Abstract
Intracellular vesicle trafficking performs essential functions in eukaryotic cells, such as membrane trafficking and delivery of molecules to their destinations. A major endocytotic route in plants is vesicle trafficking to the vacuole that plays an important role in plant salt tolerance. The final step in this pathway is mediated by the AtVAMP7C family of vesicle soluble N-ethylmaleimide-sensitive factor attachment protein receptors (v-SNAREs) that carry out the vesicle fusion with the tonoplast. Exposure to high-salt conditions causes immediate ionic and osmotic stresses, followed by production of reactive oxygen species. Here, we show that the reactive oxygen species are produced intracellularly, in endosomes that were targeted to the central vacuole. Suppression of the AtVAMP7C genes expression by antisense AtVAMP711 gene or in mutants of this family inhibited fusion of H2O2-containing vesicles with the tonoplast, which resulted in formation of H2O2-containing megavesicles that remained in the cytoplasm. The antisense and mutant plants exhibited improved vacuolar functions, such as maintenance of ΔpH, reduced release of calcium from the vacuole, and greatly improved plant salt tolerance. The antisense plants exhibited increased calcium-dependent protein kinase activity upon salt stress. Improved vacuolar ATPase activity during oxidative stress also was observed in a yeast system, in a ΔVamp7 knockout strain. Interestingly, a microarray-based analysis of the AtVAMP7C genes showed a strong down-regulation of most genes in wild-type roots during salt stress, suggesting an evolutionary molecular adaptation of the vacuolar trafficking.
Keywords: NADPH oxidase, reactive oxygen species, salt stress
The vesicle trafficking machinery is highly conserved among eukaryotes. Its major components include lipids and integral membrane proteins, such as vesicle-associated membrane proteins (VAMPs). The trafficking process is regulated by proteins that assist the vesicle budding, trafficking, and fusion with target membranes. The VAMP proteins constitute the major component of a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex, which functions in facilitating the fusion between the vesicle and the target membranes (1). The fusion is executed by formation of a biochemically stable trans-SNARE complex between the vesicle-SNARE (v-SNARE) that resides on the vesicle membrane and a homologous target-SNARE (t-SNARE) protein located in the target membrane. Plants contain an especially large set of SNAREs (2) that corresponds to existence of multiple vacuoles with different functions (3, 4). Vesicle trafficking has been traditionally viewed as a housekeeping process, but recent findings in plant, yeast, and animal cells show that it also can play an important role in stress responses (5–8).
One of the major vesicle trafficking pathways in plants is the transport of vesicles from the plasma membrane and endoplasmic reticulum to the central lytic vacuole. Recently, plants were found to contain several vacuoles with distinct functions that accumulate specific compounds (9). Major types of vacuoles include the lytic vacuole, which is analogous to animal lysosomes and to the yeast vacuole, and the protein storage vacuole, which is found in mature roots and seeds (10). The vacuoles play an important role in plant salt tolerance. The physiological and biochemical identity of vacuoles largely is determined by correct targeting of vesicles and their cargo (11). A synaptobrevin-like v-SNARE, VAMP711C, was identified in Arabidopsis vegetative vacuoles in the complex that mediated the vesicle docking to the tonoplast (12). We have shown previously that vesicle trafficking to the vacuole plays an important role in salt tolerance (8). The vacuole plays a central role in salt tolerance by sequestering sodium. These functions are facilitated by the Na+/H+ antiporter, which utilizes the proton gradient between the vacuole and the cytoplasm generated by the H+-pyrophosphatase and H+ ATPase enzymes that reside in the tonoplast (13–15).
Highly homologous VAMP7 proteins exist in yeast and animal cells and have been shown to function in the analogous part of the endocytotic pathway in the vesicle transport from late endosomes to lysosomes (16). Moreover, the VAMP7 protein was shown to play a crucial role in the onset of phagocytosis in macrophages (17). Here, we show that vesicle fusion with the vacuole plays an important role in plant tolerance to high salt. Abrupt rise in salt concentration is common during switching the irrigation to low-quality water after germination.
One of the major plant responses to abiotic stress, including salt stress, is the generation of reactive oxygen species (ROS). The major intracellular sources of ROS in plants are chloroplasts, mitochondria, and peroxisomes, although ROS also are produced by enzymatic reactions as byproducts or by the NADPH oxidase that is induced by many abiotic stresses, including salt (8, 18). Recently, induction of endosome-associated ROS generation by NADPH oxidase was described in guard cells treated with abscisic acid (19). The NADPH oxidase enzyme accepts electrons from NADPH at the cytosolic side of the membrane and donates them to molecular oxygen at the other side of the membrane, thus producing superoxide either outside of the plasma membrane or in the endosomes, as described in phagosomes (20). Once produced, the superoxide spontaneously or enzymatically dismutates to H2O2. Recently, the endogenous production of ROS from endoplasmic reticulum and its intracellular compartmentation has been proposed to act as part of the intracellular communication system that regulates the stress response (18).
Results and Discussion
To assess the role of the vacuolar trafficking during salt stress, we analyzed seedlings that were engineered to have altered expression of the major vacuole-targeted v-SNARE, AtVAMP7C (12). The AtVAMP7C protein comprises a small v-SNARE subfamily of four members with extended regions of homology, namely AtVAMP711, AtVAMP712, AtVAMP713, and AtVAMP714. The AtVAMP7C protein family is believed to form a putative tonoplast-specific SNARE–pin complex, which is thought to direct the vacuolar membrane fusion at the tonoplast. Indeed, all four AtVAMP7C proteins recently have been identified in Arabidopsis tonoplast in vivo (12).
