Abstract
Plants sense their environmental light conditions by using three photoreceptors that absorb in the UV, blue/near UV, and red/far-red spectral ranges. These photoreceptors have specific chromophore components corresponding to their absorption spectra. Phytochrome, a red/far-red light receptor, has phytochromobilin as its chromophore, whereas the blue/near UV photoreceptors cryptochrome and phototropin have a pair of flavin derivatives. Plants use these various photoreceptors to assess the surrounding light environment. Phytochrome 3 (PHY3) is a red light receptor found in some ferns, which preferentially grow under weak light. PHY3 is composed of a phytochrome chromophore-binding domain in its N-terminal portion and an almost full-length phototropin in its C-terminal half. This unusual domain organization implies that two different light-sensing systems coexist in this single photoreceptor, although these light-sensing systems usually reside in independent photoreceptors. Here, we show that PHY3 acts as a dual-channel photoreceptor that possesses both the red light-sensing system of phytochrome and the blue light-sensing system of phototropin. Furthermore, red- and blue-light signals perceived by PHY3 are processed synergistically within this single chromoprotein. These unusual properties might confer an enhanced light sensitivity on PHY3, allowing ferns to grow under a low-light canopy.
Keywords: fern, phototropism, photoreceptor, red light, blue light
Plants have developed sophisticated sensing systems of intensity, direction, duration, and spectral quality of light. Because light consists of components with different wavelengths, plants perceive light signals through various photoreceptors corresponding to wavelength range. These light signals induce various environmental responses of plants and even modulate developmental programs of them (1, 2). Many photomorphogenic responses have been described, for example, control of seed germination, hypocotyl elongation, cotyledon opening, chloroplast development, and gene expression. Among these responses, phototropic response and chloroplast photorelocation movement can be elicited in dicotyledonous plants only by blue/near UV light (3) and are mediated by phototropins (4). However, in some cryptogamic plants, both phototropic response and chloroplast photorelocation movement are induced by red light (RL) as well as blue light (BL) (5).
In the fern Adiantum, both BL- and RL-dependent phototropic responses and chloroplast photorelocation movements are well described (6–9). We showed that these RL-dependent responses of Adiantum are mediated by phytochrome 3 (PHY3) (10). The deduced amino acid sequence indicates that the N-terminal portion of PHY3 contains the chromophore-binding domain of phytochrome, and the C-terminal half shows a remarkable similarity to phototropin (11). The predicted domain organization of PHY3 implies that two different light-sensing systems coexist in this single chromoprotein, although it is not known whether PHY3 can function as a BL receptor or not. In PHY3-deficient mutants of Adiantum, RL-dependent phototropism and chloroplast photorelocation movement are phenotypically deficient, whereas BL-dependent responses appear normal (12). As in the case of dicotyledonous plants, such BL-dependent responses can be mediated by phototropins in Adiantum (13, 14). Therefore, as far as investigating PHY3-deficient mutants, it is difficult to see whether PHY3 can act as a BL receptor for these responses.
Based on its structural features, it is anticipated that PHY3 signals are relayed to phototropin signal-transduction pathways. Functional complementation tests in phototropin-deficient mutants can be used to study the photobiological properties of PHY3 in vivo and, especially, to investigate whether PHY3 has the function of a BL photoreceptor. Arabidopsis phototropin mutants, in which whole endogenous phototropins are functionally deficient, are currently available. Hence, we introduced the PHY3 gene from Adiantum into a phototropin-deficient Arabidopsis mutant (phot1–5 phot2–1) (15, 16) and observed the phototropic responses.
