Abstract
Precursor of nerve growth factor (proNGF) has been found to be proapoptotic in several cell types and mediates its effects by binding to p75 neurotrophin receptor (p75NTR) and sortilin. The proNGF molecule is processed by proteases at three dibasic sites found in the pro domain to form mature NGF (termed herein as sites 1, 2, and 3 from the proNGF N terminus). Of these processing sites, site 3, adjacent to the N terminus of mature NGF, was thought to be the major site responsible for processing of proNGF to mature NGF. We found that mutating this major processing site (site 3) resulted in a form of proNGF that was only partially stable. On introducing additional mutations in the pro domain at the other two dibasic sites, we found the stability of proNGF to increase significantly. Here we describe the construction, expression, and purification of this more stable proNGF molecule. The two consecutive basic residues at each of the three sites were mutated to neutral alanine residues. Expression was performed in stably transfected Sf21 insect cells. Purification involved strong cation-exchange chromatography and N60 immunoaffinity column chromatography. The construct with all three sites mutated (termed proNGF123) gave all proNGF with no mature NGF and was not cleaved by three proconvertases (furin, PACE-4, and PC-2) known to proteolyze proneurotrophins in vivo. This stable proNGF molecule demonstrated proapoptotic activity on rat pheocytochroma PC12 cells, PC12nnr cells, C6 glioblastoma cells, and RN22 schwannoma cells.
Keywords: p75 receptor, Sf21 stable cell line, Trk receptor, neurotrophin, apoptosis
The neurotrophin family includes structurally related proteins that promote the survival, growth, and maintenance of neurons in the central and peripheral nervous systems (1). Nerve growth factor (NGF), the first member of the family, was discovered by Levi-Montalcini and coworkers (2) over 50 years ago. Other members of the family include brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), NT-4/5, and NT-6 (1–4). Neurotrophins play a crucial role in neuronal survival, differentiation, growth, and apoptosis (5). Each neurotrophin binds to a 140-kDa tyrosine kinase receptor, Trk receptor. NGF binds to TrkA, BDNF and NT-4/5 selectively bind to TrkB, and NT-3 binds to TrkC (1, 6). In addition to a selective Trk receptor for each neurotrophin, there is a common neurotrophin receptor (NTR), p75NTR (7, 8). The two receptors, Trk and p75NTR, are structurally unrelated with neurotrophins interacting primarily with the Ig-like C2 (IgGC2) domain of the Trk receptors but with the cysteine-rich domains of the p75NTR receptor (9–11). Neurotrophin binding to the Trk receptors transduces positive signals like growth, survival, or differentiation, whereas binding to the p75NTR/Trk heterodimer can transduce positive or negative signals (12–16). Unlike the Trk receptors that possess signature tyrosine kinase motifs, p75NTR lacks any intrinsic catalytic activity. The p75NTR receptor mediates signals through a series of adaptor proteins (1, 6, 8, 17). The role of p75NTR in cell signaling, particularly apoptosis, is being found to be increasingly important.
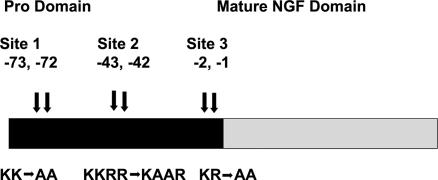
Mature NGF is a 118- to 120-aa protein. NGF exists as a 26.5-kDa noncovalent dimer with the two monomers arranged in a parallel orientation (18, 19). The monomer is a 13-kDa polypeptide with three disulfide bonds forming a cystine knot (4). Like other growth factors, mature NGF is synthesized from an immature precursor, proNGF (20). Because of homology in the pro domains of the neurotrophins, the precursor was thought to have a role in assisting folding or directing sorting to the constitutive or the regulated secretory pathway and production of the mature neurotrophin (21, 22). Interestingly, proNGF now has been found to be the higher-affinity ligand for the p75NTR receptor and also reported to be more effective in inducing apoptosis mediated by p75NTR than mature NGF in some systems (23–25), although substantial evidence has been presented that questions this observation (26, 27). The precursor of proNGF, prepro-NGF, is 31–35 kDa in size and has a hydrophobic signal peptide at its N terminus followed by the pro region and mature region. proNGF is 241 aa in length and has a molecular mass of ≈32 kDa. After translation of the NGF mRNA, posttranslational glycosylation in the pro domain and limited proteolytic cleavage at conserved dibasic sites give rise to the mature NGF. The pro region contains two sites for N-glycosylations and three separate sequences of two or more contiguous basic amino acids (21, 22, 28). Intracellular cleavage of the proNGF to produce active NGF takes place after pairs of basic amino acids of the type I precursor motif Arg-Xaa-Lys/Arg-ArgX, where Xaa is Ser, Val, and Arg for proNGF. proNGF is processed into mature NGF after the arrival of the precursor in the trans-Golgi network (22). The major cleavage site for processing of precursor to mature NGF earlier was shown to be located at −1 and −2 aa positions (relative to mature NGF) of the NGF polypeptide chain, termed site 3 in this study (Fig. 1) (29). Most of the recent research on proNGF has been done on proNGF mutated at this major site. In this article, we show that site 3 is not the only site used in processing of proNGF. Mutating site 3 alone renders proNGF partially stable and hence still prone to processing. In this study, we report that mutation of both the other two processing sites (termed sites 1 and 2), together with site 3, significantly enhances the stability of proNGF and thus renders it much more suitable for biophysical as well as cellular studies.
