Abstract
The twin-arginine translocation (Tat) pathway is a protein transport system for the export of folded proteins. Substrate proteins are targeted to the Tat translocase by N-terminal signal peptides harboring a distinctive R-R-x-Φ-Φ “twin-arginine” amino acid motif. Using a combination of proteomic techniques, the protein contents from the cell wall of the model Gram-positive bacterium Streptomyces coelicolor were identified and compared with that of mutant strains defective in Tat transport. The proteomic experiments pointed to 43 potentially Tat-dependent extracellular proteins. Of these, 25 were verified as bearing bona fide Tat-targeting signal peptides after independent screening with a facile, rapid, and sensitive reporter assay. The identified Tat substrates, among others, include polymer-degrading enzymes, phosphatases, and binding proteins as well as enzymes involved in secondary metabolism. Moreover, in addition to predicted extracellular substrates, putative lipoproteins were shown to be Tat-dependent. This work provides strong experimental evidence that the Tat system is used as a major general export pathway in Streptomyces.
Keywords: Protein transport, secondary metabolism, Tat pathway, twin arginine signal peptide, proteome
In most bacteria, the general secretory pathway (Sec) is the predominant route for protein export. Proteins exported via Sec are translocated across the membrane in an unfolded state through a membrane-embedded translocon, to which they are targeted by cleavable N-terminal signal peptides (1). More recently, a second export pathway has been described, first in thylakoid membranes of plant chloroplasts and, subsequently, in bacteria. This pathway, designated Tat (for twin-arginine translocation), transports only prefolded protein substrates (2). Unlike the ubiquitous Sec system, Tat appears to be encoded only by approximately half of prokaryotic genomes sequenced so far (3).
Proteins are targeted to the Tat pathway by tripartite N-terminal signal peptides, which superficially resemble Sec signal peptides but differ in important features. The most striking is the presence of a conserved twin-arginine motif in the n region of Tat signal peptides. This can be loosely defined as R-R-x-Φ-Φ, where Φ represents a hydrophobic amino acid (4). The consecutive arginine residues are almost invariant and are critical for transport by this pathway (5, 6). A second discriminating feature is a generally less hydrophobic h region in Tat than in Sec signal peptides (7).
Most studies of the bacterial Tat pathway have focused on the Gram-negative bacterium Escherichia coli, in which three membrane proteins, TatA, TatB, and TatC, make up the translocon (8–11). The number of Tat substrates in E. coli is unlikely to exceed 28 (reviewed in ref. 12). Approximately two-thirds of these bind redox cofactors and are important for respiratory metabolism. For the Gram-positive Bacillus subtilis, the only other bacterium to have been studied in depth, only two Tat substrates have been experimentally verified, and there are probably no more than seven in total (13, 14).
Estimates of Tat substrate numbers in other organisms are based almost entirely on analysis of genome sequences by using programs trained to recognize the specific features of Tat targeting sequences (3, 15, 16), with no experimental verification of in silico predictions other than analysis of selected signal peptides (e.g., ref. 17). Interestingly, these predictions suggest that the Tat pathway is a general protein export pathway in numerous prokaryotes. By far the largest numbers of predicted Tat substrates are in Streptomyces species, represented by the sequenced strains Streptomyces coelicolor and Streptomyces avermitilis. Streptomycetes, which encode TatA, TatB, and TatC homologs (18), are members of the high G+C Gram-positive actinomycetes. They are mycelial organisms, frequently soil-dwelling, that undergo complex morphological differentiation involving the formation of aerial hyphae and spores. Streptomycetes produce a range of diverse and important secondary metabolites, including many commercial antibiotics (19). They also are important for biotechnology because they are prolific protein secretors and are used for the commercial production of valuable proteins, such as secreted cellulases. Clearly, if these organisms use the Tat pathway extensively, they may provide an ideal system for the heterologous expression of proteins that are incompatible with secretion by the Sec pathway.
Of ≈800 predicted exported proteins in S. coelicolor, in silico predictions suggest 145–189 of these to be Tat-dependent (15, 16). In this work, we have set about identifying members of the Tat secretome of S. coelicolor experimentally. Concentrating on cell wall-associated proteins, the exported proteome of S. coelicolor M145 and a mutant defective in Tat transport were compared. Using state-of-the-art protein separation and identification technologies, in combination with a powerful previously undescribed reporter assay, a broad spectrum of 27 exported proteins were unequivocally established as being Tat-dependent. This experimental demonstration shows that the Tat pathway is used as a general protein export system in S. coelicolor.