Results and Discussion
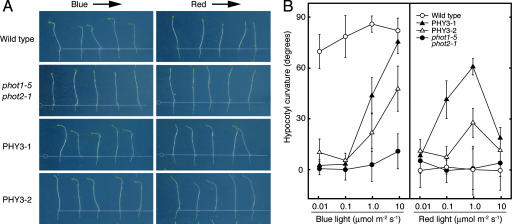
As shown in Fig. 1, transgenic Arabidopsis expressing PHY3 clearly showed a positive phototropic response to BL, demonstrating that PHY3 can function as a BL photoreceptor. This phenotypic complementation of phototropism in Arabidopsis indicates that light signals captured by PHY3 are transmitted to the endogenous phototropin signal-transduction pathway of Arabidopsis. This result further suggests that, in Adiantum, PHY3 and phototropins would be able to use the same signal-transduction pathway immediately downstream of the photoreceptor. Fig. 1 also shows an apparent RL-induced phototropic response in PHY3-expressing Arabidopsis hypocotyls. It is well known that RL alone does not induce phototropism in Arabidopsis seedlings (17); however, in these transgenic Arabidopsis seedlings, the RL signal perceived by PHY3 must have been transferred to the Arabidopsis phototropin signal-transduction pathway to induce RL-dependent hypocotyl curvature. As a result, Arabidopsis acquires the additional capability of RL-induced phototropism through PHY3.
Fig. 1.
PHY3 can mediate both RL- and BL-induced phototropic responses of phot1 phot2 transgenic Arabidopsis seedlings. (A) Phototropic response of Arabidopsis hypocotyls. Wild type, phot1–5 phot2–1, and two independent phot1–5 phot2–1 transformants expressing PHY3 (PHY3–1, PHY3–2) were grown in the dark for 3 days and irradiated by either unilateral BL (10 μmol·m−2·s−1) or unilateral RL (1.0 μmol·m−2·s−1) for 16 h before imaging. Arrows indicate the direction of lateral light irradiation. (B) Fluence rate response of hypocotyl phototropism. Phototropic responses were induced as in A. Each bar indicates a mean of 15 measurements with standard deviations.
The phototropic responses observed were fluence rate-dependent, and the optimum fluence rates were different for RL and BL irradiation (Fig. 1B). For BL, hypocotyl curvature increased as the fluence rate was increased to at least 10 μmol·m−2·s−1, but RL-dependent phototropism was attenuated >1.0 μmol·m−2·s−1. This difference suggests that phototropic responses induced by RL and BL might be mediated by different chromophores and perhaps the phytochromobilin of phytochrome (18) and flavin adenine mononucleotide (FMN) of phototropin (19). As mentioned above, the N-terminal half of PHY3 contains the chromophore-binding domain of phytochrome, and the C-terminal half shows a remarkable similarity to phototropin and contains tandemly repeated LOV (light, oxygen, or voltage) domains (LOV1, LOV2) and a Ser/Thr kinase domain (11). In vitro experiments using recombinant PHY3 demonstrated that phytochromobilin could bind to PHY3 expressed in yeast (11) and that FMN could attach to each LOV domain of PHY3 expressed in Escherichia coli (19). However, phytochromobilin shows a second absorption peak in the BL wavelength range (20) and could serve as a chromophore for a BL photoreceptor (21–24).
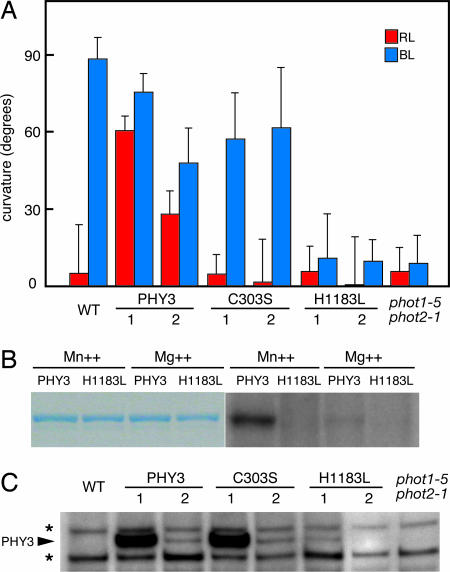
To investigate whether the BL-dependent phototropic response of transgenic Arabidopsis is mediated by FMN attached to the phototropin domain of PHY3 or by phytochromobilin bound to the phytochrome domain, we constructed a mutated form of PHY3 (C303S) that lacks phytochromobilin-binding ability and then observed the phototropic responses of Arabidopsis transformed with this construct. Cys-303 of PHY3 is the chromophore-binding site of the phytochrome domain, and when substituted to serine residue (C303S), is no longer able to bind phytochromobilin (25, 26). C303S construct-containing Arabidopsis did not show RL-dependent phototropism, indicating that this RL response must be mediated by the light-sensing system of the phytochrome domain in PHY3 (Fig. 2A). In contrast, the BL-dependent phototropic response was still observed in C303S-transformed Arabidopsis. These results clearly demonstrate that the BL signal could be received by the light-sensing system of the phototropin region of PHY3. Thus, two different light-sensing systems containing three chromophores, one phytochromobilin and two FMN, are actually functional in PHY3, and PHY3 indeed acts as a dual-channel photoreceptor. However, the signal-output mechanism of PHY3 is unknown, and it is unclear whether light signals captured by two different sensing systems of PHY3 use a common mode of transduction.
