Abstract
The synthesis of meso-diaminopimelic acid (m-DAP) in bacteria is essential for both peptidoglycan and lysine biosynthesis. From genome sequencing data, it was unclear how bacteria of the Chlamydiales order would synthesize m-DAP in the absence of dapD, dapC, and dapE, which are missing from the genome. Here, we assessed the biochemical capacity of Chlamydia trachomatis serovar L2 to synthesize m-DAP. Expression of the chlamydial asd, dapB, and dapF genes in the respective Escherichia coli m-DAP auxotrophic mutants restored the mutants to DAP prototrophy. Screening of a C. trachomatis genomic library in an E. coli ΔdapD DAP auxotroph identified ct390 as encoding an enzyme that restored growth to the Escherichia coli mutant. ct390 also was able to complement an E. coli ΔdapD ΔdapE, but not a ΔdapD ΔdapF mutant, providing genetic evidence that it encodes an aminotransferase that may directly convert tetrahydrodipicolinate to l,l-diaminopimelic acid. This hypothesis was supported by in vitro kinetic analysis of the CT390 protein and the fact that similar properties were demonstrated for the Protochlamydia amoebophila homologue, PC0685. In vivo, the C. trachomatis m-DAP synthesis genes are expressed as early as 8 h after infection. An aminotransferase activity analogous to CT390 recently has been characterized in plants and cyanobacteria. This previously undescribed pathway for m-DAP synthesis supports an evolutionary relationship among the chlamydiae, cyanobacteria, and plants and strengthens the argument that chlamydiae synthesize a cell wall despite the inability of efforts to date to detect peptidoglycan in these organisms.
Keywords: Chlamydophila, meso-diaminopimelic acid biosynthesis, peptidoglycan, Nod1, pathway holes
The synthesis of meso-diaminopimelic acid (m-DAP) is crucial for survival of most bacteria. m-DAP is the direct precursor of lysine, an amino acid essential for protein synthesis. Furthermore, m-DAP and lysine play pivotal roles in peptidoglycan (PG) synthesis by cross-linking PG glycan chains to provide strength and rigidity to the PG (1). Plants also synthesize lysine via the m-DAP pathway (2, 3). In contrast, mammalian cells neither synthesize nor use m-DAP as a substrate in any metabolic pathway, and lysine is an essential amino acid that is obtained from dietary sources (4–6). The absence of an m-DAP/lysine synthesis pathway in mammalian cells makes the enzymes of the bacterial pathway attractive targets for antimicrobial therapy.
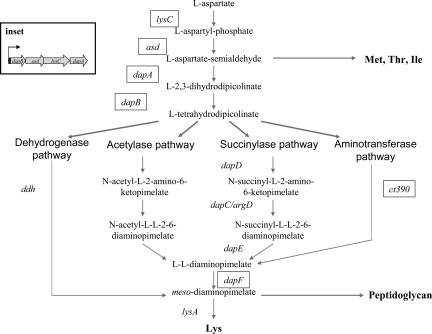
m-DAP/lysine synthesis comprises a branch of the aspartate metabolic pathway that also includes the synthesis of methionine, threonine, and isoleucine (Fig. 1). Common to the synthesis of all these amino acids is the conversion of l-aspartate to l-aspartate-semialdehyde via LysC and Asd (7, 8). The first reaction unique to m-DAP/lysine synthesis is the DapA-catalyzed condensation of l-aspartate-semialdehyde and pyruvate to generate dihydrodipicolinate, which is reduced subsequently by DapB to tetrahydrodipicolinate (THDP). Hereafter, we refer to the four-step synthesis of THDP as the upper m-DAP synthesis pathway. From THDP, three variant pathways have been defined for m-DAP synthesis in the bacteria: the succinylase, acetylase, and dehydrogenase pathways. The succinylase pathway uses succinylated intermediates and is the most widely distributed in bacteria. Genes encoding THDP succinyltransferase (dapD) and N-succinyl-l,l-DAP desuccinylase (dapE) have been characterized, whereas two enzymes, DapC and ArgD, have been shown to possess N-succinyl-l,l-DAP aminotransferase activity (9–13). An analogous pathway using acetylated intermediates has been detected biochemically in certain Bacillus spp., yet few genes in this variant have been characterized (14). The final step in both of the acyl pathways is the epimerization of l,l-diaminopimelic acid (ll-DAP) to m-DAP by DapF. A third pathway exists in which a diaminopimelic acid (DAP) dehydrogenase directly converts THDP to m-DAP. The dehydrogenase pathway is used by a small number of Gram-positive organisms, and in some cases, in conjunction with an acylase pathway (15). We term the steps that convert THDP to m-DAP as the lower m-DAP biosynthesis pathway. Once m-DAP is synthesized, it is either incorporated into PG or decarboxylated to lysine by LysA.