Results
To determine the impact of Tat-mediated protein export in S. coelicolor, marked mutations in each of the tatA, tatB, and tatC genes and unmarked in-frame deletions of tatB and tatC were constructed. The gross phenotypes of all of the S. coelicolor tat mutants were similar to those observed for the tatB and tatC mutants of S. lividans (refs. 18 and 20; Fig. 4, which is published as supporting information on the PNAS web site). In particular, the S. coelicolor tat mutants exhibited dispersed growth and were susceptible to lysis in liquid culture. Therefore, to study the extracellular proteins of these fragile strains, it was necessary to grow the cells on solid media only and focus analysis on the exported proteins associated with the cell wall.
Tat-Dependent Proteins Identified by 2D Gel Electrophoresis (2-DGE).
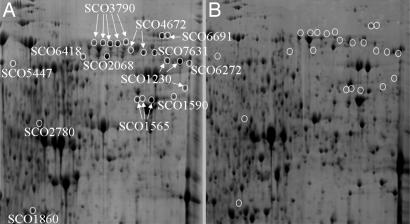
The S. coelicolor parent strain (M145) and the ΔtatC mutant strain were cultured on three standard types of solid media: R5, mannitol soya (MS), and complete medium (CM). Proteins released by washing the cell walls of the M145 and ΔtatC strains were analyzed by 2-DGE with pH gradients of 4–7. Typical 2D gel electrophoretograms for the cell wall-associated proteome of these strains cultured on R5 medium are shown in Fig. 1 and for MS and CM media in Fig. 5, which is published as supporting information on the PNAS web site. Significant differences in the staining patterns were observed between the two strains, and proteins that were present in M145 but absent from the ΔtatC strain clearly represented candidates for Tat substrates. Putative Tat-targeted proteins were identified by MALDI-TOF mass spectrometry (after in-gel digestion with trypsin); several of them are marked in Figs. 1 and 5. A total of 98 proteins that migrated with unique positions were identified in the M145 washes. In all, 37 of these proteins had identifiable N-terminal signal peptides, and 34 of those signal peptides contained RR dipeptides (Table 2, which is published as supporting information on the PNAS web site). Cross-referencing with the predicted Tat substrates encoded by the S. coelicolor genome and identified by the programs TatP and TATFIND (15, 16), suggested that 21 of this group of 37 had plausible Tat-targeting signal peptides.
Fig. 1.
Two-dimensional gel analysis of proteins from cell wall fractions of S. coelicolor M145 (A) and a ΔtatC mutant derivative (B). Strains were cultured on R5, and cell wall proteins were isolated as described. Proteins were separated in the first dimension by isoelectric focusing (pH gradient 4–7) and in the second dimension by SDS/PAGE. Protein spots that migrated to specific positions in the M145 cell wall fractions but were absent from the corresponding position in cell wall fractions of the ΔtatC strain are circled. The identities of the protein spots were determined by in-gel tryptic digestion, followed by mass spectrometry.
In addition to the 37 putative exported proteins, the remaining 61 proteins unique to the tat+ strain included ribosomal subunit proteins and proteins of the thioredoxin pathway (Table 3, which is published as supporting information on the PNAS web site). The identification of such obviously cytoplasmic proteins undoubtedly reflected contamination of the cell wall washes with cytoplasmic proteins from lysed bacteria. Cytoplasmic protein contamination in this type of analysis is almost inevitable given the S. coelicolor life cycle, which involves altruistic lysis of the mycelium to release nutrients for the continued growth of the aerial parts of the colony (21). Moreover, such cytoplasmic protein contamination has been reported during analyses of extracellular proteins from B. subtilis (22) and Mycobacterium tuberculosis (23). Indeed, cell lysis, as well as substrate modification (see below) and up-regulation of Sec substrates, also may contribute to the presence of additional extracellular proteins in the ΔtatC strain that are absent in M145 (Table 4, which is published as supporting information on the PNAS web site).
Several of the exported proteins were detected as multiple spots in the M145 sample, possibly as a result of posttranslational modification or proteolysis, and it was not uncommon for one or more of the additional spots for a particular protein to be absent from cell wall fractions of the ΔtatC strain. Therefore, because several putative extracellular proteases are predicted Tat substrates (3), the lack of a particular protein spot in the 2-DGE analysis of the ΔtatC strain might be indicative of the lack of postexport protein modification rather than a lack of Tat translocation per se.
Tat-Dependent Proteins Identified by Multidimensional Protein Identification Technology.