Fig. 2.
PHY3 can act as phototropin as well as phytochrome, and signal output from PHY3 depends on autophosphorylation activity of the Ser/Thr kinase domain. (A) Phototropic response of Arabidopsis seedlings. Wild type, two independent transgenic lines expressing either wild type PHY3 (PHY3–1, -2) or mutated-PHY3 (C303S-1, -2; H1183L-1, -2), and phot1–5 phot2–1 were grown in the dark for 3 days and irradiated by unilateral BL (10 μmol·m−2·s−1) or RL (1.0 μmol·m−2·s−1) for 16 h. Each bar indicates a mean of 15 measurements with standard deviations. (B) Autophosphorylation activity of PHY3 protein kinase domain expressed in E. coli. Affinity-purified recombinant protein of either wild-type PHY3 (PHY3) or mutated PHY3 (H1183L) was used. For phosphorylation assay, either divalent manganese (Mn++) or magnesium (Mg++) was included in a reaction buffer. Coomassie brilliant blue staining (Left) and autoradiography (Right) of the same gel are shown. (C) Western blot analysis of PHY3 protein. Microsomal membranes prepared from dark-grown transgenic Arabidopsis expressing PHY3, C303S, or H1183L mutants were probed with anti-PHY3 polyclonal antibody. An asterisk on the left indicates a nonspecific band that was cross-reacted with PHY3 polyclonal antibody.
Because BL-dependent phototropism in Arabidopsis is thought to correlate with the autophosphorylation activity of phototropin (27, 28), we introduced a mutated-PHY3 (H1183L) carrying a mutation in the Ser/Thr kinase domain into Arabidopsis and then examined the phototropic responses of transgenic lines. His-1183 is located in kinase subdomain IV and is conserved in all phototropins reported to date. To investigate in vitro autophosphorylation activity of PHY3, we expressed a range of peptide fragments containing the protein kinase domain from PHY3 in E. coli. Fig. 2B shows that the PHY3 protein kinase domain has kinase activity, and substitution of His-1183 to leucine (H1183L) results in loss of autophosphorylation activity. In this experiment, the GST-tagged PHY3 protein kinase domain in the kinase buffer containing divalent manganese (Mn++) exhibited a relatively high level of phosphorylation compared with that in the buffer containing divalent magnesium (Mg++). As shown in Fig. 2A, H1183L transgenic Arabidopsis lines did not show a phototropic response to RL or BL irradiation. Because BL and RL failed to induce a phototropic response in H1183L transgenic lines, PHY3 must require protein kinase activity for both RL- and BL-dependent phototropism. This observation suggests that light signals perceived by the two chromophores of PHY3 would be transmitted by the same mode of action, through protein kinase activity.
Expression of PHY3 or mutated-PHY3 protein in transgenic plants was analyzed by Western blotting using anti-PHY3 polyclonal antibody. Microsomal membranes were isolated from dark-grown seedlings and used for Western blot analysis. As shown in Fig. 2C, PHY3 or mutated-PHY3 protein was detected in transgenic lines, but the protein level of transgene product has varied among these lines. In all of the transgenic lines, accumulation of the PHY3 transcript was confirmed by using RT-PCR analysis (Fig. 4, which is published as supporting information on the PNAS web site). C303S transgenic lines show very different protein levels, although these lines exhibit similar degrees of BL-induced hypocotyl curvature (Fig. 2A). Similar observations have been reported in Arabidopsis phototropin transgenic studies (28, 29). When phot1 is expressed in Arabidopsis phototropin-deficient mutants, transgenic plants can restore the phot1-dependent BL responses despite the phot1 protein levels. Reduced levels of phot1, even though phot1 protein was undetectable by Western blot analysis, do not appear to impair functional responsiveness of BL. Our results could be consistent with these studies, and the level of PHY3 or mutated-PHY3 protein might not correlate with the biological responses in the transgenic plants.
