Abstract
Comparisons of gene expression between human and non-human primate brains have identified hundreds of differentially expressed genes, yet translating these lists into key functional distinctions between species has proved difficult. Here we provide a more integrated view of human brain evolution by examining the large-scale organization of gene coexpression networks in human and chimpanzee brains. We identify modules of coexpressed genes that correspond to discrete brain regions and quantify their conservation between the species. Module conservation in cerebral cortex is significantly weaker than module conservation in subcortical brain regions, revealing a striking gradient that parallels known evolutionary hierarchies. We introduce a method for identifying species-specific network connections and demonstrate how differential network connectivity can be used to identify key drivers of evolutionary change. By integrating our results with comparative genomic sequence data and estimates of protein sequence divergence rates, we confirm a number of network predictions and validate these findings. Our results provide insights into the molecular bases of primate brain organization and demonstrate the general utility of weighted gene coexpression network analysis.
Keywords: microarray, differential network analysis, selection, systems biology
Genetic evidence suggests that humans and chimpanzees diverged from a common ancestor within the past five to six million years (1). Since then, humans have acquired a remarkable set of defining characteristics, including a vastly expanded neocortex (2). The high extent of sequence homology between human and chimpanzee proteins supports the longstanding hypothesis that many phenotypic differences between the species reflect differences in the regulation of gene expression, in addition to differences in amino acid sequences (3). Several studies have used microarrays to explore differences in gene expression between human and chimpanzee brains (4–8). Despite the success of these studies (reviewed in refs. 9 and 10), it has been difficult to interpret the evolutionary significance of specific gene-expression differences between the species. For example, some expression differences may evolve neutrally and therefore have little functional consequence (11). Thus, new tools are needed that can systematically discern between gene-expression changes that are likely to be functionally significant and those that are not.
Network approaches have been used to study a variety of biological systems, bridging the gap from individual genes to systems biology by exploring the observed relationships between gene products (12–20). Here, we pioneer the use of weighted gene coexpression network analysis (WGCNA) to reveal shared and unique properties of the large-scale organization of gene expression in adult human and chimpanzee brains. We identify and visualize modules of coexpressed genes, which correspond to functionally relevant brain anatomy, and explore differences in these modules between the species. These comparisons provide a systems-level context in which to evaluate the potential impact of evolutionary changes in a particular gene's expression level or protein-coding sequence, while simultaneously identifying candidate genes that may have contributed to the emergence of uniquely human cognitive specializations. More generally, the construction of weighted gene coexpression networks represents an efficient means of translating gene-expression differences into critical functional insights relevant to understanding the nervous system.
Results
Constructing Gene Coexpression Networks in Human and Chimpanzee Brains.
We constructed gene coexpression networks from microarray data consisting of 18 human and 18 chimpanzee samples from six matched brain regions: Broca's area, anterior cingulate cortex, primary visual cortex, prefrontal cortex, caudate nucleus, and cerebellar vermis (7). For an overview of WGCNA methodology, see Figs. 5 and 6 and Supporting Text, which are published as supporting information on the PNAS web site. All possible pairwise correlations were calculated for 4,000 genes in human and chimpanzee brains in parallel and converted into measures of connection strength by taking their absolute values and raising them to a power, β (19). Summing the connection strengths for each gene with all other genes resulted in a single number (called network connectivity, or k) that represents how strongly that gene is connected to all other genes in the network. To identify modules of coexpressed genes, we searched for genes with similar patterns of connection strengths to other genes or high “topological overlap” (TO; refs. 19 and 21). We calculated TO and clustered genes on this basis for both humans and chimpanzees, identifying seven distinct gene coexpression modules in the human brain (Fig. 1; Supporting Text). Some modules appeared highly conserved between humans and chimpanzees (e.g., turquoise, yellow, and black), whereas others did not (e.g., blue and green). A summary of all genes and their modules, connectivity, and expression values can be found in Table 1, which is published as supporting information on the PNAS web site.