Fig. 1.
Schematic representation of positions of the three processing sites in the pro domain of proNGF and the mutations made at each site.
Results
Stability of proNGF Constructs.
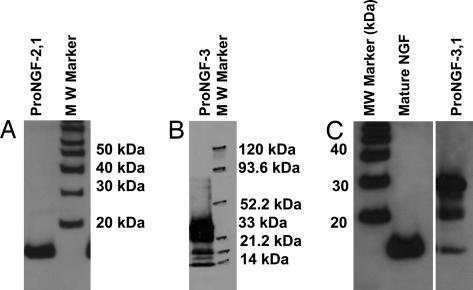
The three major protease-sensitive sites in the pro domain of proNGF are shown in Fig. 1. proNGF with sites 1 and 2 mutated resulted in mature NGF (Fig. 2A) expressed in Spodoptera frugiperda 21 (Sf21) insect cells. proNGF with mutations at site 3 resulted in proNGF, mature NGF, and intermediately processed forms of proNGF (Fig. 2B). proNGF with mutations at sites 3 and 1 resulted in proNGF, a smaller percentage of NGF, and one processing intermediate of ≈22 kDa (Fig. 2C). proNGF with sites mutated at 3 and 2 resulted in a pattern similar to proNGF mutated at sites 3 and 1 except with reduced amount of processing (data not shown). proNGF with mutations at all of the three processing sites, proNGF123, was expressed as essentially all proNGF in the stably transfected Sf21 insect cell line. Thus, mutating processing sites 1 and 2 in addition to site 3 significantly improved the stability of proNGF (Fig. 3A).
Fig. 2.
Western blot analysis of different mutants of proNGF. Sf21 insect cells were stably transfected with mutated proNGF cDNA subcloned into pIZT vector. Mutant protein was secreted into the expression medium and purified by using either weak cation-exchange (CM52) chromatography (proNGF13 and proNGF2) or strong cation-exchange (SP) chromatography (proNGF23). Western blot analysis was performed for proNGF mutated at sites 1 and 2 (A), proNGF mutated at site 3 (B), and two lanes show two different fractions of purification of mutated proNGF, proNGF mutated at sites 1 and 3 (C). Mouse β NGF (Mature NGF) was loaded as a positive control for the Western blot and size along with the molecular mass standard.
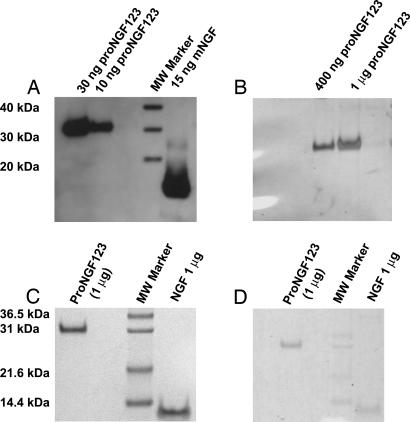
Fig. 3.