As described above, we showed that PHY3 can function as either a RL or a BL photoreceptor by irradiating with monochromatic light of different wavelengths. However, under natural conditions, light consists of a mixture of components with different wavelengths that includes both RL and BL. Because PHY3 contains two different light-sensing systems, a phytochrome system and a phototropin system, it must be able to perceive both RL and BL signals simultaneously. When light signals from these individual sensing systems are perceived simultaneously by PHY3, how are these light signals processed by a single molecule? To address this question, we simultaneously irradiated transgenic Arabidopsis expressing PHY3 with both RL and BL and analyzed the effect on phototropic responses. We applied lateral RL and BL singly or together at several different fluence rates.
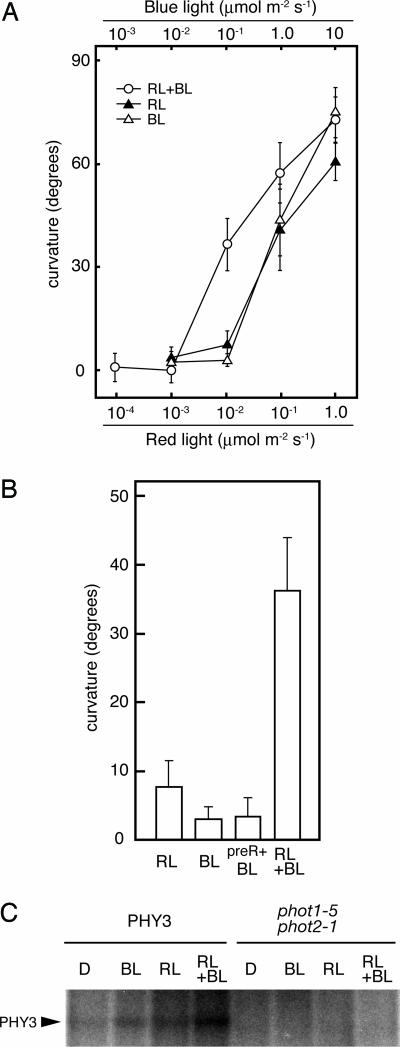
As shown in Fig. 3A, a remarkable range of hypocotyl curvature was observed when we applied 0.01 μmol·m−2·s−1 of RL and/or 0.1 μmol·m−2·s−1 of BL. With a single monochromatic light irradiation, the degrees of hypocotyl curvature induced by RL and BL were 7.7° ± 3.6° and 3.2° ± 1.8°, respectively. When RL and BL were applied simultaneously, the degree of curvature increased to 36.3° ± 7.5°. These differences in curvature are statistically significant (t test, P < 0.01), demonstrating that simultaneous irradiation with RL and BL causes greater phototropic curvature than with RL or BL alone. Moreover, simultaneous irradiation at the fluence rates described above yielded more than a simple additive effect.
Fig. 3.
RL and BL signals are processed synergistically within PHY3. (A) Fluence rate response of hypocotyl phototropism. PHY3-expressing phot1–5 phot2–1 transgenic Arabidopsis (PHY3–1) was grown in the dark for 3 days, and then light irradiation was done as follows: RL, continuous unilateral RL for 16 h; BL, continuous unilateral BL for 16 h; and RL+BL, RL and BL were irradiated simultaneously for 16 h. At the end of the lateral-light irradiation, phototropic curvatures were measured. Each bar indicates a mean of 15 measurements with standard deviations. (B) RL enhancement effect on BL-induced phototropism. Phototropic response was induced as in A. Fluence rate irradiated was 0.01 μmol·m−2·s−1 of RL and 0.1 μmol·m−2·s−1 of BL. preR+BL, exposed to 10 μmol·m−2·s−1 of RL for 58 s before being given lateral BL. Each bar indicates a mean of 15 measurements with standard deviations. (C) Autoradiograph, showing the light-dependent phosphorylation of PHY3. Microsomal membranes prepared from dark-grown transgenic Arabidopsis expressing PHY3 (PHY3–1) or Arabidopsis mutant (phot1–5 phot2–1) were given a mock irradiation (D), irradiated with BL (at a total fluence of 2.1 × 104 μmol·m−2), RL (at a total fluence of 1.1 × 104 μmol·m−2), or irradiated both BL and RL simultaneously (RL+BL). All manipulations were carried out under dim green light.