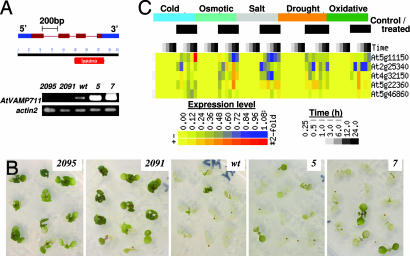
The Arabidopsis thaliana plants were transformed with sense and antisense constructs of AtVAMP711, which is the most prominent member of the vacuolar synaptobrevins (Fig. 1A) (21). Because the antisense approach could have simultaneously silenced several homologous genes, we tested the expression of individual genes from the AtVAMP7C family. Indeed, the antisense expression caused reduced transcription of the close synaptobrevin homologues, particularly in the 2091 line, but did not affect a more distant SNARE, AtVAM3 (Fig. 6A, which is published as supporting information on the PNAS web site).
Fig. 1.
Expression of AtVAMP7C gene and its effect on Arabidopsis salt tolerance. (A Lower) Semiquantitative RT-PCR analysis of the AtVAMP711 expression in wild-type (wt) and transgenic Arabidopsis lines transformed with sense (lines 5 and 7) and antisense (2091 and 2095) constructs. Total RNA was prepared from 4-week-old plants. The actin2 gene served as RNA loading control. (A Upper) The AtVAMP711 gene and protein map, indicating the synaptobrevin domain (red bar) and the exons, is shown. (B) Salt tolerance of the AtVAMP711 transgenic lines. Ten-day-old seedlings were transferred from 1/2 MS plates to 1/2 MS medium supplemented with 200 mM NaCl. Pictures were taken 6 days after transfer. Similar results also were obtained with other antisense transgenic lines (2092 and 2096). (C) Graphical display of the Arabidopsis AtVAMP7c gene family during abiotic stresses. The data are from microarrays analysis from AtGenExpress-Salt Stress (roots of seedlings grown on agar plates). The experimental details can be accessed at the Botany Array Resource database (ref. 49; http://bbc.botany.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi?pub=&dataset=atgenexp_stress).
To test the role of altered synaptobrevin expression in plant salt tolerance, the transformants were subjected to abrupt salt stress by exposing 10-day-old seedlings to 0.2 M NaCl. The treatment caused the seedlings death in 5–7 days in most of the wild-type and AtVAMP711-overexpressing plants. Treatment with 150 mM NaCl caused death 5–7 days later (data not shown). Surprisingly, a much higher survival rate occurred in plants transformed with the antisense construct (Fig. 1B). To test the role of single AtVAMP7C members in salt tolerance, we selected homozygous lines with T-DNA insertions that abrogate the synaptobrevin domain in the AtVAMP711, AtVAMP713, and AtVAMP714 genes. The mutant lines were indistinguishable from the wild-type plants when grown in 1/2 strength Murashige and Skoog (MS) medium. However, when exposed to salt stress, they exhibited increased salt tolerance (Fig. 6B), in agreement with the salt-tolerant phenotype of the antisense lines (Fig. 1B).
It remains to be elucidated whether the individual genes possess specific functions during various stresses, but the fact that mutations in each of the genes resulted in increased salt tolerance, whereas overexpression of the AtVAMP711 gene was comparable to wild type (Fig. 1B), indicates that the gene dosage/expression effect is not additive, and each gene seems to perform an essential function during salt stress, perhaps in the formation of the SNARE–pin complex. Transcriptional analysis of individual members of AtVAMP7C in the specific T-DNA mutant background showed no compensation in the expression of other genes (Fig. 6C). Interestingly, although disruption of the AtVAMP714 resulted in down-regulation of the AtVAMP713 gene, disruption of the AtVAMP713 did not affect the expression of the other AtVAMP7C members, suggesting an interrelation in the regulation of some AtVAMP7C gene expression (Fig. 6C). A scattered degree of coregulation between these genes also was observed by microarray-based analysis across different abiotic stress treatments (Fig. 1C). The data revealed a similar down-regulation in the majority of the AtVAMP7C genes during the early stages of abiotic stresses, particularly the drought, osmotic, and salt stresses.
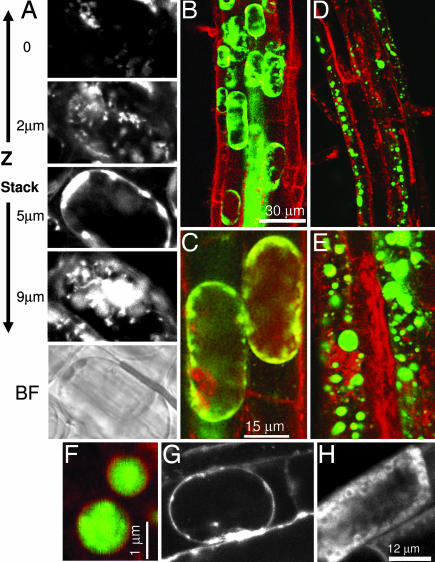
We next analyzed the accumulation of ROS in Arabidopsis roots that has been shown previously to be induced by the NADPH oxidase during salt stress (8). The generation of ROS was observed several minutes after the salt treatment in wild-type and antisense plants (Fig. 7A, which is published as supporting information on the PNAS web site) and appeared as speckles that were distributed throughout the cytoplasm, contrary to the even spreading of H2O2 that would be anticipated from diffusion. It is noteworthy that the concentration of ROS-producing speckles was low near the plasma membrane (Fig. 2A, Z stack). Double-staining of the ROS together with the cellular membranes by styryl membrane tracker dyes, FM 4-64 or FM 1-43 (4, 22), showed that the ROS-producing speckles were of endosomal origin, as can be inferred from the yellow color of the merged image (Fig. 7B). In the wild-type plants, the ROS-containing speckles appeared associated with the root cells tonoplast and seemed to fuse with it (Fig. 2 A–C).