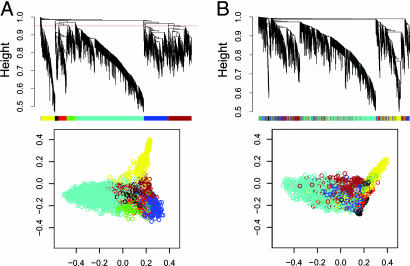
Fig. 1.
Network analysis of gene expression in human and chimpanzee brains identifies distinct modules of coexpressed genes in human (A) and chimpanzee (B). (A) Dendrograms produced by average linkage hierarchical clustering of 2,241 genes based on TO (see Supporting Text). The red line in the human dendrogram indicates the height at which the tree was cut (0.95) to define modules. Modules were assigned colors as indicated in the horizontal bar beneath the human dendrogram. Genes in the chimpanzee network are depicted by using human module colors to represent the extent of module conservation. (B) Classical multidimensional scaling plots in three dimensions (color-coded as in A) depict the relative size and cohesion of modules in humans and chimpanzees.
Gene Coexpression Modules Correspond to Brain Structures.
We explored the functional relevance of gene coexpression modules using standard heat maps of gene expression and observed that the modules identified by this analysis largely correspond to major anatomical subdivisions of the brain (Fig. 2). To provide an unbiased basis for module characterization, we performed singular value decomposition to summarize the expression levels of all genes in each module (Fig. 2). The module eigengene (i.e., the first principal component) roughly corresponds to the average of the signed normalized gene-expression values for a given sample. Except for the black module, all modules showed significant relationships to specific brain regions (Fig. 2; P < 0.01, Kruskal–Wallis test). The turquoise module is composed of genes coexpressed in cerebellum. Genes in the blue and brown modules are coexpressed in cortical samples, whereas genes comprising the yellow module are coexpressed in caudate nucleus. Genes in the green module are coexpressed in both cortex and cerebellum, whereas the red module identifies genes coexpressed in the anterior cingulate cortex and caudate nucleus. The black module likely represents white matter, because at least 11 of 25 genes with the highest connectivity in this module are involved in myelination or are expressed in glia (highlighted in Table 1). We reasoned that the large cerebellar module might obscure smaller yet biologically significant modules, so we removed the cerebellar samples from the data set and repeated the analysis, identifying a new module comprised of genes coexpressed in primary visual cortex (Fig. 2H and Table 2, which is published as supporting information on the PNAS web site). The identified gene coexpression modules thus correspond to major components of brain architecture, grounding their interpretation in a functional context.
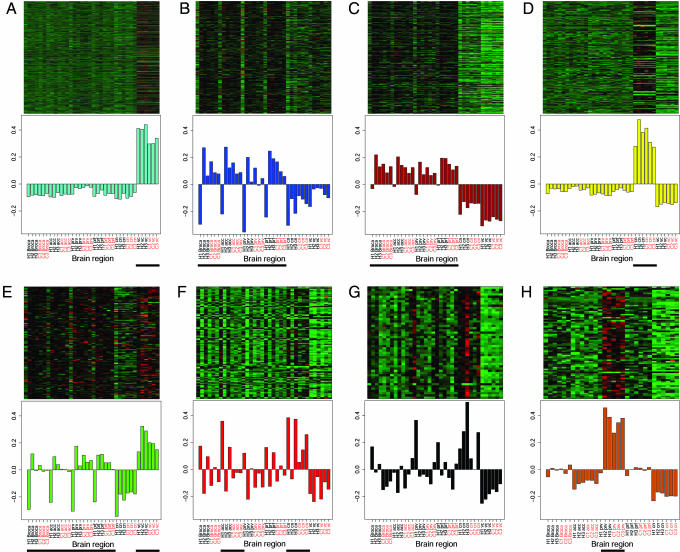
Fig. 2.