As reported, RL has some enhancement effect on BL-induced phototropic response through phytochrome-dependent modulation (30, 31). However, it is not known whether this increased curvature by simultaneous irradiation might be due to RL induction of phototropic enhancement by phytochrome or not. To investigate the effect of RL treatment on BL-induced phototropism of PHY3-expressing transgenic Arabidopsis, we preirradiated with RL given from above before lateral BL irradiation. Because the RL enhancement effect depends on the number of photons absorbed (31) regardless of incident light direction, we applied a RL pulse to achieve a fluence of 580 μmol·m−2, almost equivalent to the fluence given by lateral RL irradiation. As shown in Fig. 3B, the degree of hypocotyl curvature induced by RL preirradiation was almost equal to that without preirradiation, with no statistically significant difference (P > 0.05). On the other hand, the degree of curvature induced by BL irradiation after preirradiation with RL and induced by simultaneous irradiation with RL and BL was significantly different (P < 0.01). Therefore, the increased hypocotyl curvature induced by simultaneous irradiation with both RL and BL must not be caused by RL-dependent enhancement of phototropism. In addition, when we applied RL and BL simultaneously to the C303S transgenic line, the degree of hypocotyl curvature was almost equal to that induced by BL (Fig. 5, which is published as supporting information on the PNAS web site). This result further demonstrates that the synergistic effect observed depends on the PHY3 activity.
To address the relationship of the synergistic effect induced by simultaneous irradiation with respect to the PHY3 kinase activity, we examined in vitro phosphorylation analysis of microsomal membranes from a PHY3-expressing line (Fig. 3C). We observed both BL- and RL-potentiated phosphorylation of the 160-kDa protein in PHY3-expressing Arabidopsis seedlings. When we applied RL and BL simultaneously, an obvious increase in the phosphorylation level of the 160-kDa protein was detected compared with that of single-light irradiation. Thus, the synergistic effect induced by simultaneous irradiation is correlated with the PHY3 autophosphorylation activity. These results clearly demonstrate that two independent light-sensing systems cooperate within PHY3 and that RL and BL signals are processed synergistically by this single chromoprotein.
This synergistic effect was remarkable even at the relatively low fluence rate of monochromatic light irradiation that induced a phototropic response. In chloroplast photorelocation responses of Adiantum, sequential irradiation of RL and BL show an additive effect, and, even when the fluence rate of each light treatment is less than the threshold values, chloroplast movement is induced (8). This observation is consistent with the synergistic effect of phototropic responses described here.
PHY3 genes have been found only in the “polypodiaceous (polypod)” ferns such as Adiantum, Dryopteris, Onoclea, and Hypolepis (10). In contrast, PHY3-like gene sequences appear to be absent from the more primitive ferns such as Osmunda and Lygodium. Based on these results, we hypothesize that PHY3 genes might have arisen during the evolution of ferns. PHY3 is encoded by an uninterrupted ORF from an intronless gene, whereas other phytochrome and phototropin genes of Adiantum contain multiple introns in common with the homologous genes in other plant species. Intronless genes might readily be generated as a consequence of reverse transcription, and in the case of PHY3, we have proposed that the processed phototropin mRNA sequence might have been reverse-transcribed into the phytochrome gene locus in the course of leptosporangiate fern diversification (14).