In parallel to the traditional 2-DGE approach, the cell wall proteome of the S. coelicolor strains was analyzed by multidimensional protein identification technology (MudPIT), a sensitive modern technique used to separate and identify even low-abundance proteins from complex mixtures. MudPIT analysis of the M145 and ΔtatC strains grown on CM medium identified 308 and 279 individual proteins from the cell wall washes, respectively, of which 188 were common to both samples (Tables 2–4). Of the 120 remaining proteins exclusively present in M145 cell wall sample, 20 corresponded to proteins bearing plausible N-terminal signal peptides that contained two consecutive arginines in their sequences (including 11 that had been identified by 2-DGE analysis), strongly suggesting that this group represents the Tat substrates (Table 2).
A Tat Transport Assay Based on Agarase.
The two proteomic techniques above identified a total of 43 proteins that could have been transported by the Tat pathway (Table 2). Next, our attention turned to identifying bona fide Tat-targeting signal peptides associated with this group of proteins. Sec-dependent signal peptides are not normally recognized by the Tat machinery, and Tat-targeted proteins usually are folded and, therefore, are not normally compatible for Sec-dependent export. We therefore developed a reporter-based assay for Tat transport designed to address directly whether the group of 43 putative Tat substrates were indeed synthesized with bona fide Tat signal peptides.
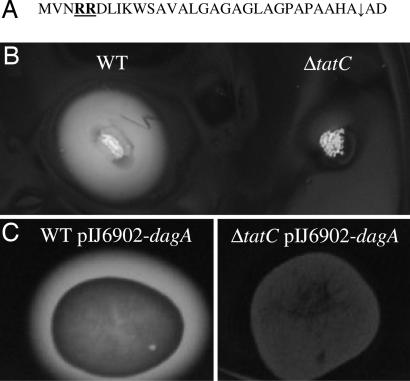
S. coelicolor M145 secretes an extracellular agarase enzyme that degrades agar to oligosaccharides that can be subsequently taken up and used as a carbon source for growth. Because of this activity, colonies of M145 are commonly observed to “sink” into agar-containing solid media. Agarase is encoded by the dagA gene, and the protein product bears an N-terminal signal peptide that contains an apparent twin-arginine motif (Fig. 2A), despite earlier reports that it interacts with components of the signal recognition particle (24). Interestingly, colonies of the S. coelicolor tat mutants failed to sink into solid media suggesting DagA is perhaps a genuine Tat substrate. Subsequent staining of the agar-containing plates with lugol, an iodine-based reagent, showed a clear halo corresponding to degradation of agar around M145, but no such halo around colonies of the tat mutants (Fig. 2B), providing strong and visual evidence that the tat mutants have no significant extracellular agarase activity.
Fig. 2.
Agarase is a Tat substrate. (A) The agarase signal peptide. The consecutive arginines of the twin-arginine motif are highlighted in bold underline. (B) S. coelicolor strains M145 (WT) and TP1 (ΔtatC::ApraR) were grown on MM-C minimal medium for 5 days and were stained with Lugol solution. (C) Strains M145 (WT) and TP4 (ΔtatC) harboring pIJ6902-dagA in single copy were grown on minimal media containing 0.5% glucose, apramycin, and thiostrepton for 3 days before staining with Lugol solution.
Transcription of dagA in S. coelicolor is highly regulated and reported to be repressed by glucose in the growth medium (25). To ensure agarase activity was expressed in the tat mutants, the dagA gene was placed under control of the tipA thiostrepton-inducible promoter and incorporated onto the chromosome in single copy at the phage φC31 attachment site. As shown in Fig. 2C, copious extracellular agarase activity was produced in the M145 derivative but no activity was detectable in the ΔtatC derivative carrying this construct. These results demonstrate S. coelicolor DagA is a Tat-dependent extracellular protein.
However, to exploit agarase as a screen for Tat-dependent protein export the ability of the DagA mature protein to be targeted via alternative export routes was first tested. The Tat-targeting signal peptide of DagA was swapped for the Sec-targeting signal peptides of three S. coelicolor proteins: SCO3053, SCO5660, and SCO6199. No extracellular agarase activity could be detected when the mature portion of the enzyme was targeted to the Sec machinery (Fig. 3; see Fig 6, which is published as supporting information on the PNAS web site), most likely as a result of folding of agarase before export. Thus, DagA can be used as a reporter exclusively for Tat-mediated protein secretion.
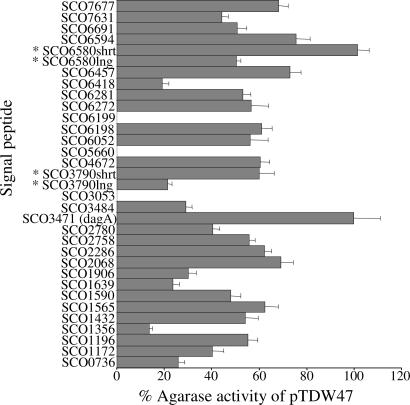
Fig. 3.