Analysis of proNGF mutated at sites 1, 2, and 3. (A) Western blot analysis of proNGF123 shows no processing. (B) Silver staining of the same samples to show purity of proNGF123 purified by using strong cation-exchange (SP) column chromatography and N-60 immunoaffinity chromatography. (C) Coomassie blue staining of proNGF123 and mature NGF. (D) Glycoprotein staining of proNGF123 as compared with the same amount of mature NGF. Mouse β NGF (NGF or mNGF) is present as a molecular mass standard.
Expression and Purification of proNGF123.
proNGF123 is constitutively produced in a stable Sf21 insect cell line. These cells are grown in spinner flasks in expression medium for laboratory scale protein production. Half of the expression medium containing protein is harvested every fourth day for purification, and the volume is replenished with fresh medium to keep the production of proNGF123 semicontinuous. Insect cells are completely healthy for at least three to four such harvests, at which time the entire medium is harvested and a new production run is initiated. The stable insect cell line is therefore a good solution to production of large quantities of stable and biologically active proNGF for structural and cellular studies.
Purification of proNGF123 involved ion-exchange chromatography and immunoaffinity chromatography. For ion exchange, a strong cation-exchanger SP fast-flow column was used. This step purified and concentrated proNGF123 ≈30-fold. This partially purified proNGF123 then was purified further to ≈99% purity and 300-fold enriched by using an immunoaffinity column made with the N-60 monoclonal antibody (30) (Fig. 3 B and C). The N60 antibody recognizes a conformational epitope on NGF, indicating proper folding of the mature NGF domain. The purity of proNGF123 by using these two steps was assessed with silver (Fig. 3B) and Coomassie blue (Fig. 3C) staining. The final yield of purified proNGF is ≈1 mg/liter of expression medium.
Bioassay of Stable proNGF123.
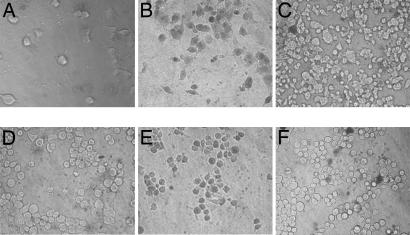
The biological activity of proNGF123 was tested on PC12 cells (TrkA+, p75NTR+, and Sortilin+), PC12nnr cells (TrkA−, p75NTR+, and Sortilin+), C6 glioma cells (TrkA− and p75NTR+), and RN22 schwannoma cells (TrkA− and p75NTR+). Mature wild-type NGF induces neurite outgrowth and stimulates survival of PC12, PC12nnr cells, C6 glioma, and RN22 schwannoma cells at concentrations ranging from 1 nM to 50 nM (Figs. 4 and 5). In contrast, proNGF123 induces cell death at the same concentrations in all of the four cell lines (Figs. 4 and 5). In the first 12 h of proNGF123 treatment, PC12 cells show surface flattening and some dendritic outgrowth, but they do not generate full neurites even after 120 h of proNGF123 treatment at any of the proNGF123 concentrations used (Fig. 4B). Trypan blue analysis 72 h posttreatment with proNGF123 shows at least 60% cell death (at 2 nM or higher concentration) in all of the four cell types, whereas NGF-treated cells show less cell death than the negative controls (15). The lack of neurites in PC12 cells, even after 120 h, suggests that the proNGF123 is stable in the cellular assay with no detectable formation of mature NGF. Because PC12 and PC12nnr cells express p75 and sortilin, this result supports the hypothesis that proposes proNGF-induced cell death through these two receptors.
Fig. 4.
proNGF123 does not support survival of PC12 cells or PC12nnr cells. PC12 (A–C) or PC12nnr (D–F) cells were plated on collagen-coated 96-well plates at a cell density of 3 × 104 cells per ml in complete DMEM containing 15% serum. Growth medium containing NGF, proNGF123, or buffer was changed every 48 h. Cell death was observed by using trypan blue staining for dead cells. (A) PC12 cells treated with 2 nM NGF demonstrate at least 85% neurite-bearing cells. (B) PC12 cells treated with 2 nM proNGF123 show ≈60% cell death. (C) PC12 cells treated with the same amount of 0.04% acetic acid as experimental cells for a negative control (<10% cell death). (D) PC12nnr cells treated with 2 nM NGF (less cell death than negative control). (E) PC12nnr cells treated with 2 nM proNGF123 show ≈60% cell death. (F) PC12nnr control cells treated with buffer as control (<10% cell death). (Magnification: ×400.) This experiment was repeated four times each with mouse β NGF and recombinant NGF with similar results.