Fig. 1.
Variant pathways for the synthesis of m-DAP/lysine in plants and bacteria. m-DAP/lysine biosynthesis genes present in chlamydiae are boxed. The genes of the upper m-DAP biosynthesis pathway are linked together in the genome of all chlamydiae (Inset) whereas the lower m-DAP homologue, dapF, is unlinked to the upper m-DAP biosynthesis pathway genes.
Bacteria of the order Chlamydiales are obligate intracellular bacteria that are pathogens of humans and animals and lack detectable PG. Despite the inability to detect PG, a nearly complete and functional PG pathway is encoded in the chlamydiae genomes (16). In addition, recent studies demonstrating the detection of chlamydiae infections by Nod1, a mammalian intracellular pattern recognition molecule that specifically detects m-DAP containing PG fragments, support the idea that PG is synthesized in these organisms (17, 18). However, the m-DAP synthesis pathway in chlamydiae is incomplete (Fig. 1). The upper m-DAP pathway is intact because lysC, asd, dapA, and dapB homologues are present in these genomes, yet no homologues of any of the lower m-DAP pathways have been annotated with the exception of dapF. Furthermore, no lysA gene has been detected in the chlamydial genomes, despite the biochemical detection of lysine decarboxylase activity in Chlamydia psittaci (19).
Here, we demonstrate that chlamydiae synthesize m-DAP via a previously undescribed lower m-DAP pathway, the aminotransferase pathway. This pathway was recently characterized in Arabidopsis thaliana and cyanobacteria (20), supporting the view that chlamydiae may share a common ancestor with the chloroplast/plant lineage (21). The data also further support the notion of PG synthesis in chlamydiae. The discovery of a variant pathway for m-DAP synthesis presents a target for the development of therapeutic agents for the treatment of chlamydial infections.
Results
C. trachomatis Asd, DapB and DapF Are Functional Enzymes Involved in m-DAP Synthesis.
Four enzymes (LysC, Asd, DapA, and DapB) comprise the upper m-DAP synthesis pathway (Fig. 1). The predicted chlamydial homologues share between 10% and 33% similarity with their respective Escherichia coli homologues (see Table 4, which is published as supporting information on the PNAS web site). To address the functionality of the upper m-DAP synthesis pathway, the C. trachomatis asd and dapB genes were studied. In E. coli, Asd catalyzes the synthesis of l-aspartate-semialdehyde, the branch point intermediate for m-DAP/lysine biosynthesis and methionine, threonine, and isoleucine biosynthesis. E. coli asd mutants are auxotrophic for all of these amino acids on minimal agar medium but only auxotrophic for DAP on LB agar. Screening a C. trachomatis serovar L2 genomic DNA library in χ1825 (ΔbioH-asd) identified five independent clones containing the intergenic DNA region 5′ of dapB and the complete dapB and asd genes of C. trachomatis (Fig. 1), which could restore growth to the E. coli mutant on rich medium in the absence of exogenous DAP. The percent efficiency of DAP complementation of one representative library plasmid was nearly 100% (Table 1). Expression of asd also restored methionine, threonine, isoleucine, and lysine prototrophy to χ1825 on minimal agar medium (data not shown), further attesting to the functionality of the chlamydial Asd as an aspartate semialdehyde dehydrogenase. DapB catalyzes the synthesis of THDP, the branch point substrate of the different lower m-DAP pathways (Fig. 1). Expression of C. trachomatis dapB under control of the ara promoter in ATM783 (ΔdapB) restored DAP prototrophy to the E. coli mutant (Table 1). Together, these results suggest that the upper m-DAP synthesis pathway in C. trachomatis is functional in synthesizing THDP.
Table 1.
Functionality of C. trachomatis serovar L2 m-DAP-synthesis homologues
| Activity | E. coli gene homologue | E. coli mutant | Complementing L2 ORF* |
|---|---|---|---|
| Aspartate semialdehyde dehydrogenase | asd | χ1825 (Δbio-asd) | CT363 (asd) |
| Dihydrodipicolinate reductase | dapB | ATM783 (ΔdapB::kan) | CT364 (dapB) |
| Tetrahydrodipicolinate N-succinyltransferase | dapD | ATM759 (ΔdapD::tet) | CT390† |
| l,l-diaminopimelate epimerase | dapF | ATM784 (ΔdapF::kan) | CT430 (dapF) |
*Complementation was determined by the ability of the E. coli mutant to grow on LB agar medium in the absence of DAP.