Modules correspond to functional subdivisions of the brain. (A–G) (Upper) Heat maps depicting expression levels for all genes (rows) in all human and chimpanzee brain regions (columns; black labels are human samples and red are chimpanzee) for each module: turquoise (A), blue (B), brown (C), yellow (D), green (E), red (F), and black (G). Red, increased expression; black, neutral expression; green, decreased expression. (Lower) Barplots of the values of the module eigengene (i.e., the first principal component) derived from singular value decomposition are displayed for each module. Black horizontal lines beneath the barplots denote indicator variables (line = 1, no line = 0). Modules were characterized as follows (Kruskal–Wallis test): cerebellum (1,001 genes, P = 0.00013; A), cortex (360 genes, P = 0.00089; B), cortex (343 genes, P = 0.0000014; C), caudate nucleus (200 genes, P = 0.00013; D), cortex and cerebellum (126 genes, P = 0.003; E), and anterior cingulate cortex and caudate nucleus (122 genes, P = 0.008; F). The black module (G), consisting of 50 genes, is a white matter module as characterized by manual inspection of its constituent genes (see text and Table 1). To assess module conservation between humans and chimpanzees, the Spearman correlations in intramodular connectivity (kin) were calculated for each module between the species: r = 0.55 (A), r = 0.30 (B), r = 0.39 (C), r = 0.51 (D), NS (E), r = 0.42 (F), and r = 0.62 (G). All correlations were highly significant (P < 10E-6), with the exception of the green module (P = 0.32). H1, human 1; C1, chimp 1, etc.; Broca, Broca's area; acc, anterior cingulate cortex; prv, primary visual cortex; prf, prefrontal cortex; cn, caudate nucleus; vc, cerebellum; NS, not significant. (H) Upon removal of the cerebellar samples from the dataset, an additional module specific to primary visual cortex was identified (P = 0.0011, Kruskal–Wallis test). The Spearman correlation in kin between humans and chimpanzees was 0.54 (P = 1.36E-6).
Gene Coexpression Relationships Are Poorly Conserved in Cerebral Cortex Relative to Subcortical Brain Regions.
The overall extent of conservation between two networks can be assessed by comparing the values of k for all genes. We observed that the extent of network conservation (the correlation between human and chimpanzee k) depends on the brain regions used to construct the network. Strikingly, the purely cortical networks showed the weakest conservation between the species; this relationship was not observed in comparisons of gene expression alone (Fig. 7, which is published as supporting information on the PNAS web site). Because the extent of network conservation might partly reflect differences in the variance of gene expression across brain regions, we assessed module conservation between the species; all modules (except primary visual cortex) were identified by analyzing coexpression relationships across the same set of samples (i.e., all six brain regions). Intramodular connectivity (kin), which is defined as the sum of a gene's connection strengths with all other genes in its module (see Supporting Text), was compared between humans and chimpanzees for all genes within their respective modules. A gene with extremely high kin in humans and low kin in chimpanzees would be strongly connected to many genes in the human module and few genes in the chimp and vice versa. As an example, consider NRG1. NRG1 is ranked 126th in terms of kin in humans and 332nd in chimpanzees of 343 genes in the brown cortical module. Although expression levels for this gene in cortex are not significantly different between the species (P = 0.30, t test; Table 1), comparison of connectivity reveals that expression of NRG1 is highly correlated (r > 0.8, Pearson), with 64 genes in this module in humans and none in chimpanzees. We observed a striking gradient in that subcortical modules showed significantly greater conservation in chimpanzees than cortical modules (see Fig. 2 and legend). The lack of conservation of gene coexpression modules in cerebral cortex is consistent with the dramatic expansion of this brain region in humans.