Fig. 2.
Intracellular production of ROS in Arabidopsis seedlings during salt stress. (A) Visualization of NaCl-induced ROS production within a single cell along Z stack. Wild-type Arabidopsis seedlings were grown on agar plates in 1/2 MS medium. Ten-day-old seedlings were exposed to 200 mM NaCl solution for 10 min, and the production of ROS was assayed by confocal microscopy with a ROS-sensitive probe H2DCFDA. Shown are four sections (0.5-μm-thick) along Z stack of a whole single representative cell from the elongation zone. Shown are layers from cell surface, cytosol, vacuole, and cytosol (image depth is indicated on the left). The bright-field (BF) image of the cell is shown at the bottom. (B–E) Simultaneous staining of intracellular membranes (FM 4-64, red) and ROS (H2DCFDA, green). Wild-type (B and C) and AtVAMP711 antisense (line 2091) plants (D and E) were treated as in A. Pictures were taken 5 min after the salt treatment. (Scale bars of B and C also apply to D and E, respectively.). (F) Magnified view of megavesicles in antisense 2091 line double-stained with FM 4-64 and H2DCFDA as described in B–E. (G and H) Single-membrane dye staining with FM 1-43 in seedlings of wild type (G) and antisense 2091 line (H) were treated as in A and stained for 10 min then analyzed by confocal microscopy. Similar results were obtained with FM 4-64 staining.
Similar results also were observed in the AtVAMP711 overexpressing lines (data not shown), but a striking ROS distribution was observed in the antisense lines, which showed ROS caged within megavesicles (Fig. 2 D–F) that did not fuse to the tonoplast and remained in the cytoplasm (Fig. 2 D and E). Importantly, although in the antisense plants no vacuoles could be seen in the double-stained images in Fig. 2 D or E, a central vacuole could be seen by viewing the individual confocal Z sections (Fig. 2H). This result further corroborates the absence of vesicle fusion with the tonoplast in the antisense cells, as apparent from the absence of the line of membrane tracker dye (compare Fig. 2 H with G).
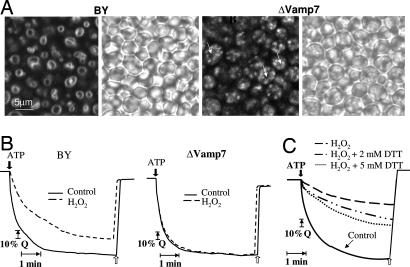
To test the impact of vesicle fusion with the vacuole, we analyzed the tonoplast functioning in a parallel yeast system. Indeed, although staining of wild-type yeast with FM 4-64 showed dye accumulation around the vacuole, a VAMP7 knockout strain BY4741ΔVamp7 showed dispersed intracellular endosomal staining (Fig. 3A). The yeast cells were stimulated with H2O2 for 1 h to allow for bulk-flow fluid-phase endocytosis (23). In the wild-type yeast, the treatment resulted in accumulation of ROS in a cytosolic ring surrounding the vacuoles but showed dispersed aggregation of ROS-containing endosomes within the cytoplasm in the ΔVamp7 mutants (Fig. 8, which is published as supporting information on the PNAS web site). To test the effect of ROS on the tonoplast function in yeast, we assayed the formation of pH gradients across the tonoplast that are generated by V-ATPase activity. H2O2 treatment significantly impaired the V-ATPase activity in the wild-type yeast, but in the knockout ΔVamp7 strain, the activity remained virtually undamaged (Fig. 3B). The effect of ROS on V-ATPase activity was verified by treatment of isolated vacuoles with H2O2 and then with DTT, which resulted in a dose-dependent restoration of the activity (Fig. 3C).
Fig. 3.
Vacuolar functioning of wild-type and ΔVamp7 yeast treated with H2O2. (A) Saccharomyces cerevisiae BY4741 (wt) and BY4741ΔVamp7 (mutant) strains were stained in 10 μM FM 4-64 for 20 min, washed twice, and analyzed by confocal microscopy as described in Materials and Methods. Bright-field images are presented. Arrows indicate central vacuoles. (B) Formation of pH gradients across the tonoplast of isolated vacuole by V-ATPase activity. Wild-type (BY4741) Saccharomyces cerevisiae and ΔVamp7 mutant strains (in the same background) were treated with 9 mM H2O2 or water (control), and acidification of the vacuole (fluorescence quenching, Q) was determined as described in Materials and Methods. The collapse of the ΔpH across the tonoplast membrane was done by addition of 50 mM K+ and 5 μM nigericin (white arrows). (C) Vacuoles were isolated from wild-type yeast and treated with 80 μM H2O2 for 15 min or with water (control). Acidification of the vacuole (fluorescence quenching) was measured as described above. Where indicated, DTT was added immediately after treatment with H2O2.