Fig. 5.
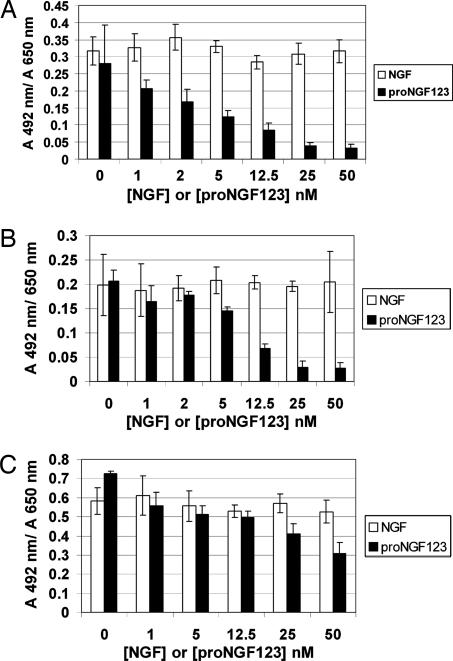
Cell-survival quantification with the XTT assay after treatment with recombinant NGF (open bars) or proNGF123 (filled bars) at the concentrations indicated for 72 h in defined DMEM. RN22 schwannoma cells (A), PC12nnr cells (B), and C6 glioma cells (C) show similar survival as negative control on being treated with various concentrations of NGF, whereas higher cell death is observed with increasing concentration of proNGF123. Values are averages ± SD for four replicates.
Survival for PC12, PC12nnr, C6 glioma, and RN22 schwannoma cells was quantitated by counting dead and live cells in trypan blue exclusion assay and 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) (XTT) cell-survival assay (Figs. 4 and 5). Concentrations of NGF and proNGF123 used varied from 1 nM to 50 nM. XTT survival assay confirmed the results of the trypan blue exclusion assay, showing that proNGF123 induces cell death in various cell types that contain p75NTR. Because of the significantly lower metabolic rate of PC12 cells than the rest of the cell lines used and the low number of cells plated, the XTT assay was more difficult to analyze for PC12 cells (data not shown). Therefore, PC12 cell survival was quantified by counting the live and dead cells in the trypan blue exclusion assay (Fig. 4 A–C).
Stability of proNGF to Proteolysis.
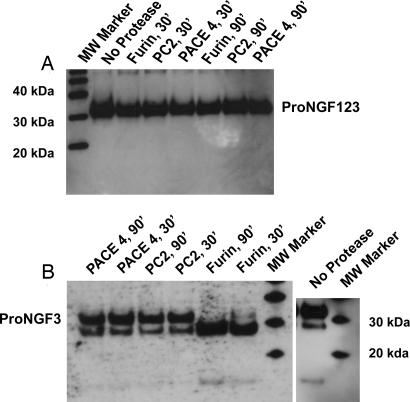
Stability of proNGF123 was tested by determining its susceptibility to proteolysis by furin, PACE-4, and PC-2 enzymes. One microgram of proNGF123 was exposed to either 2 or 4 units of each of these enzymes for 30- and 90-min durations at their optimal temperatures. The activity of these enzymes was established independently on a fluorogenic substrate peptide by the change in fluorescence upon hydrolysis (see Materials and Methods). Western blot analysis of the proteolysis experiment did not show any conversion of proNGF123 to either mature NGF or any other intermediate with any of the three proteases (Fig. 6A), thereby indicating substantial resistance of proNGF123 to degradation by naturally occurring enzymes. On the other hand, proNGF3, which was expressed as a mixture of proNGF, mature NGF, and intermediately processed forms, was stable to further processing by some proconvertases (Fig. 6B). Although furin proteolyzed full-length proNGF3 to an intermediate form of ≈26 kDa, neither furin nor any other proconvertase tested processed it to mature NGF significantly (Fig. 6B). The 26-kDa intermediate is consistent with processing of proNGF3 at site 1. This finding suggests that the stability conferred to the proNGF123 by additional mutations at sites 1 and 2 might be because of resistance to one of the other proconvertases known to be responsible for processing of neurotrophins like PC-1, PC-5, or PC-5/6B (22) and not furin, PC-2, or PACE-4. proNGF123 is not only more resistant to proteolysis than proNGF3 intracellularly, but it also is more stable for storage as observed by effect of repeated freeze–thaw cycles on each of these mutations (data not shown).
