren SJ. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thanassi DG, Saulino ET, Lombardo MJ, Roth R, Heuser J, Hultgren SJ. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren A, Branden CI. Nature. 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 12.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 13.Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 14.Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ. Proc Natl Acad Sci USA. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 16.Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, Vetsch M, Bettendorff P, Jelesarov I, Grutter MG, Wuthrich K, et al. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng TW, Akman L, Osisami M, Thanassi DG. J Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Mol Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 20.Lee YM, Almqvist F, Hultgren SJ. Curr Opin Pharmacol. 2003;3:513–519. doi: 10.1016/j.coph.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Svensson A, Larsson A, Emtenas H, Hedenstrom M, Fex T, Hultgren SJ, Pinkner JS, Almqvist F, Kihlberg J. ChemBioChem. 2001;2:915–918. doi: 10.1002/1439-7633(20011203)2:12<915::AID-CBIC915>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Emtenäs H, Åhlin K, Pinkner JS, Hultgren SJ, Almqvist F. J Comb Chem. 2002;4:630–639. doi: 10.1021/cc020032d. [DOI] [PubMed] [Google Scholar]
- 23.Emtenäs H, Taflin C, Almqvist F. Mol Diversity. 2003;7:165–169. doi: 10.1023/b:modi.0000006800.46154.99. [DOI] [PubMed] [Google Scholar]
- 24.Emtenäs H, Alderin L, Almqvist F. J Org Chem. 2001;66:6756–6761. doi: 10.1021/jo015794u. [DOI] [PubMed] [Google Scholar]
- 25.Åberg V, Hedenström M, Pinkner JS, Hultgren SJ, Almqvist F. Org Biomol Chem. 2005;3:3886–3892. doi: 10.1039/b509376g. [DOI] [PubMed] [Google Scholar]
- 26.Pemberton N, Åberg V, Almstedt H, Westermark A, Almqvist F. J Org Chem. 2004;69:7830–7835. doi: 10.1021/jo048554y. [DOI] [PubMed] [Google Scholar]
- 27.Hedenström M, Emtenäs H, Pemberton N, Åberg V, Hultgren SJ, Pinkner JS, Tegman V, Almqvist F, Sethson I, Kihlberg J. Org Biomol Chem. 2005;3:4193–4200. doi: 10.1039/b511857c. [DOI] [PubMed] [Google Scholar]
- 28.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 29.Schilling JD, Martin SM, Hung CS, Lorenz RG, Hultgren SJ. Proc Natl Acad Sci USA. 2003;100:4203–4208. doi: 10.1073/pnas.0736473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Toole GA, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 31.Merritt EA, Murphy MEP. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 33.Thanassi DG, Stathopoulos C, Dodson K, Geiger D, Hultgren SJ. J Bacteriol. 2002;184:6260–6269. doi: 10.1128/JB.184.22.6260-6269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama M, Vetsch M, Puorger C, Jelesarov I, Glockshuber R. J Mol Biol. 2003;330:513–525. doi: 10.1016/s0022-2836(03)00591-6. [DOI] [PubMed] [Google Scholar]
- 35.Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Y-C, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 37.Veselovsky AV, Ivanov YD, Ivanov AS, Archakov AI, Lewi P, Janssen P. J Mol Recognit. 2002;15:405–422. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]
- 38.Gauthier A, Robertson ML, Lowden M, Ibarra JA, Puente JL, Finlay BB. Antimicrob Agents Chemother. 2005;49:4101–4109. doi: 10.1128/AAC.49.10.4101-4109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 40.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Navaza J, Saludjian P. Macromol Crystallogr A. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Kleywegt GJ, Henrick K, Dodson EJ, van Aalten DMF. Structure (London) 2003;11:1051–1059. doi: 10.1016/s0969-2126(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 44.Slonim LN, Pinkner JS, Branden CI, Hultgren SJ. EMBO J. 1992;11:4747–4756. doi: 10.1002/j.1460-2075.1992.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.