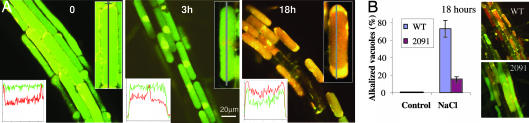
One of the essential functions of the plant tonoplast is generation and maintenance of ΔpH between cytosol and vacuole. The ΔpH is critical for removal of salt from the cytosol by serving as the driving force for the vacuolar antiporters, including the major sodium/proton antiporter, AtNHX1 (13). The function of the tonoplast that keeps the vacuolar interior acidic, versus a neutral/slightly basic environment in the cytosol, is shared with the plasmalemma. The disruption of the H+ gradient between the vacuole and the cytoplasm is indicative of a loss of tonoplast function. To follow the intracellular dynamics of the pH during the salt stress, we used a fluorescent pH probe, carboxy-SNARF (24), which produced comparable pH values to other methods (25). In the wild-type plants a 3-h exposure to salt stress caused a minor change in ΔpH, but overnight treatment resulted in vacuolar alkalization in 80% of cells of the root-hair zone, causing disruption in the ΔpH (Fig. 4A and Table 1). In contrast, in the AtVAMP711 antisense lines the overnight treatment caused only little vacuolar alkalization, as evident from the overall acidic color of the root cells' vacuoles (Fig. 4B Right), as well as by calculation of the percentage of cells with the acidic vacuoles (Fig. 4B).
Fig. 4.
Changes in vacuolar pH during salt stress in Arabidopsis root cells. (A) The intracellular pH in wild-type seedlings was analyzed by using the carboxy-SNARF probe, which shifts its emission from green to red when alkalinized (24). Confocal microscope images were taken in the root-hair zone cells after 0, 3, and 18 h from exposure to 0.2 M NaCl (see Table 1). Merged images of simultaneous emission of green and red filters are given (570 nm and 655 nm, respectively). (Insets) Inserted graphs indicate relative fluorescence (y axis) of each filter along the gray longitudinal line of the cells (x axis) shown and quantified by ImagePro program. (B) Percentage of alkalized vacuoles in wild-type and AtVAMP711C antisense (2091) lines during salt stress. Shown is a representative triplicate experiment, n = 10, ± SE. (Insets) Representative green/red merged images of wild-type and antisense seedlings after 18 h from the beginning of stress.
Table 1.
Intracellular pH values
| Cell compartment | Wild type |
2091 | ||
|---|---|---|---|---|
| 0 h | 3 h | 18 h | 18 h | |
| Vacuole | 5.97 ± 0.05 | 6.17 ± 0.05 | 7.44 ± 0.04 | 6.1 ± 0.05 |
| Cytosol | 6.95 ± 0.02 | 6.97 ± 0.02 | 6.97 ± 0.03 | 7.0 ± 0.04 |
Averaged intracellular pH values ± SE at 0, 3, and 18 h after beginning of stress (n = 30). The pH was calculated from green/red fluorescence ratio in each cell (such as presented in Fig. 4A Insets). The ratio reflects pH according to buffer calibration values.
It is noteworthy that the overnight treatment did not cause cytosolic acidification in the wild-type plants (Table 1), suggesting that the change in the vacuolar pH was caused by reduction in the functioning of the tonoplast proton pumps (PPase and AtPase) and not by proton leakage from the vacuole. This suggestion is supported by the V-ATPase measurements in yeast that show reduced activity by H2O2 treatment (Fig. 3 B and D).
Interestingly, although the disruption of ΔpH has a negative effect on the tonoplast proton antiporters (13), Yamaguchi et al. (26) recently have showed that the Na+/H+ activity is increased with rising pH values, through pH-dependent changes in its C terminus that resides in the vacuolar lumen and affects its interaction with a vacuolar calmodulin-like protein. Thus, as long as some ΔpH still exists, the vacuolar alkalinization that occurs during salt stress actually positively modifies the catalytic activity of the Na+/H+ antiporter, which partially helps to counteract the worsening intracellular conditions.
Another crucial tonoplast function is storage of intracellular calcium in the vacuoles, which constitute the major intracellular Ca2+ store in plants (27). The release of Ca2+ from the vacuole triggers various stress-associated signaling pathways, such as drought, salt, and cold (28, 29). Moreover, elevated Ca2+cyt concentration triggers induction of programmed cell death (30, 31). Furthermore, Ca2+cyt influx was shown to be mediated by ROS via activation of Ca2+-permeable channels in soybean cell culture and in Arabidopsis guard cells (31, 32). The impaired tonoplast functioning in regulation of the vacuolar ΔpH during salt stress prompted us to analyze potential leakage of Ca2+ into the cytosol.