†CT390 was identified by screening a genomic library of C. trachomatis L2.
Whereas the upper m-DAP pathway appears to be functional, it is not evident whether the lower m-DAP synthesis pathway is present in chlamydiae based on the genome sequences (Fig. 1 and Table 4). The only lower m-DAP pathway gene that is clearly predicted in chlamydiae is dapF. To test for function, the chlamydial dapF gene was expressed in ATM784 (ΔdapF). This mutant is not completely auxotrophic for DAP on rich medium because E. coli can form colonies by incorporating ll-DAP in place of m-DAP into PG (22). The basal level of suppression of DAP auxotrophy by ll-DAP incorporation into the PG of ATM784 carrying pBAD18 alone was 0.43%. Expression of C. trachomatis dapF in ATM784 increased DAP complementation by >20-fold, indicating that the chlamydial DapF is functional (Table 1).
C. trachomatis ORF ct390 Encodes an Aminotransferase That Complements DAP Auxotrophy in a ΔdapD ΔdapE Double Mutant of E. coli.
To determine how the chlamydiae metabolize THDP, the C. trachomatis genomic library was screened in E. coli strain ATM759 (ΔdapD) to search for clones that are able to restore the mutant to DAP prototrophy. Five plasmid classes were identified and common to all plasmid inserts was a complete ORF, ct390 (see Fig. 4, which is published as supporting information on the PNAS web site). One library clone (pC1) contained the complete ct390 ORF flanked by partial ct389 and ct391 sequences. This plasmid restored DAP prototrophy to ATM759 on rich medium to almost 100% (Table 1). A subclone of ct390 with its native promoter in pUC19 restored DAP prototorophy to ATM759 (data not shown), indicating that neither CT389 nor CT391 were required for complementation.
CT390 shows homology to aminotransferases, particularly several homologues from plants, and was annotated as AspC, an aspartate aminotransferase (21, 23). Although aspartate aminotransferases exhibit broad substrate specificity, overexpression of E. coli aspC in ATM759 did not restore DAP prototrophy, suggesting that the substrate specificity of AspC is not broad enough to function in the m-DAP synthesis pathway (Table 2). Similarly, expression of CT390 in ATM808 (ΔaspC ΔtyrB) did not restore growth of the E. coli mutant on minimal medium in the absence of either aspartate or tyrosine (Table 2). These results suggest that CT390 is not an aspartate aminotransferase.
Table 2.
Activity of CT390 in different E. coli m-DAP and Asp/Tyr auxotrophic mutants
| E. colistrain and mutant genotype | Auxotrophic requirement | Plasmid | Complementation (+/−) |
|---|---|---|---|
| ATM759 | m-DAP | pUC18 | − |
| (ΔdapD::tet) | pNEA16 (dapDEc) | + | |
| pC1 (CT390) | + | ||
| pNEA18 (aspCEc) | − | ||
| ATM808 | Aspartate | No vector | − |
| (ΔaspC ΔtyrB::kan) | pNEA18 (aspCEc) | + | |
| pC1 (CT390) | − | ||
| ATM8088 | Tyrosine | No vector | − |
| (ΔaspCΔtyrB::kan) | pNEA18 (aspCEc) | + | |
| pC1 (CT390) | − |
Besides sharing homology to aminotransferases, CT390 is weakly similar (13%) to a cystathionine β-lyase in Treponema denticola. Interestingly, cystathionine can replace m-DAP in the PG structure (24). However, the complementation evidence suggests that CT390 produces ll-DAP or m-DAP and not another diamine compound because it complemented ATM759 on minimal medium (data not shown). dap mutants require m-DAP for both PG and lysine synthesis (Fig. 1). Although different diamine compounds can replace m-DAP in PG, lysine is synthesized solely from m-DAP.
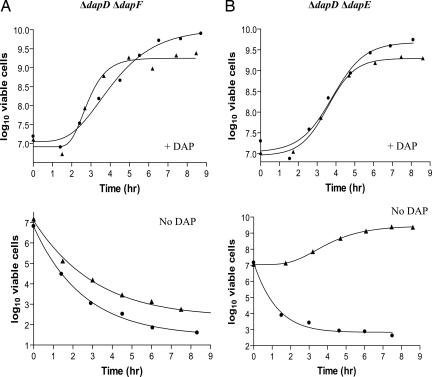
Whether CT390 produces m-DAP or ll-DAP directly from THDP was examined by testing for complementation of E. coli mutants deleted for either dapD and dapF or dapD and dapE. ct390 failed to complement ATM764 (ΔdapD ΔdapF) but was able to complement ATM769 (ΔdapD ΔdapE) (Fig. 2). Because dapF interconverts m-DAP and ll-DAP, the complementation results indicate that CT390 produces ll-DAP specifically.