Export of agarase mediated by S. coelicolor signal peptides. The y axis shows the signal peptides from a range of S. coelicolor cell wall-associated proteins (listed in Tables 2 and 4). The x axis gives a measure of agarase export from each fusion protein compared with agarase bearing its native signal peptide (set at 100%). Each construct carries the native agarase promoter and ribosome-binding site, and, therefore, the activity should be a measure of the efficiency of export directed by each particular signal peptide. The assay was verified by using signal peptides from SCO3053, SCO5660, and SCO6199, which were included as negative controls because they lack twin arginines in the signal peptide and were detected in the cell wall fractions of both M145 and the ΔtatC mutant by MudPIT analysis. None of these signal peptides mediated extracellular agarase activity. The signal peptides from the following proteins also were tested and were found to be negative in this assay: SCO0432, SCO0474, SCO0494, SCO0930, SCO1230, SCO1396, SCO1824, SCO1948, SCO1968, SCO2226, SCO2383, SCO2446, SCO2591, SCO2637, SCO2786, SCO2821, SCO3456, SCO4010, SCO4142, SCO4152, SCO4884, SCO4885, SCO5074, SCO5113, SCO5447, SCO5461, SCO6009, SCO6644, SCO6738, and SCO7399. ∗, two versions of each of these signal peptides (designated long and short), representing two alternative start sites, were tested (see Tables 2 and 4).
Fusions of the signal peptides of each of the putative 43 Tat-targeted proteins identified by the proteomic techniques to mature agarase were constructed. The fusion proteins were expressed in a nonagarase-producing host strain (Streptomyces lividans 1326) and then assessed for the production of agarase activity by using the lugol-based plate test. In all, 25 of the 43 signal peptides clearly directed Tat-dependent secretion of DagA, consistent with their being bona fide Tat signal peptides (Fig. 3 and Table 1; a representative sample of these also was tested in a S. lividans tatC strain (26), further demonstrating their Tat dependence; Fig. 6). Of these 25, TATFIND predicted 20 and TatP predicted 18 to be Tat signal peptides. Both prediction programs failed to identify two of the Tat-active signal peptides (SCO2286 and SCO3790) because of their unusual length; however, in silico N-terminal truncation of these signal peptides restored the ability of both programs to recognize them as Tat-targeting signal peptides. Moreover, it is clear when comparing the results from the agarase assays (Figs. 3 and 6) that secretion efficiency varies significantly with signal peptide primary structure. Thus, comparisons of heterologous protein translocation by using only a single Tat or Sec signal peptide, as recently performed for hTNFalpha and hIL10 in S. lividans (27), may not reflect the true potential of each pathway for protein transport.
Table 1.
S. coelicolor Tat-dependent proteins identified in this work
| Protein | Description |
|---|---|
| SCO0736 | Putative secreted protein |
| SCO1172 | Putative amidase (putative secreted protein) |
| SCO1196 | Putative secreted protein |
| SCO1356*† | Putative iron sulfur protein (putative secreted protein) |
| SCO1432 | Putative membrane protein |
| SCO1565 | Putative glycerophosphoryl diester phosphodiesterase |
| SCO1590 | Putative secreted protein (transglycosidase) |
| SCO1639† | Putative secreted peptidyl-prolyl cis-trans isomerase protein |
| SCO1906 | Putative secreted protein (PhoX phosphatase) |
| SCO2068 | Putative secreted alkaline phosphatase |
| SCO2286 | Putative alkaline phosphatase |
| SCO2758 | β-N-acetylglucosaminidase (putative secreted protein) |
| SCO2780† | Putative secreted protein (cd01146, FhuD, Fe3+-siderophore binding domain FhuD) |
| SCO2786 | β-N-acetylhexosaminidase (Transglycosidase) |
| SCO3484† | Putative secreted sugar-binding protein |
| SCO3790 | Conserved hypothetical protein of PhoX type |
| SCO4672 | Putative secreted protein |
| SCO6052 | Putative membrane protein |
| SCO6198 | Putative secreted protein |
| SCO6272* | Putative secreted FAD-binding protein |
| SCO6281* | Putative FAD-binding protein |
| SCO6457 | Putative β-galactosidase (Galactose mutarotase-like) |
| SCO6580 | Hypothetical protein (Xylose isomerase-like) |
| SCO6594 | Putative secreted protein |
| SCO6691 | Putative phospholipase C |
| SCO7631 | Putative secreted protein |
| SCO7677 | Putative secreted solute-binding protein |
*Cofactor containing.
†Putative lipoproteins.