Fig. 6.
Proteolytic treatment of proNGF123 (A) and proNGF3 (B) with furin, PACE-4, and PC-2. Furin was bought from New England Biolabs, whereas PACE-4 and PC-2 were expressed by using baculoviral system. Activities of purified PACE-4 and PC-2, relative to that of furin, were determined by using the fluorogenic substrate peptide Boc-R-V-R-R-AMC and a TECAN plate reader. One microgram of proNGF123 or proNGF3 was incubated with 4 units of each of the convertases for 30–90 min at ambient temperatures and then analyzed by Western blotting.
Discussion
An existing challenge for structural and cellular studies with proNGF is the production of a sufficient quantity of stable and biologically active proNGF. Previous studies directed toward understanding the mechanism of processing of prepro-NGF to mature NGF and its secretion demonstrate that the major site for processing of prepro-NGF to mature NGF is site 3 (Fig. 1) (29). In our study, we verified that site 3 is the major site for processing when mutated individually because mutating site 1 or 2 without mutating site 3 results in all mature NGF, whereas mutating site 3 results in proNGF, mature NGF, and intermediately processed forms of proNGF. Contrasting reports in the literature have indicated that proNGF and NGF have opposite effects of death versus survival (25) or similar effects on survival through TrkA (27). Hence, the studies using preparations containing a mixture of the two species are difficult to interpret. On mutating sites 1 and 2 in concert with site 3, we found that the resulting proNGF, proNGF123, is very stable, has essentially no processing (Fig. 3), and is therefore ideal for studies aimed at assessing the biological role of proNGF from that of mature NGF.
For producing sufficient quantity of the stable proNGF, we report construction of stably transfected Sf21 insect cell line and setup for semicontinuous production. This setup yields 1–1.5 mg of proNGF, after purification, per liter of expression medium. A relatively simple two-step purification scheme, as described in Materials and Methods, yields a >99% pure protein preparation. Although proNGF-3,1 binds to the weak cation-exchanger CM52, proNGF-3,2 and proNGF123 do not bind well to CM52 and, therefore, the SP resin had to be used with the latter two. proNGF123 has fewer positively charged amino acid residues, which explains its requirement for the SP strong cation exchanger. Although proNGF-3,1 and proNGF-3,2 have the same number of positively charged residues mutated to the neutral alanines, they must be shielded differently to provide a different pI and binding affinity for CM52. This difference might be of importance in trying to determine which residues are more exposed than others.
Bioactivity of proNGF123 has been demonstrated by using neurite outgrowth and survival assays in PC12, PC12nnr, C6 glioma, and RN22 schwannoma cells. PC12 cells have p75, TrkA, and sortilin receptors; PC12nnr cells have p75NTR and sortilin receptors; and both C6 glioma and RN22 schwannoma cells are known to be p75+, TrkA− (31, 32). Concentrations of proNGF123 were varied from 1 to 50 nM to ensure that minor contaminants/activities were not affecting the interpretations. The optimum concentration appeared to be ≈2 nM. proNGF123 induces cell death in all of the four cell types. A similar result has not been reported in PC12 cells with proNGF mutated only at site 3. The lack of such a report may be attributed to potential processing of proNGF3 by processing enzymes in either the PC12 cells themselves or the media used to grow the PC12 cells.
Wild-type proNGF, expressed in bacteria, is produced as proNGF without any processing. Although it is not processed inside the bacteria, it is present in inclusion bodies and, upon refolding, is susceptible to proteolysis. Also, bacterial proNGF does not have posttranslational modifications, like glycosylations (21). proNGF123, expressed in insect cells, is folded and glycosylated inside eukaryotic cells and, hence, is closest to the native proNGF. The stability of proNGF123 makes it suitable for biophysical and physiological studies. Furthermore, these mutations also should be helpful in animal model studies where proteases for proNGF processing are present.
Prohormone convertases like furin and PACE-4 have been shown to process proNGF to mature NGF (22, 29). The stability of proNGF123 was tested by using purified furin, PACE-4, and PC-2. As seen in Fig. 6A, proNGF123 is completely resistant to processing by the three enzymes. proNGF3, when tested for stability by using these three enzymes, did not show significant processing to mature NGF, although it was processed by furin to smaller intermediate of ≈26 kDa. This result suggests that proNGF123 may be resistant to additional processing enzymes involved in the conversion of proNGF to mature NGF such as PC5/6B, PC-5, PC-1, etc. (22). The exact processing enzyme(s) responsible for the stability of proNGF123 remain(s) to be determined.