Fig. 2.
Complementation of ΔdapD::tet ΔdapF::kan and ΔdapD::tet ΔdapE::kan double mutants by ct390. (A and B) ATM764 (ΔdapD::tet ΔdapF::kan; A) and ATM769 (ΔdapD::tet ΔdapE::kan; B) carrying pUC19 (circles) or pC1 (triangles) were grown with (Upper) or without (Lower) DAP as described. At various times, the bacteria were diluted and plated onto LB agar with DAP to determine the number of viable cells.
Enzyme Kinetics.
CT390 was expressed in E. coli as a fusion with a His-Tag and purified by Ni-affinity chromatography to further examine its function. To gain a better understanding of the chlamydial orthlogs of CT390, the gene product of pc0685 from Protochlamydia amoebophila also was expressed as a recombinant protein. Spectral analysis of pure CT390 and PC0685 revealed the presence of an absorbance feature centered at ≈420 nm, indicative of the presence of pyridoxal phosphate (PLP; data not shown). All known transaminases use PLP as a cofactor. The amino acid sequences of CT390 and PC0685 both show canonical PLP binding sites (Lys-236 in CT390 and Lys-246 in PC0685). Most aminotransferases catalyze reversible reactions. To determine whether this is true for CT390 and PC0685, their activity was studied by using coupled assay systems for measurement of the catabolic reverse reaction and the biosynthetically relevant forward reaction. Like their ortholog from the plant Arabidopsis thaliana (20), the chlamydial enzymes functioned much more efficiently in the reverse direction than in the forward direction (Table 3 and Fig. 5, which is published as supporting information on the PNAS web site). Despite the unfavorable forward activity, the complementation data clearly show that the chlamydial aminotransferases are able to synthesize ll-DAP under in vivo conditions. A high ratio of glutamate to 2-oxoglutarate (2-OG) in the cytoplasm would favor the forward activity in vivo as was proposed for the Arabidopsis aminotransferase (20).
Table 3.
Kinetic properties of CT390
| Enzyme |
Vmax, μmol·min−1·mg−1 of protein |
Ratio R/F |
Km, μM |
Km, mM |
|||
|---|---|---|---|---|---|---|---|
| Reverse | Forward | LL-DAP | THDP | 2-OG | Glu | ||
| CT390 | 0.58 ± 0.02 | 0.01 ± 0.001 | 58 | 116 ± 11 | 19 ± 4 | 2.1 ± 0.2 | 4.0 ± 1.0 |
| PC0685 | 1.84 ± 0.15 | 0.09 ± 0.004 | 20 | 6 ± 2 | 5 ± 1 | 1.1 ± 0.3 | 0.4 ± 0.2 |
Reverse activity is defined as THDP synthesis from LL-DAP and 2-OG and forward activity is defined as LL-DAP synthesis from THDP and Glu. Ratio R/F = Vmax in the reverse assay divided by the forward activity. The substrate titrations used to calculate the kinetic constants are shown in Fig. 6. All assays were performed under initial velocity conditions. Kinetic constants were calculated by nonlinear regression, and the experimental variation is given in standard error.
The kinetic constants of the chlamydial aminotransferases differed, with PC0685 showing slight to significantly higher apparent affinities for the substrates. The kinetic constants for both chlamydial enzymes were within the same range as reported for the plant ortholog from Arabidopsis (20). Considering the high similarity of the enzymes from plants and chlamydiae, it was of interest to examine the substrate specificities of the enzymes. Table 5, which is published as supporting information on the PNAS web site, shows that both chlamydial enzymes display a broader substrate specificity than the Arabidopsis enzyme in that they were able to use m-DAP as an amino donor nearly as effectively as ll-DAP and were also able to use lysine, ornithine, and cystathionine (structural analogs of DAP) with reduced efficiency. By contrast, the Arabidopsis enzyme was unable to use these amino donors or used them very inefficiently. To further examine its substrate specificity, CT390 was tested with a range of amino acceptor compounds and either glutamate or ll-DAP as amino donors (Table 6, which is published as supporting information on the PNAS web site). By comparison to the ability to interconvert THDP and ll-DAP, CT390 was unable to transfer the amino group from glutamate to oxaloacetate (aspartate aminotransferase) or phenylpyruvate (phenylalanine aminotransferase), but showed low-level ability to transfer an amino group from ll-DAP to either pyruvate (alanine aminotransferase) or indole 3-pyruvate (l-tryptophan aminotransferase). This result supports the complementation data, indicating that CT390 lacks aspartate aminotransferase activity. In total, the enzyme activity analyses indicate that CT390 and PC0685 are transaminases that can synthesize ll-DAP from THDP, confirming the in vivo complementation results. The ability to use m-DAP for the reverse reaction distinguishes CT390 from the recently characterized plant ll-DAP aminotransferase, which is specific for ll-DAP (20). Although the basis for the relaxed substrate specificity is presently not known, it is unlikely to be of physiological significance because d-THDP would be produced from m-DAP rather than l-THDP. d-THDP would be a metabolic dead-end because it could be converted back to only m-DAP. Moreover ll-DAP must be the normal reaction product of the enzyme presented with l-THDP, as is supported experimentally by the results of the complementation experiments (Fig. 2A and 2B).