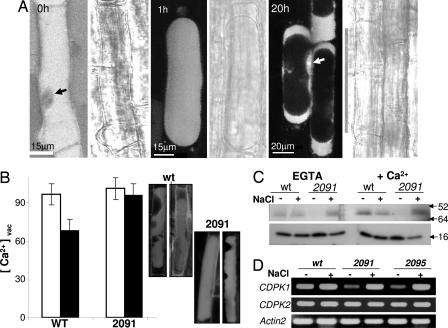
The vacuolar Ca2+ dynamics in Arabidopsis root cells during salt stress were assayed with Fluo-4 acetoxymethyl ester (Fluo-4-AM) that detects free cytosolic Ca2+. In the nonstressed seedlings, the dye was seen predominantly in the vacuoles, which occupy most of the mature cell volume (Fig. 5A). In comparison, a much lower Ca2+ concentration was detected in the nuclei (Fig. 5A, black arrow), in agreement with published data (27). Salt stress caused plasmolysis and vacuolar shrinkage within several minutes, but Ca2+ remained caged in the vacuole >1 h after the onset of stress. The intracellular Ca2+ distribution began to change 4–6 h later, leading to a dramatic Ca2+ redistribution after 20 h in NaCl, producing a major shift of the vacuolar Ca2+ into the cytosol (Fig. 5A). To quantify the vacuolar Ca2+ dynamics during the salt stress, we analyzed the content of the vacuolar Ca2+ at an intermediate stage of 10 h after salt treatment. Although the vacuoles of wild-type plants released >35% of their Ca2+ at this point, there was no significant change in vacuolar calcium in the antisense transgenics (Fig. 5B).
Fig. 5.
Dynamics of vacuolar Ca2+ in root cells during salt stress. (A) Confocal microscope images of intracellular Ca2+ fluorescence by using Fluo-4-AM dye in the root-hair zone of wild-type cells after 0, 1, and 20 h after treatment with 0.2 M NaCl. A bright-field image is attached to each fluorescent image at the right. Arrows indicate nuclei. Dye loading is described in Materials and Methods. (B) Quantification of the translocation of vacuolar Ca2+ into the cytosol 10 h after beginning of stress in wild-type (wt) and AtVAMP711 antisense (2091) seedlings. Confocal Z sections (1-μm-thick) of each vacuole were projected to quantify the whole-vacuolar Fluo-4-AM intensity. (Left) Z sections, projections, and fluorescence measurements were done with ImagePro (n = 25 ± SE). Black bars are from salt-treated seedlings, and white bars are control. (Right) Ca2+ staining in representative vacuoles at 10 h. Note the vacuolar shrinkage. (C) The CDPK activity was assayed by in-gel kinase method in roots of 3-week-old wild-type and AtVAMP711 antisense (2091) seedlings. Plants were treated with 0.2 M NaCl for 3 h. Kinase activity was assayed in protein extracts in the presence of 1 nM CaCl2 or 2.5 mM EGTA as described in ref. 33. Numbers indicate mass in kDa. (D) Semiquantitative RT-PCR analysis of AtCDPK1 (At1g18890) and AtCDPK2 (At1g35670) expression during salt stress (200 mM NaCl for 2 h) in roots of wild-type (wt) and AtVAMP711 antisense (2091 and 2095) lines. Total RNA was prepared from 21-day-old roots of 20 plants in each treatment. The actin2 gene served as loading control.
To assay the possible effects of Ca2+ fluctuations on the Ca2+-dependent signal transduction, we compared the calcium-dependent protein kinase (CDPK) activity in wild-type and antisense plants by in-gel kinase activity assay (33). Interestingly, although the salt stress reduced the activity of a 60-kDa CDPK in wild-type plants, it strongly induced its activity in the 2091 antisense line from undetectable levels in 1/2 MS medium (Fig. 5C). In addition to the CDPK activity, we analyzed the expression of two CDPK genes. In nonstimulated seedlings, the CDPK1 gene showed very low transcription in the antisense plants, as compared with the wild-type plants. The gene underwent strong induction upon exposure to salt (Fig. 5D). Parallel analysis of the CDPK2 gene expression showed similar expression levels in wild-type and antisense plants and was not affected by the salt stress.
Together, our results show that blocking the fusion of endosomes with the vacuolar tonoplast in the antisense AtVAMP711 plants during salt stress improved maintenance of the ΔpH and retained the free Ca2+ in the vacuoles. Both these processes depend primarily on the activity of the tonoplast V-ATPase and v-H+ pyrophosphatase (34). Our results suggest that the impaired tonoplast functioning may be caused by the local oxidative stress, resulting from fusion of the ROS-carrying vesicles with the tonoplast. Indeed, H2O2 caused a reduction of V-ATPase activity in wild-type yeast, but not in the ΔVAMP7 mutant, that could be restored by antioxidant treatment. In plants, the vacuolar ΔpH also is maintained by the v-H+ pyrophosphatase, which contains several cysteine residues that are critical for its activity, and is accessible for H2O2-dependent oxidation, resulting in its inactivation (35). Thus, inhibition of the vesicle fusion with the vacuoles may preserve the tonoplast function. Furthermore, increased salt tolerance of the antisense plants also may be aided by other mechanisms, such as altered CDPK activity, as proposed by Hrabak et al. (36), or by subcellular ROS-dependent signaling that depends on the targeting of ROS-containing vesicles (18).
Concluding Remarks
In summary, our results underscore the importance of the subcellular localization of ROS in Arabidopsis root cells. The traffic destination of the ROS-producing endosomes during salt stress is regulated by the v-and/or t-SNAREs (2, 37). In the wild-type plants, delivery of the ROS-carrying vesicles to the tonoplast results in continuous oxidative bombarding of the membrane that affects the functioning of the tonoplast pumps and channels, thus causing a decrease of ΔpH and an increase in cytosolic Ca2+. These results may explain the earlier studies that showed activation of the voltage-dependent slow vacuolar Ca2+-permeable channels by ROS (38).