In light of recent studies showing a proapoptotic role for proNGF in neuronal death (23, 25), a study of the structural and functional aspects of receptor–ligand interactions at the surface of the cell is important. In this study, we provide evidence for successful production of a stable biologically active mutated form of proNGF on a laboratory scale by using a stably transfected insect cell line. The proapoptotic activity on both PC12 and PC12nnr cells, as well as the neuronal tumor cell lines C6 and RN22, demonstrates the importance of the proNGF123 molecule; elucidating its specific actions will be intriguing. Stable proNGF123, produced and purified by using methods described above, is a valuable tool for biophysical and structural studies to gain insights into proNGF–p75NTR complex formation, which may ultimately help to design pharmaceutical agents to interfere with this apoptotic complex.
Materials and Methods
Cell Cultures.
PC12 (from Lloyd A. Greene, Columbia University, New York, NY), PC12nnr (from Phil Barker, McGill University, Montreal, CA), C6 glioma (from ATCC, Manassas, VA), and RN22 schwannoma (from Bruce Carter, Vanderbilt University, Nashville, TN) cells were grown in DMEM (Cellgro; Voigt Global Distribution, Kansas City, MO) supplemented with 10% horse serum, 5% FBS, 4.5 mg/ml glucose, 4.0 mM l-glutamine, 100 units/ml penicillin, 100 pg/ml streptomycin, and 0.25 pg/ml amphotericin-B25 at 37°C in a humid atmosphere containing 5% CO2. The cells were subcultured every 72 h at a ratio of 1:3. Treatments of PC12, PC12nnr, C6 glioma, and RN22 schwannoma cells were carried out in the same medium as used for subculturing immediately after plating in collagen-coated 96-well plates.
Sf21 insect cells were grown in Grace's insect cell medium (GIBCO, Carlsbad, CA) supplemented with 10% FBS at 27°C. They were subcultured every 72 h at a ratio of 1:4. Stably transfected Sf21 insect cells expressing proNGF mutants were grown in Grace's insect cell medium supplemented with 10% FBS and 400 μg/ml zeocin. Transfected Sf21 cells were grown at 27°C and subcultured every 48 h at a ratio of 1:2.
Plasmid Constructs and Production of Stable Transfectants.
Mouse β prepro-NGF (241 aa) cDNA was subcloned in insect cell vector pIZT/his/V5 (Invitrogen, Carlsbad, CA) at the KpnI–AgeI restriction sites of the polylinker region. The native stop codon in the cDNA of mouse β NGF was included to avoid the His-6 residue tag in the vector. Sequential site-directed mutagenesis was performed with separate primers for each site mutation. All constructs were analyzed by sequence analyses to verify the mutations and the correct reading frame. Plasmids with mutated cDNAs were amplified in Escherichia coli and purified by using Qiagen (Valencia, CA) maxi prep kit for transfection-grade DNA.
Stable transfectants of Sf21 cells were generated by transforming the cells with pIZT vector, which contained mutated proNGF cDNAs, by using lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were selected by using 500 μg/ml zeocin in the medium. Zeocin concentration used for selecting the stable transfectants was calculated by performing a kill curve on nontransfected insect cells.
Expression and Purification.
For expression of wild-type and all mutated proNGFs, stably transfected insect cells were grown in spinner flasks in Excel-401 medium with 100 μg/ml zeocin added every 72 h for ≈4 days. After 4 days, expression medium was centrifuged for clarification and loaded onto cation-exchange columns. An SP fast-flow column (Amersham–GE, Uppsala, Sweden) was used for proNGF123 and proNGF mutated at sites 3 and 2, whereas a CM52 weak cation-exchanger column was used for the rest of the proNGF mutants and wild-type (recombinant) NGF. Two washes were performed with 20 mM Tris (pH 8.0) and 50 mM Tris (pH 9.0). The proNGF was eluted with 400 mM NaCl, 50 mM Tris (pH 9.0). Fractions containing proNGF then were loaded onto an N60 immunoaffinity column, washed with 400 mM NaCl, 50 mM Tris (pH 9.0), and 20 mM Tris (pH 8.0). proNGF was eluted with 0.02 M glycine (pH 2.6), and the pH was restored to 7.0 with 2 M Tris. The degree of purification of proNGF after these two columns is ≈99% and 300-fold. Purified protein was used for all experiments. Recombinant (wild-type) mature NGF was used as control in some experiments as indicated in the figure legends.