Homologs of CT390 are highly conserved. Orthologous sequences are found in all of the chlamydial species whose genomes have been sequenced. The most divergent representative is P. amoebophila ORF pc0685, which shares 42.6% amino acid sequence identity with CT390. PC0685 is the most closely related ortholog to Arabidopsis ll-DAP aminotransferase in the microbial protein sequence databases (56% identity). P. amoebophila also resembles C. trachomatis in lacking the orthologs of enzymes for the lower DAP pathway other than dapF (see Table 4). To further analyze the chlamydial DAP pathway, the function of PC0685 was analyzed. PC0685 complemented E. coli strain ATM769 (ΔdapD::tet ΔdapE::kan), demonstrating that it is able to produce ll-DAP in vivo. In vitro, PC0685 displayed ll-DAP aminotransferase activity and, like CT390, could use m-DAP as a substrate (see Table 5).
DAP Synthesis Genes Are Expressed During Chlamydial Cell Division.
The organization of genes encoding the upper m-DAP synthesis pathway in C. trachomatis predicts that they form an operon of dapB, asd, lysC, and dapA (Fig. 1). Furthermore, the genomic context of CT390 suggests that it may be in an operon with CT391, a secreted protein of unknown function, and dapF is predicted to be in an operon with glyA, clpB, and another gene of unknown function (see Fig. 6, which is published as supporting information on the PNAS web site). These gene orders are conserved within the members of the Chlamydiaceae family, but not the closely related Parachlamydiaceae family. Interestingly, in P. amoebophila, the genomic context of pc0685 suggests that it might exist as an operon along with dapB and dapA and two other genes, recD and pc0688, a putative phospholipase D. RT-PCR analysis confirmed the polycistronic structure of mRNAs derived from the predicted C. trachomatis operons (see Fig. 6).
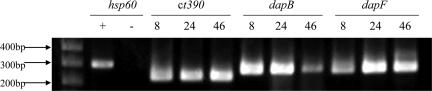
The PG structure of nearly all Gram-negative bacteria incorporate m-DAP into the third position of the PG-peptide. Recent biochemical data have shown that the chlamydiae have the capacity to synthesize PG despite a lack of biochemical detection of PG from these organisms (16). Thus, expression of the m-DAP synthesis genes would be required during intracellular growth and division when other PG synthesis genes are expressed (25–27). As seen for other PG synthesis genes, expression of dapB, dapF, and ct390 mRNA was detected as early as 8 h after infection (Fig. 3).
Fig. 3.
m-DAP synthesis gene expression patterns during Chlamydia development. Mouse L2 fibroblast cells were infected as described. RNA was isolated at 8, 24, and 46 h after infection. cDNA was amplified by using primers internal to hsp60, Ct390, dapB, and dapF. +, DNA control; −, −RT control.
Discussion
The annotation of >350 bacterial genomes has provided a wealth of knowledge regarding variations in microbial physiology. In the absence of a system of genetic manipulation, annotation of the genome sequences of nine strains representing five species of chlamydiae and the closely related P. amoebophila has facilitated greatly our understanding of the unique biology of these organisms. However, annotation of gene products depends on similarity-based comparative analyses that often fail to assign function or assign incorrect function to many genes, resulting in the generation of incomplete biosynthesis pathways (28). Assembly of biosynthetic pathways in the chlamydiae from genome sequencing data predicts that pathways for synthesis of peptidoglycan, folates, coenzymes, and amino acids, such as tryptophan and m-DAP/lysine are incomplete. Studies by several groups support the notion that many incomplete chlamydial pathways are functional (19, 27, 29–32). In some instances, investigators have reasoned that chlamydiae may bypass the “holes” in the biosynthetic pathways by acquiring key intermediates or final products from the host. In the case of the incomplete m-DAP/lysine pathway, lysine could be acquired from the host but not m-DAP because mammalian cells do not synthesize or use m-DAP. Using genetic and biochemical tools, we have identified CT390 as the “hole filler” in the incomplete m-DAP pathway of chlamydiae and Protochlamydia. ct390 originally was annotated as aspC, although it has no AspC activity. We propose that ct390 and its respective chlamydiae homologues be renamed dapL-encoding ll-DAP aminotransferase (EC 2.6.1.83).