The remaining 18 signal peptides of the 43 that contained consecutive arginine residues could not mediate secretion of active agarase, and of these, TATFIND predicted 0 and TatP predicted only 1 to be a genuine Tat-targeting signal peptide (Table 2). These 18 signal peptides therefore were considered not to be Tat-targeting signals and the corresponding passenger proteins are not detected in the cell wall fraction of the ΔtatC strain presumably because of pleiotropic effects.
Of the exported proteins identified in cell wall analyses of the ΔtatC strain (Table 4), two (SCO0736 and SCO7677) were predicted by both TATFIND and TatP programs to contain Tat-targeting signal peptides. These signal peptides were subjected to the agarase test, and both conferred extracellular agarase activity to the S. lividans host strain, bringing the total of verified Tat-targeting signal peptides to 27 (Fig. 3). The apparent presence of these final two proteins in the ΔtatC strain cell wall might have been due to minor contamination of washes because of cell lysis.
Finally, rare examples of Tat-dependent signal peptides lacking consecutive arginine residues have been reported (e.g., ref. 28). A total of seven exported proteins were identified in the cell wall fraction of M145 that did not contain obvious twin-arginine motifs in their signal peptides (Table 2). The agarase test was applied to six of these seven signal peptides, but none were found to direct export of active agarase.
Discussion
Taken altogether, this work has unambiguously identified 27 Tat-dependent proteins in S. coelicolor. This number represents ≈30% of all of the exported proteins that we detected in the cell wall fraction of the tat+ strain and clearly demonstrates that the Tat pathway is a major protein translocation route. The agarase reporter assay developed here and used to validate possible Tat-targeting signal peptides is particularly powerful because it is not only facile and rapid but also semiquantitative and, therefore, provides a ready measure of transport efficiency for a particular Tat-targeting signal peptide. It is also anticipated that the agarase reporter system will facilitate the exploitation of the Tat pathway for heterologous protein production.
The identified Tat-exported proteins listed in Table 1 represent a broad spectrum of functional classes: several are predicted to be involved in phosphate and carbohydrate metabolism, nutrient transport, and lipid metabolism. However, unlike the well characterized E. coli system, only 3 of the 27 Tat substrates detected here are likely to be cofactor-containing and, remarkably, 2 of these (SCO6272 and SCO6281) are associated with a type I modular polyketide synthase gene cluster, indicating a role in secondary metabolism and, therefore, are not expected a priori to be exported. Substrates of the twin-arginine translocation pathway have hitherto not been found associated with a secondary metabolite gene cluster.
Other notable Tat substrates identified here include two proteins involved in peptidoglycan metabolism, SCO0736 and SCO1172, the latter being a probable cell wall amidase. Considering the connectivity between nonexport of amidases and the integrity of the cell envelope in E. coli tat mutants (29), the identification of SCO1172 as a Tat substrate may well account for the fragility of the Streptomyces tat mutants. Moreover, as with the E. coli model, it may perhaps be possible to “rescue” the fragile phenotype of the Streptomyces tat mutants by increased expression of a Sec-dependent amidase. This likely would enable a further proteomic analysis of the “Tat secretome” in this organism and the assignment of Tat-targeted proteins not associated with the cell wall.
A major surprise was that 4 of the 27 Tat substrates are annotated as lipoproteins (16) (Table 1). Again, this in vivo data demonstrates the Tat dependence of putative lipoproteins and implies that class I and class II cellular signal peptidases can recognize and cleave Tat-targeting signal peptides. We note that very recently an outer membrane-localized dimethyl sulfoxide reductase, a Tat substrate that is also strongly predicted to be a lipoprotein, has been described in Shewanella oneidensis (30).
Overall, these data demonstrate that the Tat pathway can export a diverse range of proteins and further reinforces the contention that the Tat pathway is a truly general protein export system in this organism. The ability of the Streptomyces Tat pathway to export proteins requiring various anti- and possibly posttranslocation modifications, as well as two of the largest single Tat substrates ever reported (SCO6198 and SCO6457, at 116 and 146 kDa, respectively), underscores the potential of the Tat system in Streptomycetes for heterologous protein production.
Materials and Methods
Strains, Growth Conditions, and Gross Phenotypic Analysis.