Purified protein was analyzed by SDS/PAGE and subsequent Coomassie blue or silver staining. Concentration of the purified protein was calculated by determining absorbance at 280 nm and by using 1.6 absorbance units/mg per ml as the absorptivity coefficient in Beer–Lambert's equation because of the similarity of proNGF with NGF.
Cellular Assays.
Neurite outgrowth and live-cell assays were performed with PC12 cells, PC12nnr cells, C6 glioma, and RN22 schwannoma cells on collagen-coated 96-well plates. PC12, PC12nnr cells, C6 glioma, and RN22 schwannoma cells were grown in 0.5% serum containing DMEM for 24 h. Each of the four cell lines then was washed, resuspended in defined medium (DMEM supplemented with 4 mM glutamine, 20 nM progesterone, 100 μM putrescine, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin-B), diluted to ≈30,000 cells per ml, and 100 μl of cell suspension was added to each well of collagen-coated 96-well plates. Cells then were treated with mature NGF or proNGF plus buffer for 72 h in defined DMEM. Fresh media were replaced every 48 h. After 72 h, neurites were counted and a live-cell assay was performed with trypan blue exclusion. Two hundred single cells were counted for calculating percentage of live cells and neurite-bearing cells per well.
The XTT survival assay was performed for PC12nnr, C6 glioma, and RN22 schwannoma cells at the end of 72 h as per manufacturer's instructions (Roche Applied Biosciences, Indianapolis, IN). Both of these assays also were performed in complete DMEM-supplemented 4 mM glutamine, 10% horse serum, 5% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin-B with same results. Assays performed in complete DMEM were analyzed after 6 days of treatment with NGF or proNGF.
Other Reagents.
PACE-4 and PC-2 enzymes were made by using the baculoviral expression system (both the baculoviruses were a generous gift from Margaret Fahnestock, McMasters University, Hamilton, ON, Canada). Furin was obtained from New England Biolabs (Ipswich, MA). Activities of purified PACE-4 and PC-2, relative to that of furin, were determined by using the fluorogenic substrate peptide Boc-R-V-R-R-AMC (BACHEM Bioscience Inc., King of Prussia, PA) and a TECAN GENios plate reader (33).
Acknowledgments
We thank Dr. Margaret Fahnestock, (McMasters University, Hamilton, ON, Canada) for the PACE-4 and PC-2 baculoviruses and Dr. Bruce Carter (Vanderbilt University, Nashville, TN) for RN22 schwannoma cells. We also thank Tammi Fritz, Vijayakanth Pagadala, Sidharth Mahapatra, Hrishikesh Mehta, and Sang B. Woo for helpful advice during this project. This work was supported by U.S. Public Health Service, National Institutes of Health Grant NS24380.