In bacteria, m-DAP has two fates: direct decarboxylation to lysine and incorporation into PG. Although DAP decarboxylase activity in C. psittaci lysates has been reported, genome sequencing failed to identify a lysA homologue in the genome of this species (G. Myers, personnel communication). Similarly, our attempts to genetically complement an E. coli ΔlysA mutant failed to identify a chlamydial gene that encodes DAP decarboxylase activity (data not shown). It is likely that the chlamydiae obtain lysine from the mammalian host and synthesize m-DAP solely for PG synthesis. Incorporation of m-DAP in chlamydial PG is supported further by the findings that Nod1 is able to detect chlamydial infections (17, 18).
A surprising finding in the genomes of chlamydiae is the presence of a high number of plant-like genes. This discovery led Brinkman et al. (21) to propose an evolutionary relationship between the chlamydiae and the cyanobacteria, the endosymbiont ancestor of chloroplasts, because horizontal gene transfer does not appear to be occurring in these organisms. Interestingly, the same “hole” in the chlamydial m-DAP pathway also is present in the m-DAP/lysine synthesis pathway of plants and cyanobacteria (3). Recently, a novel ll-DAP-aminotransferase was characterized and shown to complete the m-DAP synthesis pathway in plants and cyanobacteria. Our data provide additional support for an evolutionary relationship between chlamydiae and cyanobacteria-chloroplasts.
Chlamydiae cause significant disease worldwide in both humans and animals. C. trachomatis is the most prevalent cause of bacterial sexually transmitted infections as well as the leading cause of preventable infectious blindness. C. pneumoniae infections have been associated with coronary heart disease and atherosclerosis. Chlamydophila spp. are responsible for a wide variety of clinically and economically important diseases in poultry and livestock. Because m-DAP/lysine synthesis is unique to plants and bacteria, compounds that target this pathway are attractive candidates as herbicides and antimicrobials. Our studies suggest that inhibitors that directly target ll-DAP-aminotransferases could have applications as chlamydiae-specific antibiotics.
Materials and Methods
Bacterial Strains and Growth Conditions.
Detailed descriptions of the E. coli strains and plasmids used in this study are listed in Table 7, which is published as supporting information on the PNAS web site. Strains were grown in M9 minimal with glucose or LB medium with aeration or on agar. Medium was supplemented with ampicillin (25 or 100 μg/ml), kanamycin (50 μg/ml), tetracycline (5 μg/ml), arabinose (0.2%), and DAP (racemic mixture; 100 μg/ml; Sigma, St. Louis, MO) as needed, and cultures were incubated at 30°C or 37°C.
All E. coli DAP mutants were constructed by using the one-step linear recombination method (33). P1L4 was grown on each strain and used to transduce mutations into E. coli MG1655. m-DAP synthesis genes from either C. trachomatis serovar L2 or E. coli MC4100 were PCR-amplified, ligated into pGEM-T (Promega, Madison, WI), subcloned into pBAD18, pUC18, or pUC19, and sequenced. Primers used for chlamydial gene cloning were based on the published C. trachomatis serovar D sequence. Details on the construction of plasmids expressing ct390 and pc0685 as well as purification of CT390 and PC0685 can be found as Supporting Text, which is published as supporting information on the PNAS web site.
Construction of C. trachomatis Serovar L2 DNA Library.
Genomic DNA was isolated from renografin-purified C. trachomatis serovar L2 elementary bodies by using the Qiagen Genomic-tip 100/G kit. A partial Sau3AI digest of DNA was size fractionated by sucrose gradient and fragments between 1.5 and 3 kb were ligated into BamHI-digested pUC19 and transformed into DH5α. Transformants were selected on LB agar containing ampicillin and pooled to isolate plasmid DNA. The library represents 3-fold coverage of the genome at 99.9% confidence and an average insert size of 2.3 kb.
Plating Efficiencies and Viability Curves.
Efficiency of growth on agar medium was performed by diluting overnight (O/N) cultures into LB broth with ampicillin and DAP and growing to an OD600 of 0.5 to 1.0. Cultures were washed in buffered saline gelatin and plated on LB agar or M9 minimal agar containing ampicillin with and without DAP and arabinose when needed. DapF complementation studies were performed by using O/N (stationary phase) cultures grown in LB with ampicillin, DAP, and arabinose. Viability curves were obtained by diluting O/N cultures to an OD600 of 0.03 in LB broth containing ampicillin with and without DAP. At indicated times, dilutions of the cultures were plated onto LB agar supplemented with DAP.