Derivatives of S. coelicolor strain M145 (19, 31) and Streptomyces lividans 1326 (31) were used. Construction of the M145 strains with antibiotic-marked and unmarked in-frame deletions of tatC and tatB followed published protocols and is described in full in Supporting Text, which is published as supporting information on the PNAS web site. Strains were cultured on the following standard laboratory growth media: minimal medium (31), CM (32), MS (33), or R5 (34) for growth on plates and Tryptone soya broth (TSB) or Yeast extract-malt extract (YEME) for liquid growth (31). Antibiotics were used at a final concentration of 50 μg/ml unless otherwise stated. When cultured on solid MS, the S. coelicolor tat mutant strains formed very small colonies that appeared to hypersporulate (Fig. 4A). Conversely, on the hyperosmotic solid R5 medium (Fig. 4B), tat mutants failed to sporulate and produced very little of the blue-pigmented antibiotic actinorhodin. In TSB liquid medium, the tat mutants grew in a very dispersed manner compared with M145, which normally grows as pellets. Consistent with the pronounced developmental defect noted on R5 agar, tat mutant strains failed to grow in liquid YEME medium (which contains 34% sucrose) unless sucrose was excluded. S. coelicolor tat mutant mycelium were markedly more fragile than M145 mycelium and were particularly prone to lysis on shaking.
Plasmid Construction.
The primer sequences used to assemble a reporter construct based on the promoter and structural gene for agarase, dagA, are listed in Table 5, which is published as supporting information on the PNAS web site. pTDW46 (based on the shuttle vector pSET152; ref. 35) carries an insert consisting of a copy of the dagA gene plus its promoter, where the region encoding the signal peptide has been replaced with aadA, the streptomycin resistance gene from pIJ778 (36). The aadA gene can be removed from this plasmid by digestion with NdeI and BamHI and replaced with DNA fragments encoding different signal peptides. Thus, each of these clones carries the dagA ribosome-binding site, with identical spacing between the ribosome-binding site and the start codon. A list of oligonucleotide primers, the signal peptides they amplify and the subsequent derivatives of pTDW46 carrying these signal peptides are listed in Table 5.
pIJ6902dagA carries a C-terminally myc epitope-tagged derivative of agarase under control of the thiostrepton-inducible promoter PtipA. The agarase gene was amplified by using primers kddagaf and kddagamyc (Table 5), digested with NdeI and BamHI, and subsequently cloned into similarly digested pIJ6902 (37). pIJ6902dagA was transferred subsequently to the φC31 site on the chromosome of M145 and TP4 as described in ref. 35.
Isolation of Cell Wall-Associated Proteins and 2D Electrophoresis.
Proteins were prepared for 2D gel analysis or MudPIT as follows. S. coelicolor strains M145 or TP1 were grown by inoculating 106 spores onto sterile cellophane disks placed on the surface of CM, R5, or MS media. After incubation for 48 h at 30°C, the biomass was scraped from the disks, dispersed in 5 M LiCl solution, and left on ice for 30 min. The suspensions subsequently were vortexed for 2–3 min, and the biomass was removed by centrifugation (1,800 × g for 5 min) followed by passage through a 0.45-μm filter. Proteins then were precipitated from the LiCl solution by adding trichloroacetic acid to a final concentration of 20%, incubating on ice for 30 min, and centrifuging at 1,800 × g for 15 min. After centrifugation, the mixture had settled into two phases. The upper phase was removed and discarded, and water was added to the lower phase to bring it to the original volume. At this point, the solution turned cloudy and it again was centrifuged at full speed for 15 min, after which the precipitated proteins formed a pellet. The pellet was washed three times with −20°C acetone and then air-dried.
For 2D gel analysis, the protein samples were resuspended in IEF sample buffer [8 M urea/0.5% CHAPS/0.2% DTT/0.5% IPG buffer, pH 4–7 (Amersham Biosciences, Little Chalfont, U.K.)/0.002% bromophenol blue], and protein concentration was determined by using the Bio-Rad (Hercules, CA) DC protein assay. The proteins in the sample were separated in the first dimension by their differing isoelectric points. Samples of protein (500–1,000 μg) were loaded onto Amersham Biosciences 18-cm Immobiline Drystrip pH 4–7 or pH 6–11 isoelectric focusing gels and subsequently were resolved by electrophoresis for 33 kVh by using an Amersham Biosciences Ettan IPGphor isoelectric focusing unit. Focused strips were treated as described in ref. 38 and separated in the second dimension by SDS/PAGE with Amersham Biosciences DALT 12.5% gels. The gels were stained by using a colloidal Coomassie blue stain and scanned with a Proxpress Proteomic Imaging System scanner for later comparison. Proteins spots of interest were excised from the gel, digested with trypsin, and identified by MALDI-TOF peptide mass fingerprint analysis as described in ref. 38.
Multidimensional Protein Analysis.