The localization of ROS-carrying vesicles with the tonoplast also may contribute to understanding of some aspects of root water relations observed during salt stress. One of the physiological effects of salt stress in plants is a decrease in the hydraulic conductivity of roots, which was shown to be regulated by a decrease in the expression of aquaporins in roots (39). Because H2O2 has been recently shown to cause a reduction in root water transport (40, 41), the presence of H2O2 on the tonoplast of root cells during salt stress, shown here, may add another level of regulation by oxidative gating of the tonoplast intrinsic proteins (TIPs).
In addition to the effect of ROS on the tonoplast functioning, the localization of the ROS-containing vesicles in the cytoplasm may have other effects on ROS-dependent signal transduction through subcellular modifications of signaling proteins, as recently suggested by Rhee (42) in animal systems and by Fedoroff (18) in plants. Because in plants the generation of ROS is induced by many different stimuli that are associated with various environmental stresses, the intracellular targeting and localization of ROS add another level in the regulation in ROS signaling.
The alteration of the intracellular vesicle trafficking also can influence the signal transduction pathways, as shown here for CDPK activity, whereby the suppression of AtVAMP711gene expression affected the CDPK gene expression and kinase activity even in nonstressed plants and absence of ROS. Altered vacuolar trafficking also may affect the composition of vacuolar contents. Proteomic analysis of the vacuoles in the trafficking mutants will link the trafficking gene function with the cargo delivery.
Earlier work by Sanderfoot et al. (43) on the complementary t-SNARE (syntaxin) family in Arabidopsis plants showed that the disruption of individual t-SNARE members caused a lethal phenotype, indicating that each member of those genes performs essential functions even in nonstressed conditions. A similar analysis of the AtVAMP7C genes (which interact with the syntaxins) showed that the v-SNARE genes are not essential during normal growth. On the contrary, the disruption of individual members of the v-SNARE family improved salt tolerance, as opposed to the t-SNAREs. Interestingly, the proteins that interact with the SNARE–pin complex include the GTP binding proteins of the AtRabG3 (AtRab7) class (44), which are homologous to yeast small GTPase Ypt7p that also were shown to function in salt-stress tolerance (8).
Finally, the microarray analysis of the AtVAMP7C gene expression in Arabidopsis roots showed a strong down-regulation of all members except ATVAMP714 gene during abiotic stresses and particularly during salt stress (Fig. 1C), suggesting an evolutionary molecular adaptation of the vacuolar trafficking, which as we show here can protect the seedlings from stress-caused damage. Thus, the regulation of vesicle trafficking during stress may be more complex than expected.
Materials and Methods
Biological Material and Treatments.
The AtVAMP711 (At4g32150) was cloned into the BamHI site of the binary pMD1 transformation vector (8) after amplification of the cDNA by using the primers 5′-GGATCCATCGCGATTCTGTA-3′ and 5′-GGATCCTTAAAATGCAAGATGG-3′ and standard PCR conditions. Direction of the insertion was verified by restriction analysis and by sequencing. For control we used the pBI121 from Clontech (San Diego, CA). Sense/antisense constructs of VAMP711 (6) were transformed into Arabidopsis Col ecotype plants and expressed under 35S promoter. Seedlings were selected on kanamycin and transferred to 1/2 MS plates supplemented with 0.15/0.2 M NaCl. The percent of dead/bleached seedlings was determined 5–7 days after transfer. Seedlings were kept in 1/2 MS ± NaCl also during the microscope studies.
Saccharomyces cerevisiae BY4741 (wild type) and BY4741ΔVamp7 (mutant) strains were from the mutant deletion EUROSCARF collection. All strains were grown overnight in YPD medium (1% yeast extract/1% peptone/2% glucose; Sigma, St. Louis, MO) at 30°C to OD600 0.5. Yeast was treated for 1 h with 9 mM H2O2 or an equal volume of sterile H2O for control condition before harvest.
Analysis of Gene Expression.
AtVAMP711 gene expression in the transgenic plants was analyzed by RT-PCR. RNA was extracted as described in ref. 8. The reverse-transcriptase analysis was performed with a SuperScript III kit (Invitrogen, Carlsbad, CA) using 300 ng of RNA sample.
The VAMP711 transcription was tested with the following primers: reverse (5′-ATCGACAAGAAGCTCCAAAC-3′) and forward (5′-ACCATCGCCAAACAGATCC-3′). CDPK1 (At1g18890) was tested with the following primers: forward (5′-GTGTGGAGTGCCGGAGTTAT-3′) and reverse (5′-CCAAGTTGTGAACCGACCTT-3′). CDPK2 (At1g35670) was tested with the following primers: forward (5′-GATGAACAAGCAGCACCAGA-3′) and reverse (5′-TCCCGTCATTGTCAAGATCA-3′). The actin2 gene was used as a loading control for all experiments: forward (5′-AGTGGTCGTACAACCGGTATGT-3′ and reverse (5′- GATGGCATGGAGGAAGAGAA-3′). In all RT-PCR experiments, the expression was assayed in the linear phase (23–26 cycles).
Confocal Microscopy.
Arabidopsis roots were analyzed in an MRC-1024 confocal microscope (Bio-Rad, Hercules, CA) with a 40× oil immersion objective (N.A. 1.3). Excitation was from argon laser. The transmitted light (bright-field) images were recorded as well. All images were scanned by using identical conditions (laser power, gain of photomultiplier tube, iris size, and zoom) and quantified with ImagePro software package (Media Cybernetics, Silver Spring, MD).