Enzyme Assays.
ll-DAP and m-DAP were synthesized and purified as described in ref. 34. Corynebacterium glutamicum DAP dehydrogenase (CgDdh) was produced as a recombinant protein from plasmid pET28-CgDDH (from D. I. Roper, University of Warwick, Coventry, U.K.) in BL21(DE3). CgDdh was expressed to ≈90% of the soluble protein and converted m-DAP to THDP at a rate of 20 μmol·min−1·mg−1 protein at 30°C so it was not further purified for use in enzyme assays.
Enzyme assays were carried out at 30°C essentially as described in Hudson et al. (20) and were monitored with a Beckman DU800 spectrophotometer. Kinetic constants were determined by varying substrate concentrations while keeping the cosubstrate level constant at the concentration described in Table 3 and Fig. 6 legends. Kinetic data were analyzed by nonlinear regression analysis by using GraphPad (San Diego, CA) Prism Version 4.03.
Pure ll-DAP aminotransferase activity was measured in 1 ml containing 100 μmol HepesKOH (pH 7.6), 0.3 μmol NADPH, 50 μmol NH4Cl, 0.5 μmol ll-DAP, 5 μmol 2-OG, 25 μg CgDdh, and pure CT390 or PC0685. The background rate was measured in a reaction lacking either ll-DAP or 2-OG.
Quantitative assay of the physiological forward reaction was carried out in a two-step reaction. In the first step, THDP was generated from m-DAP, and aminotransferase activity was measured in the second step. THDP was generated in 0.95 ml containing 100 μmol HepesKOH (pH 7.6), 0.5 μmol NADP+, 0.5 mM m-DAP, 25 μg of CgDdh, 0.3 μmol thio-NAD+, 0.3 μmol CoA, and 0.5 μmol glutamate. The prereaction was run to completion and monitored by the increase in absorbance at 340 nm resulting from NADPH formation. Aminotransferase and 200 μg of 2-OG dehydrogenase (0.625 μmol·min−1·mg−1 of protein) then were added, bringing the volume to 1 ml, and then thioNADH formation was monitored at 398 nm. The background rate was measured in a reaction lacking either glutamate or THDP (m-DAP not added to the prereaction).
A semiquantitative assay of the reverse activity was used for measuring ll-DAP aminotransferase activity in crude protein extracts. Soluble proteins were extracted from E. coli by sonication in 100 mM HepesKOH (pH 7.6) followed by buffer exchange with an Amicon Ultra 30,000 MWCO filter (Millipore, Bedford, MA). The reaction contained in 1 ml of 100 μmol HepesKOH (pH 7.6), 0.5 μmol ll-DAP (or other amino donor as indicated in Table 5 legend), 2.0 μmol 2-OG (and other amino acceptors as indicated in Table 6), 1.25 mg O-aminobenzaldehyde, and protein extract. Reactions were monitored at 440 nm. Background rate was measured in a reaction lacking ll-DAP or 2-OG.
RT-PCR.
L2 mouse fibroblast cells were infected with C. trachomatis serovar L2 at a multiplicity of infection of 5 (for 8-h time points) and 1 (for 24- and 46-h time points). Total RNA was extracted by using TRIzol Reagent (Invitrogen, Carlsbad, CA), treated with DNaseI, and reverse transcribed with a gene specific primer or random hexamers by using the Thermoscript RT-PCR System (Invitrogen). cDNA was analyzed for operon and gene expression with Taq polymerase (Qiagen, Valencia, CA).
Supplementary Material
Acknowledgments
We thank Rachel Binet and Scotty Merrell for thoughtful discussions. Protochlamydia amoebophila UWE25 genomic DNA was a kind gift from Matthias Horn (Universität Wien, Wien, Austria). This work was supported by National Institute of Allergy and Infectious Diseases Grant AI044033 (to A.T.M), National Institute of General Medical Sciences Predoctoral Fellowships GM55145 and GM069264 (to A.O.H.), and National Science Foundation Grant IBN-0449542 (to T.L. and C.G.).
Glossary
Abbreviations
- DAP
diaminopimelic acid
- ll-DAP
l,l-diaminopimelic acid
- m-DAP
meso-diaminopimelic acid
- PG
peptidoglycan
- THDP
tetrahydrodipicolinate
- 2-OG
2-oxoglutarate.
Footnotes
The authors declare no conflict of interest.