PHY3-like genes were recently found in the alga Mougeotia (MsNEO1 and MsNEO2) (32). Interestingly, both MsNEO1 and MsNEO2 contain multiple introns in their phytochrome and phototropin regions. In addition, the cysteine residue of the LOV2 domain that plays an important role in light sensing of phototropin (28) is not conserved in either MsNEO1 or MsNEO2 (32), suggesting that MsNEOs would not function like the BL photoreceptor phototropin. This evidence suggests that PHY3 and MsNEO genes arose independently during evolution (32) and that each photoreceptor established its own functional properties independently.
Many polypod ferns preferentially grow in low-light environments typical under the canopy of dense forests. These ferns would need to have developed some adaptive mechanisms for living in such shady habitats. It has been proposed that PHY3 might be involved in proliferation of polypod ferns under low-light conditions because it is a RL receptor that can regulate phototropin-mediated responses (10, 33). The ability to use RL signals for these responses might confer some advantage when growing in low-light environments. In this report, we demonstrate that PHY3 is a photoreceptor in which both phytochrome and phototropin photosensory systems coexist. These two different light-sensing systems cooperate within this single chromoprotein and confer on PHY3 the potential to process RL and BL signals synergistically. This intramolecular effect could result in an increased light sensitivity of PHY3 when it perceives both RL and BL signals simultaneously. Therefore, in addition to broadening the wavelength range corresponding to phototropin-mediated responses, PHY3 might also enable ferns to sense low-light signals in the natural light environment. It is consistent with our previous observations that wild-type Adiantum seedlings are two orders of magnitude more sensitive to white light in phototropic responses than PHY3-deficient mutants (10). These findings imply that ferns having PHY3 would acquire enhanced light sensitivity and would be able to increase fitness under low-light canopy conditions. Furthermore, this dual-channel photoreceptor might play a role in promoting diversification of polypod ferns under low-light environmental niches such as the subcanopy of angiosperm forests. The results presented here also demonstrate that PHY3 can function effectively even in seed plants, by using both RL and BL signals. The PHY3-type photoreceptor might provide a promising clue to improve light sensitivity in a wide variety of plants and have many downstream applications.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana Columbia ecotype carrying the glabrous1 (gl1) mutation (34) was used for all of the experiments reported here. The gl1 mutant is the parental line of the phot1–5 phot2–1 (nph1–5 cav1–1) double mutant (15, 16, 35), thus we used this mutant line as a wild-type control.
Plant Transformation.
The full length Adiantum PHY3 coding sequence (11) was amplified by PCR and cloned into pCR4Blunt-TOPO plasmid vector (Invitrogen, Carlsbad, CA). In the course of the cloning, we accidentally obtained the H1183L mutated fragment, and thereafter we applied this fragment as the H1183L mutation. The C303S mutation was generated by using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The nucleotide sequence of each construct was confirmed by sequencing. All of the constructs were inserted between cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase (NOS) terminator of pBI H1-IG plasmid vector (36). The Arabidopsis phot1–5 phot2–1 double mutant was transformed with the plasmids described above by the floral-dip method (37) using the Agrobacterium tumefaciens EHA101 strain. The transformants were screened on Murashige–Skoog medium containing 10 μg·ml−1 hygromycin. For each construct, T2 generation segregated ≈3:1 for hygromycin-resistance was established. T3 self-progeny of homozygous T2 plants were used for experiments.
Measurement of Phototropic Response.
Measurement of hypocotyl curvature was performed as described (4). Surface-sterilized seeds were planted in square Petri dishes on half-strength Murashige–Skoog medium with 1.0% agar (wt/vol). Seedlings were grown vertically in darkness for 3 days and then exposed to unilateral light for 16 h. After illumination, the images of the seedlings were captured by using an image scanner (GT-8700F; Seiko Epson, Nagano, Japan), and curvatures were measured by using the ImageJ program (http://rsb.info.nih.gov/ij). For phototropic enhancement experiments, preirradiation of RL was done by a RL pulse given from above before the lateral light irradiation. BL and RL was supplied by either BL-emitting diode (maximum wavelength of 470 nm with a bandwidth of 30 nm) or RL-emitting diode (maximum wavelength of 660 nm with a bandwidth of 20 nm) (LED-mB, LED-mR; Tokyo Rikakikai, Tokyo, Japan). The fluence rate of the light source was adjusted by using RND plastic films (RDS, Tokyo, Japan). Fluence rates were measured by using a LI-1000 photometer with LI-190SA quantum sensor (LI-COR Biosciences, Lincoln, NE).