To further analyze the functional significance of differences in modules between humans and chimpanzees, we used GenMAPP 2.0 (22) to identify overrepresented gene ontology (GO) categories (23) in human gene coexpression modules. We focused on genes with higher kin in humans relative to chimpanzees to identify biological pathways that have taken on greater importance during recent human brain evolution (Table 3, which is published as supporting information on the PNAS web site). Interestingly, the majority of overrepresented GO categories with higher kin in humans were found in cortical modules, particularly the blue module, which was very poorly conserved in chimpanzees. In the blue module, overrepresented GO categories included protein transporter activity (13 genes), the microtubule cytoskeleton (9 genes), and ion transporter activity, including 11 members of the electron transport chain (ETC). The increased kin observed for these genes is especially intriguing in light of previous work suggesting accelerated evolution of the ETC in anthropoid primates (24).
Visualization of Modules Enables Rapid Identification of Hub Genes and Human-Specific Network Connections.
To identify specific “hub” genes (genes with central positions within modules), we depicted the strongest connections in each module using VisANT (ref. 25; Fig. 3A; see also Figs. 8–10, which are published as supporting information on the PNAS web site). To determine whether these connections were also present in the chimpanzee brain network, we assessed module conservation by viewing only those connections for which the human TO was far greater than the chimpanzee TO (see Materials and Methods). These connections represent gene coexpression relationships that are present in the human brain but essentially absent in chimpanzee (Fig. 3B; see also Figs.9 and 10B). It is interesting to note that the percentages of human-specific connections in each module recapitulate known evolutionary hierarchies, e.g., 17.4% in cortex; (Fig. 3B), 7.8% in caudate nucleus, and 4.5% in cerebellum (Figs. 8 and 9). Furthermore, many of these connections converge on the same hub genes within a module. Although a large difference in connectivity for a particular gene can, in principle, reflect evolutionary changes affecting that gene or all of its neighbors, parsimony strongly favors the former possibility. It is therefore likely that these genes have experienced significant change during recent human/chimpanzee evolution, e.g.: LDOC1 (Fig. 3B), EYA1 and LECT1 (Fig. 9), and PGAM2 (Fig. 9).
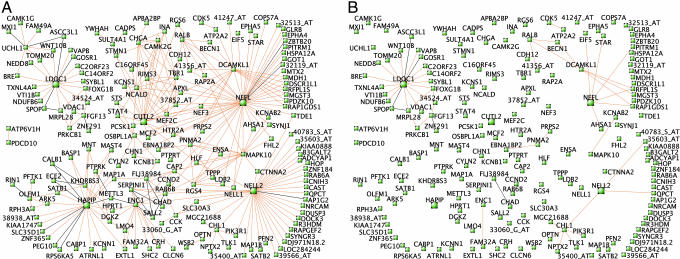
Fig. 3.
Module visualization identifies hub genes and human-specific connections. (A) Three hundred pairs of genes with the greatest TO in humans are depicted for cortex (brown module). Genes with expression levels that are negatively correlated are connected by black lines. Where gene symbols are unknown, Affymetrix probe set IDs are shown (e.g., 37158_at). (B) Connections from A that are present in humans but absent in chimpanzees (see Materials and Methods).
Differential connectivity (DC) may reflect disparate types of evolutionary change, including changes in gene expression, splicing, mRNA stability, or protein-coding sequence. We observed that EYA1, LECT1, and PGAM2, all of which possess significantly higher kin in humans, are also expressed significantly higher in their respective brain regions in humans; however, LDOC1, which also possesses significantly higher kin in humans, was not differentially expressed (Table 1). To explore this issue more broadly, we compared differential expression (DE) and DC for all genes assigned to modules [excluding the black module, which was not characterized by specific samples (Fig. 2)]. There is a modest but highly significant correlation between DE and DC (r = 0.32; Fig. 4A), indicating that DE explains only ≈10% of the variance in DC in these networks.
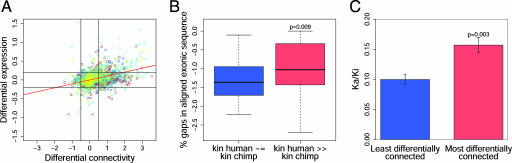
Fig. 4.