References
- 1.van Heijenoort J. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 2.Vogel HJ. Proc Natl Acad Sci USA. 1959;45:1717–1721. doi: 10.1073/pnas.45.12.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson AO, Bless C, Macedo P, Chatterjee SP, Singh BK, Gilvarg C, Leustek T. Biochim Biophys Acta. 2005;1721:27–36. doi: 10.1016/j.bbagen.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Harb OS, Abu Kwaik Y. Infect Immun. 1998;66:1898–1903. doi: 10.1128/iai.66.5.1898-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns-Keliher LL, Portteus A, Curtiss R., III J Bacteriol. 1997;179:3604–3612. doi: 10.1128/jb.179.11.3604-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cersini A, Salvia AM, Bernardini ML. Infect Immun. 1998;66:549–557. doi: 10.1128/iai.66.2.549-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutton CA, Southwood TJ, Turner JJ. Mini Rev Med Chem. 2003;3:115–127. doi: 10.2174/1389557033405359. [DOI] [PubMed] [Google Scholar]
- 8.Cox RJ, Sutherland A, Vederas JC. Bioorg Med Chem. 2000;8:843–871. doi: 10.1016/s0968-0896(00)00044-4. [DOI] [PubMed] [Google Scholar]
- 9.Velasco AM, Leguina JI, Lazcano A. J Mol Evol. 2002;55:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 10.Bukhari AI, Taylor AL. J Bacteriol. 1971;105:844–854. doi: 10.1128/jb.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs TM, Schneider B, Krumbach K, Eggeling L, Gross R. J Bacteriol. 2000;182:3626–3631. doi: 10.1128/jb.182.13.3626-3631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann M, Tauch A, Eggeling L, Bathe B, Mockel B, Puhler A, Kalinowski J. J Biotechnol. 2003;104:199–211. doi: 10.1016/s0168-1656(03)00156-1. [DOI] [PubMed] [Google Scholar]
- 13.Cox RJ, Wang PSH. J Chem Soc [Perkin 1] 2001;1:2006–2008. [Google Scholar]
- 14.Weinberger S, Gilvarg C. J Bacteriol. 1970;101:323–324. doi: 10.1128/jb.101.1.323-324.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw-Reid CA, McCormick MM, Sinskey AJ, Stephanopoulos G. Appl Microbiol Biotechnol. 1999;51:325–333. doi: 10.1007/s002530051398. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ, Maurelli AT. Trends Microbiol. 2006;14:70–77. doi: 10.1016/j.tim.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Opitz B, Forster S, Hocke AC, Maass M, Schmeck B, Hippenstiel S, Suttorp N, Krull M. Circ Res. 2005;96:319–326. doi: 10.1161/01.RES.0000155721.83594.2c. [DOI] [PubMed] [Google Scholar]
- 18.Welter-Stahl L, Ojcius DM, Viala J, Girardin SE, Liu W, Delarbre C, Philpott DJ, Kelly KA, Darville T. Cell Microbiol. 2006;8:1047–1057. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 19.Moulder JW, Novosel DL, Tribby IC. J Bacteriol. 1963;85:701–706. doi: 10.1128/jb.85.3.701-706.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson AO, Singh BK, Leustek T, Gilvarg C. Plant Physiol. 2006;140:292–301. doi: 10.1104/pp.105.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkman RSL, Blanchard JL, Cherkasov A, Av-Gay Y, Brunham RC, Fernandez RC, Finlay BB, Otto SP, Ouellette BFF, Keeling PJ, et al. Genome Res. 2002;12:1159–1167. doi: 10.1101/gr.341802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx D, Michaud C, Richaud C, Blanot D, van Heijenoort J. J Bacteriol. 1988;170:2031–2039. doi: 10.1128/jb.170.5.2031-2039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, et al. Science. 2004;304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 24.Mengin-Lecreulx D, Blanot D, van Heijenoort J. J Bacteriol. 1994;176:4321–4327. doi: 10.1128/jb.176.14.4321-4327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. Proc Natl Acad Sci USA. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Sandlin RC, Maurelli AT. J Bacteriol. 2003;185:1218–1228. doi: 10.1128/JB.185.4.1218-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osterman A, Overbeek R. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Maurelli AT. Mol Microbiol. 2005;57:41–52. doi: 10.1111/j.1365-2958.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- 30.Hesse L, Bostock J, Dementin S, Blanot D, Mengin-Lecreulx D, Chopra I. J Bacteriol. 2003;185:6507–6512. doi: 10.1128/JB.185.22.6507-6512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan H, Brunham RC, McClarty G. J Clin Invest. 1992;90:1803–1811. doi: 10.1172/JCI116055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. J Biol Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilvarg C. J Biol Chem. 1959;234:2955–2959. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.