Preparation of Recombinant Protein and in Vitro Phosphorylation Analysis.
By using both Adiantum PHY3 and PHY3 (H1183L) sequence as templates, the coding sequence corresponding to PK domain of PHY3 (amino acids 1038–1465) was amplified by PCR and was inserted into the pGEX-6P-1 (Amersham Biosciences, Piscataway, NJ) to express as a GST fusion protein. The resultant plasmids were transferred into E. coli Rosetta cells (Novagen, San Diego, CA), and recombinant proteins were prepared according to the manufacturer's protocols. Soluble protein extracts were affinity-purified by using a glutathione Sepharose column, and purified GST-PHY3PK and GST-H1183LPK were used for in vitro phosphorylation analyses. GST-PHY3PK or GST-H1183LPK was incubated in a kinase buffer (37.5 mM Tris·Mes, pH 7.5/5.3 mM MnCl2(or MgCl2)/150 mM NaCl/1 mM EGTA/1 mM DTT/5 mM ε-aminocaproic acid/1 mM benzamidine/1 μg·ml−1 leupeptin/1 μg·ml−1 antipain) at 25°C for 30 min in the presence of [γ-32P]ATP. The reaction mixture was then subjected to SDS/PAGE, proteins were visualized by Coomassie brilliant blue staining, and the phosphorylation levels of GST fusion proteins were estimated by autoradiography of the gel.
Western Blot Analysis.
Microsomal membrane fractions were prepared from 3-day-old, dark-grown Arabidopsis seedlings as described (38). All of the manipulations for microsomal membrane preparation were performed at 4°C under dim green light. Microsomal membranes from Arabidopsis (40 μg) were heated in NuPAGE LDS sample buffer (Invitrogen), electrophoresed on a NuPAGE 3–8% Tris-acetate gel (Invitrogen), and blotted onto the PVDF membrane. Rabbit polyclonal antibody to synthetic peptide mixture corresponding to amino acid residues 1–14, 574–590, and 1437–1450 of PHY3 was used as a primary antibody. Anti-rabbit IgG conjugated to horseradish peroxidase (Amersham Biosciences) was used as a secondary antibody, and Western blots were analyzed by using the chemiluminescence method (ECL Advance Western Blotting Detection kit, Amersham Biosciences).
In Vitro Phosphorylation Analysis of Microsomal Membrane Fractions.
In vitro autophosphorylation analysis of microsomal membranes from Arabidopsis were performed as described (38), with a slight modification. Twenty micrograms of microsomal membranes were dissolved in phosphorylation buffer [37.5 mM Tris·HCl, pH 7.5/5.3 mM MgSO4/150 mM NaCl/1 mM EGTA/1 mM DTT/5 mM ε-aminocaproic acid/1 mM benzamidine/1× complete protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN)] containing 0.5% (vol/vol) Triton X-100. Samples were irradiated at a total fluence of 2.1 × 104 μmol·m−2 of BL and/or 1.1 × 104 μmol·m−2 of RL on ice. After the irradiation or mock irradiation as dark control, 1.25 μl of 5-fold-diluted [γ-32P]ATP (111 terabecquerels/mmol; Amersham Biosciences) with unlabeled ATP (10 μM) was added to each phosphorylation reaction (total volume of reactions was 10 μl after the addition of ATP). Reactions were allowed to incubate at room temperature for 2 min and an equal volume of 2× SDS sample buffer was added to stop the reaction. All manipulations were carried out under dim green light.
Supplementary Material
Acknowledgments
We thank Jane Silverthorne for critical reading of the manuscript; Hiroko Kawai-Toyooka for discussion about H1183L mutation; Akeo Kadota for advice about this work; and Kayoko Hara for technical assistance. This work was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.K. and M.W.).