H2O2 was detected by using 10 μM 2′,7′-dichlorofluorescin diacetate (H2DCFDA) probe (Molecular Probes/Invitrogen, Carlsbad, CA). The dye was loaded for 10 min, washed twice in the same buffer, and analyzed by using 485 ± 10 nm/535 ± 10 nm excitation/emission as described in refs. 8 and 45. Intracellular membranes were detected by using the styryl dyes FM 1-43 and FM 4-64 (Molecular Probes/Invitrogen, Carlsbad, CA). Both dyes were loaded at 10 μM concentration for 10 min and washed twice in the same buffer. Fluorescent emission was collected by confocal microscope at 655 nm and 695 nm, respectively.
Intracellular pH was detected by carboxy-SNARF (Molecular Probes/Invitrogen, Carlsbad, CA), whose emission is shifted from green to red when alkalinized (24). Emission of green and red filters was collected (570 nm and 655 nm, respectively) simultaneously. Relative fluorescence of the collected images' intensity was quantified by using ImagePro software. Cellular pH values were calculated from the green/red ratio after a calibration assay in which SNARF fluorescence of the green (570 nm) and red (655 nm) filters was detected in different pH buffer solutions, and the green/red ratio of each pH was determined.
Intracellular Ca2+ was assayed by the Fluo-4-AM probe (Molecular Probes/Invitrogen, Carlsbad, CA). The dye was loaded at 25 μM in the presence of pluronic acid (Molecular Probes/Invitrogen, Carlsbad, CA) and washed as described in ref. 46. Quantification of vacuolar Ca2+ was performed by projecting whole-vacuole Z stack images (0.5-μm-thick) in each cell and analyzing the fluorescence intensity of the integrated image by ImagePro software.
Isolation of Intact Vacuoles from Yeast.
Intact vacuoles were isolated as described by Ohsumi and Anraku (47). The intact vacuoles were collected at the top of the gradient and resuspended in 5 mM Tris-Mes (pH 7.5). Protein concentrations were determined by the Bio-Rad (Hercules, CA) DC Protein Assay according to the manufacturer's protocol.
Transport Assays.
The fluorescence quenching of acridine orange was used to monitor the establishment and dissipation of vacuolar-inside acidic pH gradients according to ref. 48. Intact vacuoles (50 μg of vacuolar protein) were used for each assay. Vacuoles were added to a buffer containing 50 mM tetramethyl ammonium chloride, 5 μM acridine orange, 5 mM Tris-Mes (pH 7.5), and 3.125 mM MgSO4. The vacuolar ATPase was activated by the addition of 5 mM Tris-ATP, and time-dependent fluorescence changes were monitored on a PerkinElmer (Wellesley, MA) fluorescence spectrophotometer with excitation and emission wavelengths of 495 and 540 nm, respectively, and a slit width of 5 nm with a 1% transmittance filter. When a steady-state pH gradient was established, 50 mM KCl and 5 μM nigericin were added, resulting in the collapse of the ΔpH across the tonoplast membrane to allow normalization of the curves to 100% quench.
Analysis of CDPK Activity.
Ten-day-old Arabidopsis roots from 30–40 seedlings were ground in liquid nitrogen, thawed in 2 vol of extraction buffer [50 mM Hepes (pH 7.4), 5 mM EDTA, 5 mM EGTA, 5 mM DTT, 10 mM NaF, 10 mM Na3VO4, 50 mM β-glycerophosphate, and plant protease inhibitor mixture; Sigma, St. Louis, MO]. After centrifugation at 15,000 × g for 20 min at 4°C, the pellet was resuspended in 2 vol of elution buffer [20 mM Hepes (pH 7.4), 1 mM MgCl2, 1% Triton X-100, 1 mM NaF, 1 mM Na2VO4, 5 mM β-glycerophosphate, and plant protease inhibitor mixture]. The resulting crude Triton X-100-solubilized membrane extracts were stored at −70°C. The kinase activity was determined exactly as described in ref. 33.
RT-PCR Analysis and Epifluorescent Microscopy.
For details of RT-PCR analysis and epifluorescent microscopy, see Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Prof. Eduardo Blumwald (University of California, Davis) for his help with the analysis of vacuolar function in yeast and Dr. Hagai Karchi (Evogene Ltd.) for production of the AtVAMP7C lines. The research was funded by the Israel Science Foundation.