DC between humans and chimpanzees reflects differences in gene expression and protein structure. (A) DE vs. DC for 2,152 genes expressed in brain. DE is defined as log10(mean gene expression [human]/mean gene expression [chimp]) in the brain region(s) corresponding to each gene's module (as defined in Fig. 2). DC is defined as log10(kin [human]/kin [chimp]). Colors denote modules. The Spearman correlation between DE and DC is 0.32 (P < 2.20E-16; linear least-squares regression line in red). The pairs of vertical and horizontal lines have been arbitrarily drawn to illustrate the utility of DC as a means of stratifying differentially expressed genes. (B) Genes exhibiting DC show evidence of genomic rearrangements between humans and chimpanzees. For each module, 10 genes with kin human ∼ = kin chimp, and 10 genes with kin human ≫ kin chimp were selected; genes with at least one gap in their aligned exonic sequence were compared (n = 46 [kin human ≫ kin chimp] and n = 44 [kin human ∼ = kin chimp]). Data were highly skewed and log-transformed. The mean gap percentage in aligned exonic sequence was ≈3-fold higher in DC genes (P = 0.009, Wilcoxon test), suggesting that genomic rearrangements contribute to DC. (C) Genes exhibiting DC show accelerated protein sequence divergence between humans and chimpanzees. Rates of protein sequence divergence (Ka/Ki) were obtained for 1,168 genes from ref. 8. These genes were ranked according to the absolute value of DC between humans and chimpanzees as defined in A. Mean Ka/Ki was significantly higher for the most differentially connected genes (top quintile, n = 234; μ = 0.157) compared with the least differentially connected genes (bottom quintile, n = 233; μ = 0.100; P = 0.003, Wilcoxon test). (Scale bars indicate SE.)
In Silico Validation of Network Predictions.
Genome sequence data enable rapid in silico validation of network predictions. For example, LDOC1, which is a human-specific hub, has been interrupted by an inversion in chimpanzees (University of California, Santa Cruz Genome Browser), effectively abolishing the entire “submodule” anchored by LDOC1 in cerebral cortex (Fig. 3B). To further investigate the contribution of genomic rearrangements to DC, we compared the mean percentage of “gaps” in aligned exonic sequence for genes with significantly higher kin in humans and genes with approximately equal kin in both species (University of California, Santa Cruz Genome Browser). Gaps may represent sequencing gaps or evolutionary events such as insertions, deletions, or inversions. If genomic rearrangements contribute to DC, genes exhibiting DC should have a higher mean gap percentage than genes with equal kin. Comparison of the two groups suggested such a trend; on average, 20% of aligned exonic sequence among differentially connected genes consisted of gaps, compared with only 6% for genes with equal connectivity. However, the difference was not significant (P = 0.09, Wilcoxon test). Among those genes with higher connectivity in humans and at least one gap, on average 30% of aligned exonic sequence consisted of gaps; for genes with approximately equal connectivity and at least one gap, the average was 10% (P = 0.009; Fig. 4B and Table 4, which is published as supporting information on the PNAS web site). Therefore, as has recently been described (26, 27), indels and genomic rearrangements appear to be primary movers in human and chimpanzee genome evolution, and their effects are reflected in the evolution of gene coexpression networks.
Relating DC to Protein Sequence Divergence Rates.