Samples of precipitated protein (1 mg), prepared as described above, were dissolved by addition of 100 μl of 100 mM Tris-HCl pH 8.0/200 μl of 0.4% RapiGest (Waters, Elstree, U.K.) (39) in 20 mM Tris·HCl, pH 8.0/10 μl of 40 mM EDTA in water/40 μl of 45 mM DTT in 100 mM Tris·HCl, pH 8.0. Once the pellets had dissolved, the samples were heated in a water bath at 90°C for 30 min, then cooled to room temperature. Forty microliters of 100 mM iodoacetamide in 100 mM Tris·HCl (pH 8.0) was added, and the sample was incubated in the dark for 15 min. Ten micrograms of trypsin (modified sequencing grade porcine, Promega, Southampton, U.K.) in 10 μl of 20 mM Tris·HCl (pH 8.0) was added to each sample, which was incubated at 37°C. After 16 h, an additional 10 μg of trypsin was added, and the samples were incubated for an additional 8 h. The RapiGest was denatured before MudPIT mass spectrometry by adding 40 μl of 500 mM HCl to give a concentration between 30 and 50 mM (pH ≤ 2) followed by incubation at 37°C for 45 min. The cloudy samples containing hydrolyzed detergent were centrifuged at 13,000 rpm for 10 min in a BioFuge (Heraeus), and the supernatants were removed carefully for chromatographic separation.
Full details of the chromatographic separation, mass spectrometry, and database searching can be found in Supporting Text. Briefly, samples were loaded onto a biphasic column comprising a strong cation exchange phase (SCX) and a reverse phase. Peptides were eluted stepwise from the SCX phase by using increasing concentrations of salt onto the reverse phase. A reverse-phase gradient then was generated, and peptides were eluted into a Q-ToF2 mass spectrometer (Micromass, Manchester, U.K.). The data from each reverse-phase gradient was combined and searched by using MASCOT (Matrix Science, London, U.K.) (40).
Agarase Assay.
Tat dependence of S. coelicolor agarase secretion was demonstrated by growing strains M145 and TP4 harboring either pIJ6902 or pIJ6902dagA on minimal medium containing glucose (31), which strongly represses expression of the native agarase (25). Approximately 104 spores of each strain were spotted onto minimal medium containing glucose, additionally supplemented with apramycin and thiostrepton (to induce expression of the myc-tagged agarase) and grown at 37°C for 72 h, after which they were stained with Lugol solution (Sigma, St. Louis, MO) for 45 min.
Plasmids encoding signal peptide-agarase fusion proteins (listed in Table 5) were transferred by mating into the S. lividans tat+ strains 1326 and 10–164 (as 1326, msiK−) and the 10–164 isogenic tatC mutant (26), all of which lack native agarase. Plates containing mature spores of the resultant strains, along with S. coelicolor M145 and TP4, were used to inoculate, with a dissecting needle, MM-C minimal medium (10 g of Agar/1 g of (NH4)2SO4/0.5 g of K2HPO4/0.2 g of FeSO4/7H2O in 1 liter) lacking a carbon source other than agar. The inoculated plates were grown for 5 days at 30°C, after which they were stained with Lugol solution (Sigma) for 45 min before photography on a light box. Relative activities were estimated by determining the diameters of the zones of clearing by using the measure tool of the image manipulation program GIMP (open source software distributed under a GNU general public license at www.gimp.org). One sample was included in every batch of results as a standard to which the diameters of the zones of clearing were adjusted. Each zone of clearing was measured twice with each measurement being at right angles to the other. The mean of the two measurements was used to calculate R2 (R = radius) as an estimate of the concentration of agarase. All measurements then were expressed as a percentage of the activity of pTDW47, which is the reporter pTDW46 with the native agarase signal peptide reintroduced. Nine measurements were made for each data point, and the standard error of the mean was calculated for each point.
Supplementary Material
Acknowledgments
We thank R. Morosoli (Université du Québec, Ville de Laval, QC, Canada) for providing the S. lividans tatC strain; M. Bibb, D. Hopwood, and F. Sargent for advice and help with the manuscript; and A. Hesketh, M. Elliot, M. Hutchings, M. Buttner, and other members of the Streptomyces groups at John Innes Centre as well as B. Berks and J. Willey for technical advice and helpful discussion. Support was provided by National Institutes of Health Cell and Molecular Biology Training Grant T32-GM07229, American Heart Association Predoctoral Fellowship 0415376U (to K.C.D.), and Department of Energy Grant DE-FG02-01ER15169 (to M.P.). This project also was supported by Commission for the European Union Project LHSG-CT-2004-005257, Biotechnology and Biological Sciences Research Council Grants EGH16082 and 208/G18887, and a grant-in-aid (to J.I.C.), and by the Medical Research Council via a Senior Non-Clinical Fellowship award (to T.P.).