Abbreviations
- BL
blue light
- FMN
flavin adenine mononucleotide
- RL
red light.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants. 2nd Ed. Dordrecht, The Netherlands: Kluwer Academic; 1994. [Google Scholar]
- 2.Schäfer E, Nagy F. Photomorphogenesis in Plants and Bacteria. 3rd Ed. Dordrecht, The Netherlands: Springer; 2006. [Google Scholar]
- 3.Sachs J. Bot Z. 1864:353–358. [Google Scholar]
- 4.Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wada M, Kadota A. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:169–191. [Google Scholar]
- 6.Wada M, Sugai M. In: Photomorphogenesis in Plants, 2nd Ed. Kendrick RE, Kronenberg GHM, editors. Dordrecht, The Netherlands: Kluwer Academic; 1994. pp. 783–802. [Google Scholar]
- 7.Kagawa T, Wada M. J Plant Res. 1994;107:389–398. [Google Scholar]
- 8.Kagawa T, Wada M. Planta. 1996;198:488–493. [Google Scholar]
- 9.Kagawa T, Wada M. Plant Physiol. 1999;119:917–923. doi: 10.1104/pp.119.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai H, Kanegae T, Christensen S, Kiyosue T, Sato Y, Imaizumi T, Kadota A, Wada M. Nature. 2003;421:287–290. doi: 10.1038/nature01310. [DOI] [PubMed] [Google Scholar]
- 11.Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh K-C, Lagarias JC, Wada M. Proc Natl Acad Sci USA. 1998;95:15826–15830. doi: 10.1073/pnas.95.26.15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadota A, Wada M. Plant Cell Physiol. 1999;40:238–247. [Google Scholar]
- 13.Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M. Plant Cell Physiol. 2004;45:416–426. doi: 10.1093/pcp/pch045. [DOI] [PubMed] [Google Scholar]
- 14.Kanegae T, Wada M. In: Photomorphogenesis in Plants and Bacteria, 3rd Ed. Schäfer E, Nagy F, editors. Dordrecht, The Netherlands: Springer; 2006. pp. 515–536. [Google Scholar]
- 15.Huala E, Oeller PW, Liscum E, Han I-S, Larsen E, Briggs WR. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 16.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 17.Liscum E, Briggs WR. Plant Physiol. 1996;112:291–296. doi: 10.1104/pp.112.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagarias JC, Rapoport H. J Am Chem Soc. 1980;102:4821–4828. [Google Scholar]
- 19.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rüdiger W, Thümmler F. In: Photomorphogenesis in Plants, 2nd Ed. Kendrick RE, Kronenberg GHM, editors. Dordrecht, The Netherlands: Kluwer Academic; 1994. pp. 51–69. [Google Scholar]
- 21.Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagarias DM, Crepeau MW, Maines MD, Lagarias JC. Plant Cell. 1997;9:675–688. doi: 10.1105/tpc.9.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinomura T, Uchida K, Furuya M. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boylan M, Quail PH. Proc Natl Acad Sci USA. 1991;88:10806–10810. doi: 10.1073/pnas.88.23.10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boylan M, Douglas N, Quail PH. Plant Cell. 1994;6:449–460. doi: 10.1105/tpc.6.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 28.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 29.Doi M, Sigenaga A, Emi T, Kinoshita T, Shimazaki K-I. J Exp Bot. 2004;55:517–523. doi: 10.1093/jxb/erh044. [DOI] [PubMed] [Google Scholar]
- 30.Hangarter RP. Plant Cell Environ. 1997;20:796–800. doi: 10.1046/j.1365-3040.1997.d01-124.x. [DOI] [PubMed] [Google Scholar]
- 31.Stowe-Evans EL, Luesse DR, Liscum E. Plant Physiol. 2001;126:826–834. doi: 10.1104/pp.126.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. Proc Natl Acad Sci USA. 2005;102:13705–13709. doi: 10.1073/pnas.0504734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallon S, Lupia R. Nature. 2004;428:553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- 34.Koonneef M, Dellaert LWM, van der Veen JH. Mutant Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K-I. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto H, Sakamoto K, Tomizawa K-I, Nagatani A, Wada M. Plant Physiol. 1997;115:79–85. doi: 10.1104/pp.115.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 38.Liscum M, Briggs WR. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.