To further explore whether DC reflects changes in coding sequence in addition to changes in gene expression, we cross-referenced our data with estimates of protein sequence divergence (Ka/Ki) for 1,168 genes (ref. 8; Table 5, which is published as supporting information on the PNAS web site). Ka measures the rate of nonsynonymous nucleotide substitutions, whereas Ki measures the rate of nucleotide substitutions in interspersed repeats within a 250-kb window centered around each gene. Low Ka/Ki values suggest strong purifying selection, whereas elevated Ka/Ki values suggest positive selection or relaxation of constraint (8). We detected a significant correlation between Ka/Ki and DC across all genes (r = 0.06, P = 0.026), similar to the correlation reported between Ka/Ki and DE (8). To determine whether this relationship was more pronounced for the most differentially connected genes, we stratified genes into quintiles on the basis of DC. Mean Ka/Ki was significantly higher for the most differentially connected genes (μ = 0.157) compared with the least differentially connected genes (μ = 0.100; P = 0.003, Wilcoxon test; Fig. 4C). Similar results were seen when comparing the Ka/Ks ratio (Ks measures the rate of synonymous nucleotide substitutions; ref. 8). Mean Ka/Ks was significantly higher for the most differentially connected genes (μ = 1.056) compared with the least differentially connected genes (μ = 0.366; P = 0.005). Differential network connectivity can thus serve as a unifying principle for disparate types of evolutionary change, including changes that alter protein-coding sequence and changes that affect gene expression. By comparing gene coexpression networks in the brains of different species, functional changes that affect network connections can be identified and their effects on other genes explored.
Discussion
Unlocking the full potential of microarray data requires new analytic approaches that move beyond single-gene comparisons and systematically identify meaningful relationships between gene products. Network depictions can provide immediate functional insights by revealing relationships between genes and biological processes. Comparative network analysis can also prioritize genes for further study on the basis of DC, an emerging theme supported by studies in lower organisms that a gene's connectivity is a measure of functional relevance (15, 16).
We applied a recently developed methodology (WGCNA) to construct weighted gene coexpression networks in human and chimpanzee brains, revealing modules of genes that represent systems-level molecular correlates to neuroanatomical structures. It is notable that many genes with the highest intramodular connectivity in humans are conserved in chimpanzee brain, underscoring the shared molecular bases of primate brain organization. However, important differences exist between human and chimpanzee gene coexpression networks, particularly in cerebral cortex, a pattern that is strikingly consistent with the rapid expansion of this brain region on the human lineage. This distinction among brain regions along evolutionary hierarchies was not detected on the basis of gene expression differences alone. By comparing human and chimpanzee genes in terms of connectivity, we introduce an approach to identify key drivers of evolutionary change. Although our analysis was human-centric (i.e., module definitions were derived from the structure of the human brain gene coexpression network), future work could adopt a reciprocal point of view and define modules in the brains of chimpanzees or other primate species.
Modules Consist of Functionally Related Genes.
WGCNA identified modules of coexpressed genes that correspond to brain regions, successfully recapitulating one aspect of the basic functional organization of the brain. However, modules are not a simple reflection of genes that are differentially expressed across brain regions. To illustrate this point, we used standard criteria [a minimum fold change of 1.3 and a P value <0.001 (t test)] to identify differentially expressed genes in human cerebellum, caudate nucleus, or cortex, and observed that 29% of genes in the cerebellar module, 40% of genes in the caudate nucleus module, and 72% of genes in the two cortical modules were not identified as differentially expressed. The presence of these genes in their respective modules indicates that they are part of a group of genes that is highly coexpressed, and that identification of such groups cannot be made purely on the basis of DE.
Nor are modules simple representations of the input samples. For example, the presence of hub genes such as CNP, MAG, MAL, PLP1, OLIG2, and MOG in the black module suggests this module is related to white matter. Two modules (green and red) consist of genes that are coexpressed in multiple brain regions. The green module (cortex + cerebellum) is enriched for genes involved in ubiquitin-dependent protein catabolism, including C13orf22, FBXO21, PSMA2, PSMC2, PSMD7, UBE2D3, USP46, and USP9X (Table 3). The absence of this module in caudate nucleus suggests regional variation in the ubiquitin-proteasome system, which has important implications for the study of neurodegenerative disorders.
The red module, consisting of genes that are coexpressed in anterior cingulate cortex and caudate nucleus, mirrors the established physical connectivity between these brain regions. Defects in glutamatergic transmission in frontostriatal systems are thought to contribute to several neuropsychiatric disorders (28), so it is notable that three genes involved in glutamate metabolism are found in this module: GLUL, GLUD1, and GLUD2. Each shows higher connectivity in human brain, consistent with an important adaptive role in human higher cognitive functions subserved by frontostriatal systems. This work raises the possibility that brain regions comprising interconnected neural circuits may share common modules of coexpressed genes, a hypothesis that can be explored in future studies.