Abbreviations
- NGF
nerve growth factor
- Trk
tyrosine kinase
- NTR
neurotrophin receptor
- Sf21 cells
Spodoptera frugiperda 21 cells
- XTT
2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bibel M, Barde YA. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- 2.Levi-Montalcini R. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 3.McDonald NQ, Hendrickson WA. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 4.McInnes C, Sykes BD. Biopolymers. 1997;43:339–366. doi: 10.1002/(SICI)1097-0282(1997)43:5<339::AID-BIP2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Barbacid M. J Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- 6.Bibel M, Hoppe E, Barde YA. EMBO J. 1999;18:616–622. doi: 10.1093/emboj/18.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronfman FC, Fainzilber M. EMBO Rep. 2004;5:867–871. doi: 10.1038/sj.embor.7400219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao MV. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin AN, Bitler CM, Welcher AA, Shooter EM. J Biol Chem. 1992;267:8352–8359. [PubMed] [Google Scholar]
- 10.Wiesmann C, de Vos AM. Cell Mol Life Sci. 2001;58:748–759. doi: 10.1007/PL00000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiesmann C, Ultsch MH, Bass SH, de Vos AM. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 12.Eide FF, Lowenstein DH, Reichardt LF. Exp Neurol. 1993;121:200–214. doi: 10.1006/exnr.1993.1087. [DOI] [PubMed] [Google Scholar]
- 13.Friedman WJ. J Neurosci. 2000;20:6340–6346. doi: 10.1523/JNEUROSCI.20-17-06340.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman WJ, Greene LA. Exp Cell Res. 1999;253:131–142. doi: 10.1006/excr.1999.4705. [DOI] [PubMed] [Google Scholar]
- 15.Lad SP, Peterson DA, Bradshaw RA, Neet KE. J Biol Chem. 2003;278:24808–24817. doi: 10.1074/jbc.M212270200. [DOI] [PubMed] [Google Scholar]
- 16.Mufson EJ, Lavine N, Jaffar S, Kordower JH, Quirion R, Saragovi HU. Exp Neurol. 1997;146:91–103. doi: 10.1006/exnr.1997.6504. [DOI] [PubMed] [Google Scholar]
- 17.Kong H, Kim AH, Orlinick JR, Chao MV. Cell Death Differ. 1999;6:1133–1142. doi: 10.1038/sj.cdd.4400587. [DOI] [PubMed] [Google Scholar]
- 18.Maness LM, Kastin AJ, Weber JT, Banks WA, Beckman BS, Zadina JE. Neurosci Biobehav Rev. 1994;18:143–159. doi: 10.1016/0149-7634(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 19.McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL. Nature. 1991;354:411–414. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- 20.Server AC, Shooter EM. Adv Protein Chem. 1977;31:339–409. doi: 10.1016/s0065-3233(08)60221-1. [DOI] [PubMed] [Google Scholar]
- 21.Rattenholl A, Lilie H, Grossmann A, Stern A, Schwarz E, Rudolph R. Eur J Biochem. 2001;268:3296–3303. doi: 10.1046/j.1432-1327.2001.02232.x. [DOI] [PubMed] [Google Scholar]
- 22.Seidah NG, Benjannet S, Pareek S, Savaria D, Hamelin J, Goulet B, Laliberte J, Lazure C, Chretien M, Murphy RA. Biochem J. 1996;314:951–960. doi: 10.1042/bj3140951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington AW, Leiner B, Blechschmitt C, Arevalo JC, Lee R, Morl K, Meyer M, Hempstead BL, Yoon SO, Giehl KM. Proc Natl Acad Sci USA. 2004;101:6226–6230. doi: 10.1073/pnas.0305755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee R, Kermani P, Teng KK, Hempstead BL. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 25.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, et al. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 26.Peng S, Wuu J, Mufson EJ, Fahnestock M. J Neuropathol Exp Neurol. 2004;63:641–649. doi: 10.1093/jnen/63.6.641. [DOI] [PubMed] [Google Scholar]
- 27.Fahnestock M, Yu G, Michalski B, Mathew S, Colquhoun A, Ross GM, Coughlin MD. J Neurochem. 2004;89:581–592. doi: 10.1111/j.1471-4159.2004.02360.x. [DOI] [PubMed] [Google Scholar]
- 28.Kliemannel M, Rattenholl A, Golbik R, Balbach J, Lilie H, Rudolph R, Schwarz E. FEBS Lett. 2004;566:207–212. doi: 10.1016/j.febslet.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 29.Heymach JV, Jr, Kruttgen A, Suter U, Shooter EM. J Biol Chem. 1996;271:25430–25437. doi: 10.1074/jbc.271.41.25430. [DOI] [PubMed] [Google Scholar]
- 30.Nanduri J, Vroegop SM, Buxser SE, Neet KE. J Neurosci Res. 1994;37:433–444. doi: 10.1002/jnr.490370402. [DOI] [PubMed] [Google Scholar]
- 31.Weis C, Wiesenhofer B, Humpel C. J Neurooncol. 2002;56:59–67. doi: 10.1023/a:1014410519935. [DOI] [PubMed] [Google Scholar]
- 32.Gentry JJ, Casaccia-Bonnefil P, Carter BD. J Biol Chem. 2000;275:7558–7565. doi: 10.1074/jbc.275.11.7558. [DOI] [PubMed] [Google Scholar]
- 33.Fahnestock M, Zhu W. DNA Cell Biol. 1999;18:409–417. doi: 10.1089/104454999315295. [DOI] [PubMed] [Google Scholar]