Abbreviations
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- v-SNARE
vesicle-SNARE
- t-SNARE
target-SNARE
- VAMP
vesicle-associated membrane protein
- ROS
reactive oxygen species
- MS
Murashige and Skoog
- CDPK
calcium-dependent protein kinase
- Fluo-4-AM
Fluo-4 acetoxymethyl ester
- H2DCFDA
2′,7′-dichlorofluorescin diacetate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Cell Struct Funct. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- 2.Sanderfoot AA, Assaad FF, Raikhel NV. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassham DC, Raikhel NV. Curr Opin Cell Biol. 2000;12:491–495. doi: 10.1016/s0955-0674(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 4.Jurgens G. Annu Rev Cell Dev Biol. 2004;20:481–504. doi: 10.1146/annurev.cellbio.20.082503.103057. [DOI] [PubMed] [Google Scholar]
- 5.Cavalli V, Vilbois F, Corti M, Marcote MJ, Tamura K, Karin M, Arkinstall S, Gruenberg J. Mol Cell. 2001;7:421–432. doi: 10.1016/s1097-2765(01)00189-7. [DOI] [PubMed] [Google Scholar]
- 6.Levine A, Belenghi B, Damari-Weisler H, Granot D. J Biol Chem. 2001;276:46284–46289. doi: 10.1074/jbc.M107375200. [DOI] [PubMed] [Google Scholar]
- 7.Kargul J, Gansel X, Tyrrell M, Sticher L, Blatt MR. FEBS Lett. 2001;508:253–258. doi: 10.1016/s0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- 8.Mazel A, Leshem Y, Tiwari BS, Levine A. Plant Physiol. 2004;134:118–128. doi: 10.1104/pp.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okita TW, Rogers JC. Annu Rev Plant Biol. 1996;47:327–350. doi: 10.1146/annurev.arplant.47.1.327. [DOI] [PubMed] [Google Scholar]
- 10.Vitale A, Raikhel NV. Trends Plants Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- 11.Maeshima M. Ann Rev Plant Physiol Plant Mol Biol. 2001;52:469–497. doi: 10.1146/annurev.arplant.52.1.469. [DOI] [PubMed] [Google Scholar]
- 12.Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaxiola RA, Fink GR, Hirschi KD. Plant Physiol. 2002;129:967–973. doi: 10.1104/pp.020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apse MP, Aharon GS, Snedden WA, Blumwald E. Science. 1999;258:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. Genes Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Advani RJ, Yang B, Prekeris R, Lee KC, Klumperman J, Scheller RH. J Cell Biol. 1999;146:765–775. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun V, Fraisier V, Raposo G, Hurbain I, Sibarita J-B, Chavrier P, Galli T, Niedergang F. EMBO J. 2004;23:4166–4176. doi: 10.1038/sj.emboj.7600427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedoroff N. Ann Bot (London) 2006;98:289–300. doi: 10.1093/aob/mcl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park K-Y, Jung J-Y, Park J, Hwang J-U, Kim Y-W, Hwang I, Lee Y. Plant Physiol. 2003;132:92–98. doi: 10.1104/pp.102.016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato TK, Overduin M, Emr SD. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- 21.Uemura T, Sato MH, Takeyasu K. FEBS Lett. 2005;579:2842–2846. doi: 10.1016/j.febslet.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 22.Emans N, Zimmermann S, Fischer R. Plant Cell. 2002;14:71–86. doi: 10.1105/tpc.010339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vida TA, Emr SD. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albrechtova JTP, Heilscher S, Leske L, Walczysko P, Wagner E. Plant Cell Environ. 2003;26:1985–1994. [Google Scholar]
- 25.Kurkdjian A, Guern J. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:271–303. [Google Scholar]
- 26.Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E. Proc Natl Acad Sci USA. 2005;102:16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders D, Pelloux J, Brownlee C, Harper JF. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight H, Trewavas AJ, Knight MR. Plant J. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 29.Sanders D, Brownlee C, Harper JF. Plant Cell. 1999;11:691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam E, Pontier D, del Pozo O. Curr Opin Plant Biol. 1999;2:502–507. doi: 10.1016/s1369-5266(99)00026-6. [DOI] [PubMed] [Google Scholar]
- 31.Levine A, Pennell R, Alvarez M, Palmer R, Lamb CJ. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 32.Murata Y, Pei ZM, Mori IC, Schroeder J. Plant Cell. 2001;13:2513–2523. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romeis T, Piedras P, Jones JDG. Plant Cell. 2000;12:803–815. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Yang H, Ann Peer W, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 35.Zhen RG, Kim EJ, Rea PA. J Biol Chem. 1994;269:23342–23350. [PubMed] [Google Scholar]
- 36.Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blatt MR, Leyman B, Geelen D. New Phytologist. 1999;144:389–418. doi: 10.1046/j.1469-8137.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 38.Carpaneto A, Cantu AM, Gambale F. FEBS Lett. 1999;442:129–132. doi: 10.1016/s0014-5793(98)01642-1. [DOI] [PubMed] [Google Scholar]
- 39.Boursiac Y, Chen S, Luu D-T, Sorieul M, van den Dries N, Maurel C. Plant Physiol. 2005;139:790–805. doi: 10.1104/pp.105.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aroca R, Amodeo G, Fernandez-Illescas S, Herman EM, Chaumont F, Chrispeels MJ. Plant Physiol. 2005;137:341–353. doi: 10.1104/pp.104.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye Q, Steudle E. Plant Cell Environ. 2006;29:459–470. doi: 10.1111/j.1365-3040.2005.01423.x. [DOI] [PubMed] [Google Scholar]
- 42.Rhee SG. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 43.Sanderfoot AA, Pilgrim M, Adam L, Raikhel NV. Plant Cell. 2001;13:659–666. doi: 10.1105/tpc.13.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siniossoglou S, Pelham HRB. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- 45.Schopfer P, Plachy C, Frahry G. Plant Physiol. 2001;125:1591–1602. doi: 10.1104/pp.125.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walczysko P, Wagner E, Albrechtova JTP. Cell Calcium. 2000;28:23–32. doi: 10.1054/ceca.2000.0132. [DOI] [PubMed] [Google Scholar]
- 47.Ohsumi Y, Anraku Y. J Biol Chem. 1981;256:2079–2082. [PubMed] [Google Scholar]
- 48.Blumwald E, Rea PA, Poole RJ. Methods Enzymol. 1987;148:115–123. [Google Scholar]
- 49.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.