Differences Between Humans and Chimpanzees Implicate Key Drivers of Evolutionary Change.
Comparisons of human and chimpanzee brains on the basis of gene connectivity led to the striking observation that the overall conservation of gene coexpression modules between the species recapitulates evolutionary hierarchies, with white matter > cerebellum > caudate nucleus > caudate nucleus + anterior cingulate cortex > cortex, a relationship not evident from DE analysis. The correlation in kin between humans and chimpanzees in the primary visual cortex module was intermediate to the other modules, suggesting that interspecies module conservation may be greater in primary sensory cortex than in regions considered representative of association cortex.
The blue cortical module, which is nearly absent in chimpanzees, contains a number of genes involved in energy metabolism, including 11 members of the ETC. Previous work has shown that several proteins in the ETC, including three members of this module (COX5A, COX6A2, and UQCRFS1), have experienced accelerated evolution in anthropoid primates (24, 29, 30). Categories of genes that have high TO with ETC genes in human cerebral cortex, but not chimpanzee, include mitochondrial distribution and morphology (e.g., IMMT and DNM1L), synapse formation and vesicle docking (e.g., DTNAI and RAB3A), and cytoskeletal regulation (e.g., ABI2, CYFIP2, and MAP1B). It is likely that the dramatic increase in parallel processing power engendered by the expansion of the neocortex in humans has made concomitant demands upon energy metabolism; consequently, it is of significant interest to couple this process genetically to hallmarks of cortical activity such as cytoskeletal remodeling and synaptic plasticity. This module also contains several human-specific hub genes of unknown function, such as FGF12, SLC30A9, ANKMY2, and KIAA1279, which, given their network centrality, likely play important, yet underappreciated roles in human cortical function.
Materials and Methods
Choice of Genes, Generation of Weighted Gene Coexpression Networks, and Identification of Modules.
An overview of WGCNA methodology is presented in Fig. 5. The dataset used for network construction consisted of 36 Affymetrix (Santa Clara, CA) HGU95Av2 microarrays surveying gene expression with 12,625 probe sets in three adult humans and three adult chimpanzees across six matched brain regions: Broca's area, anterior cingulate cortex, primary visual cortex, prefrontal cortex, caudate nucleus, and cerebellar vermis (ref. 7; for additional information, including a description of how samples were processed, see ref. 7). After eliminating probes with sequence differences between the species, all arrays were scaled to the same average intensity, and quantile normalization was performed. Four thousand probe sets were selected for network analysis based on high variance in human brain relative to a nonneural tissue (lung). From these, 2,241 probe sets with the highest k were clustered on the basis of TO to identify modules of coexpressed genes. For additional details, see Supporting Text.
Functional Annotation of Hub Genes and Modules.
GenMAPP 2.0 (ref. 22; www.genmapp.org) was used to search among hub genes and modules for enrichment of functional categories of genes defined by the Gene Ontology Consortium (23) (www.geneontology.org). The significance of each enriched category was also assessed on the basis of DC between humans and chimpanzees (see Table 3).
Module Visualization and Differential Network Analysis.
Aproximately 300 pairs of genes with the greatest TO in humans were depicted for each module by using VisANT (ref. 25; visant.bu.edu). The “Relaxing” layout algorithm was used to confer partial network structures, which were then manually adjusted for clarity. Genes with expression levels that are negatively correlated are connected by black lines; all other genes are positively correlated. To identify pairs of genes with high TO in humans (H) and low TO in chimpanzees (C) in a given module, for each pair of genes i and j we defined the human specificity measure (HSij) as follows:
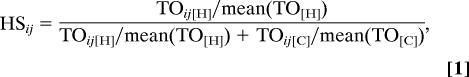 |
where mean(TO) is the mean pairwise TO value in a given module for human or chimpanzee, respectively. Connections for which the value of this ratio exceeded 0.8 were deemed present in humans and absent in chimpanzees. Given that mean(TO[HUMAN]) > mean(TO[CHIMP]) for all modules, this method is conservative.