Abbreviations
- CM
complete medium
- 2-DGE
2D gel electrophoresis
- MS
mannitol soya medium
- MudPIT
multidimensional protein identification technology
- Sec
general secretory pathway
- Tat
twin-arginine transport.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Manting EH, Driessen AJM. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 2.Berks BC, Palmer T, Sargent F. Adv Microb Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 3.Dilks K, Rose RW, Hartmann E, Pohlschröder M. J Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks BC. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 5.Halbig D, Wiegert T, Blaudeck N, Freudl R, Sprenger GA. Eur J Biochem. 1999;263:543–551. doi: 10.1046/j.1432-1327.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 6.Stanley NR, Palmer T, Berks BC. J Biol Chem. 2000;257:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 7.Cristóbal S, de Gier J-W, Nielsen H, von Heijne G. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 9.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogsch E, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 11.Sargent F, Stanley NR, Berks BC, Palmer T. J Biol Chem. 1999;274:36073–36083. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 12.Palmer T, Sargent F, Berks BC. Trends Microbiol. 2005;13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Pop O, Martin U, Abel C, Muller JP. J Biol Chem. 2002;277:3268–3273. doi: 10.1074/jbc.M110829200. [DOI] [PubMed] [Google Scholar]
- 14.Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl J .-M. Mol Microbiol. 2004;54:1319–1325. doi: 10.1111/j.1365-2958.2004.04341.x. [DOI] [PubMed] [Google Scholar]
- 15.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. Mol Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 16.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. BMC Bioinformatics. 2005;6:167–175. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Jacques PE, Ghinet MG, Brzezinski R, Morosoli R. Microbiology. 2005;151:2189–2198. doi: 10.1099/mic.0.27893-0. [DOI] [PubMed] [Google Scholar]
- 18.Schaerlaekens K, Schierova M, Lammertyn E, Geukens N, Anne J, Van Mellaert L. J Bacteriol. 2001;183:6727–6732. doi: 10.1128/JB.183.23.6727-6732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 20.Schaerlaekens K, Van Mellaert L, Lammertyn E, Geukens N, Anne J. Microbiology. 2004;150:21–31. doi: 10.1099/mic.0.26684-0. [DOI] [PubMed] [Google Scholar]
- 21.Manteca A, Fernandez M, Sanchez J. Res Microbiol. 2006;157:143–152. doi: 10.1016/j.resmic.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. Genome Res. 2001;11:1483–1502. doi: 10.1101/gr.182801. [DOI] [PubMed] [Google Scholar]
- 23.Rosenkrands I, Weldingh K, Jacobsen S, Hansen CV, Florio W, Gianetri I, Andersen P. Electrophoresis. 2000;21:935–948. doi: 10.1002/(SICI)1522-2683(20000301)21:5<935::AID-ELPS935>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 24.Palacin A, de la Fuente R, Valle I, Rivas LA, Mellado RP. Microbiology. 2003;149:2435–2442. doi: 10.1099/mic.0.26313-0. [DOI] [PubMed] [Google Scholar]
- 25.Servin-Gonzalez L, Jensen MR, White J, Bibb M. Microbiology. 1994;140:2555–2565. doi: 10.1099/00221287-140-10-2555. [DOI] [PubMed] [Google Scholar]
- 26.Faury D, Saidane S, Li H, Morosoli R. Biochem Biophys Acta. 2004;1699:155–162. doi: 10.1016/j.bbapap.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Schaerlaekens K Lammertyn E, Geukens N, De Keersmaeker S, Anne J, Van Mellaert L. J Biotechnol. 2004;112:279–288. doi: 10.1016/j.jbiotec.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Ignatova Z, Hörnle C, Nurk A, Kasche V. Biochem Biophys Res Comm. 2002;291:146–149. doi: 10.1006/bbrc.2002.6420. [DOI] [PubMed] [Google Scholar]
- 29.Ize B, Stanley NR, Buchanan G, Palmer T. Mol Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 30.Gralnick JA, Vali H, Lies DP, Newman DK. Proc Natl Acad Sci USA. 2006;103:4669–4674. doi: 10.1073/pnas.0505959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norfolk, UK: The John Innes Found; 2000. [Google Scholar]
- 32.Hopwood DA. Bacteriol Rev. 1967;31:373–403. doi: 10.1128/br.31.4.373-403.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. Appl Microbiol Biotechnol. 1989;31:272–277. [Google Scholar]
- 34.Thompson CJ, Ward JM, Hopwood DA. Nature. 1980;286:525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- 35.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 36.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih C-J, Kao CM, Buttner MJ, Cohen SN. Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 38.Hesketh AR, Chandra G, Shaw AD, Rowland JJ, Kell DB, Bibb MJ, Chater KF. Mol Microbiol. 2002;46:917–932. doi: 10.1046/j.1365-2958.2002.03219.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu YQ, Gilar M, Lee PJ, Bouvier ES, Gebler JC. Anal Chem. 2003;75:6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 40.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.