Genomic Sequence Comparisons.
All genomic sequence comparisons were made by using the University of California, Santa Cruz genome browser's May 2004 (human) and November, 2003 (chimp) assemblies. “Net” alignments were downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg17/vsPanTro1. Genes in each module were ranked by the absolute value of log10(kin[human]/kin[chimp]), and the top and bottom 10 genes from each module were selected (140 genes in all). For each gene, the total length of its coding sequence was determined by summing the lengths of all nonoverlapping exons and the fraction represented by “gaps” in the human/chimpanzee net alignments was calculated. To compare DC to estimated rates of protein sequence divergence, we crossreferenced our results with Ka/Ki and Ka/Ks values for 1,168 genes (1,330 probe sets; ref. 8). Genes were stratified on the basis of DC as described above (|log10(kin[human]/kin[chimp])|).
Supplementary Material
Acknowledgments
We thank the individuals who produced the raw data analyzed in this study (ref. 7). We thank our colleagues Todd Preuss and Mario Cáceres for continuing collaborative discussions and for help in identifying probes with sequence differences between humans and chimpanzees. We also thank Kelsey Martin and Alcino Silva for critical reading of early versions of the manuscript and Giovanni Coppola, Tao Shi, and Stephanie Tsung for valuable discussions. This work was supported by the James S. McDonnell Foundation 21st Century Collaborative Award/Bridging Brain, Mind and Behavior, National Institute of Neurological Disorders and Stroke/National Institute of Mental Health (NIMH) Grant NS52108 (to D.H.G.) and NIMH Grants MH60233 (to D.H.G.) and 1U19AI063603-01 (to S.H.).
Abbreviations
- WGCNA
weighted gene coexpression network analysis
- ETC
electron transport chain
- DE
differential expression
- DC
differential connectivity
- TO
topological overlap.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Chen FC, Li WH. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deacon TW. The Symbolic Species: The Co-evolution of Language and the Brain. New York, NY: Norton; 1997. [Google Scholar]
- 3.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 4.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 5.Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Proc Natl Acad Sci USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 9.Khaitovich P, Enard W, Lachmann M, Paabo S. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 10.Preuss TM, Cáceres M, Oldham MC, Geschwind DH. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 11.Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. PLoS Biol. 2004;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabási AL, Oltvai ZN. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 13.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 14.Yook SH, Oltvai ZN, Barabási AL. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- 15.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 16.Carter SL, Brechbuhler CM, Griffin M, Bond AT. Bioinformatics. 2004;20:2242–2250. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- 17.Stuart JM, Segal E, Koller D, Kim SK. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 18.van Noort V, Snel B, Huynen MA. EMBO Rep. 2004;5:280–284. doi: 10.1038/sj.embor.7400090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Horvath S. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 20.Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Mol Biol Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- 21.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 22.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman LI, Wildman DE, Schmidt TR, Goodman M. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Hu Z, Mellor J, Wu J, Yamada T, Holloway D, Delisi C. Nucleic Acids Res. 2005;33:W352–7. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium, The Chimpanzee Sequencing and Analysis Nature. 2005;437:69–87. [Google Scholar]
- 27.Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 28.Javitt DC. Mol Psychiatry. 2004;9:984. doi: 10.1038/sj.mp.4001551. 97:979. [DOI] [PubMed] [Google Scholar]
- 29.Doan JW, Schmidt TR, Wildman DE, Goodman M, Weiss ML, Grossman LI. J Bionenerg Biomembr. 2005;37:35–41. doi: 10.1007/s10863-005-4121-2. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt TR, Goodman M, Grossman LI. Gene. 2002;286:13–19. doi: 10.1016/s0378-1119(01)00800-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.