Abstract
Methyldiazene (HN N
N CH3) isotopomers labeled with 15N at the terminal or internal nitrogens or with 13C or 2H were used as substrates for the nitrogenase α-195Gln-substituted MoFe protein. Freeze quenching under turnover traps an S = ½ state that has been characterized by EPR and 1H-, 15N-, and 13C-electron nuclear double resonance spectroscopies. These studies disclosed the following: (i) a methyldiazene-derived species is bound to the active-site FeMo cofactor; (ii) this species binds through an [-NHx] fragment whose N derives from the methyldiazene terminal N; and (iii) the internal N from methyldiazene probably does not bind to FeMo cofactor. These results constrain possible mechanisms for reduction of methyldiazene. In the Chatt–Schrock mechanism for N2 reduction, H atoms sequentially add to the distal N before N-N bond cleavage (d-mechanism). In a d-mechanism for methyldiazene reduction, a bound [-NHx] fragment only occurs after reduction by three electrons, which leads to N-N bond cleavage and the release of the first NH3. Thus, the appearance of bound [-NHx] is compatible with the d-mechanism only if it represents a late stage in the reduction process. In contrast are mechanisms where H atoms add alternately to distal and proximal nitrogens before N-N cleavage (a-mechanism) and release of the first NH3 after reduction by five electrons. An [-NHx] fragment would be bound at every stage of methyldiazene reduction in an a-mechanism. Although current information does not rule out the d-mechanism, the a-mechanism is more attractive because proton delivery to substrate has been specifically compromised in α-195Gln-substituted MoFe protein.
CH3) isotopomers labeled with 15N at the terminal or internal nitrogens or with 13C or 2H were used as substrates for the nitrogenase α-195Gln-substituted MoFe protein. Freeze quenching under turnover traps an S = ½ state that has been characterized by EPR and 1H-, 15N-, and 13C-electron nuclear double resonance spectroscopies. These studies disclosed the following: (i) a methyldiazene-derived species is bound to the active-site FeMo cofactor; (ii) this species binds through an [-NHx] fragment whose N derives from the methyldiazene terminal N; and (iii) the internal N from methyldiazene probably does not bind to FeMo cofactor. These results constrain possible mechanisms for reduction of methyldiazene. In the Chatt–Schrock mechanism for N2 reduction, H atoms sequentially add to the distal N before N-N bond cleavage (d-mechanism). In a d-mechanism for methyldiazene reduction, a bound [-NHx] fragment only occurs after reduction by three electrons, which leads to N-N bond cleavage and the release of the first NH3. Thus, the appearance of bound [-NHx] is compatible with the d-mechanism only if it represents a late stage in the reduction process. In contrast are mechanisms where H atoms add alternately to distal and proximal nitrogens before N-N cleavage (a-mechanism) and release of the first NH3 after reduction by five electrons. An [-NHx] fragment would be bound at every stage of methyldiazene reduction in an a-mechanism. Although current information does not rule out the d-mechanism, the a-mechanism is more attractive because proton delivery to substrate has been specifically compromised in α-195Gln-substituted MoFe protein.
Keywords: diazene, reduction, intermediate, dinitrogen
The reduction of dinitrogen (N2) to two ammonia (NH3) molecules (N2 fixation) represents the exclusive pathway for input of N2 into the global biogeochemical N cycle and therefore is a reaction essential to all life (1). The majority of all N2 fixation occurs by the action of microbes that contain the enzyme nitrogenase. The Mo-based nitrogenase catalyzes the reduction of N2 to two ammonia molecules (2) according to the optimal reaction shown in Eq. 1, requiring multiple electrons, protons, the hydrolysis of MgATP, and the evolution of H2.
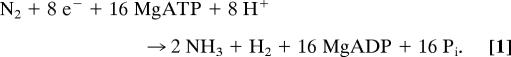 |
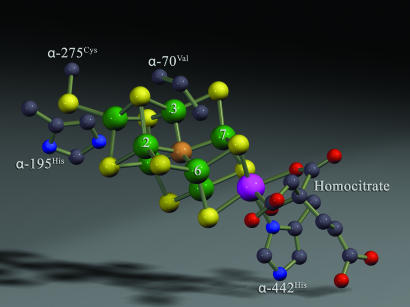
N2 binding and reduction by nitrogenase occurs at a complex metal cofactor contained in the MoFe protein component protein called FeMo cofactor (3, 4). The structure of this active-site cofactor has been disclosed by the high-resolution x-ray crystal structures of the MoFe protein by the Rees group (5–8) and others (9, 10) (Fig. 1). Recent studies using combined genetic, biochemical, and spectroscopic approaches have localized a binding site for N2 and nonphysiological alkyne substrates to a specific Fe-S face of FeMo cofactor (11) defined by Fe atoms 2, 3, 6, and 7 (numbering based on Protein Data Bank ID code 1M1N). Further, a strategy has been devised to trap intermediate states during the reduction of the alkyne substrates propargyl alcohol (12) and acetylene (13, 14) in high concentration and to show by electron nuclear double resonance (ENDOR) spectroscopy that they contain reduced substrate bound to FeMo cofactor (13–15). The ENDOR spectroscopic studies reveal that the product alkene most likely is bound side-on to a single Fe atom of FeMo cofactor. Such side-on binding of alkenes (and alkynes) to metals is consistent with binding modes observed in metal complexes outside of proteins (16). An analogous study has characterized an intermediate trapped during proton reduction (17).
Fig. 1.
A view of the FeMo cofactor highlighting Fe atoms 2, 3, 6, and 7 (numbering from Protein Data Bank ID code 1M1N), along with MoFe protein amino acids ligands (α-275Cys and α-442His) and the side chains of α-195His and α-70Val. The figure was generated by using the Protein Data Bank ID code 1M1N. Fe is green, S is yellow, Mo is purple, C is gray, O is red, and N is blue.
The mechanism for N2 reduction to ammonia by nitrogenase and the nature of the N2 reduction intermediates bound to FeMo cofactor during this reaction remain obscure (11). In contrast, a mechanism for N2 reduction at mononuclear Mo metal complexes is well advanced, with contributions from the early work of Chatt et al. (18) and Pickett (19) and more recently from Yandulov and Schrock (20) and Schrock (21, 22). Determination of the mechanism of N2 reduction by such Mo-based complexes included the structure determination of a number of intermediates along the reaction pathway. By analogy to the mechanism of these inorganic metal complexes, there is evidence that N2 reduction catalyzed by nitrogenase proceeds through intermediate states generated by the sequential addition of protons and electrons to N2 bound to FeMo cofactor (23), forming nitrogenous species at the reduction levels of diazene and hydrazine (Eq. 2, where M represents FeMo cofactor).
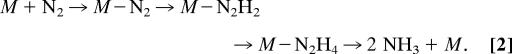 |
In support of such a mechanism is the observation that hydrazine is both a nitrogenase substrate (24, 25), being reduced to ammonia, and a minor product that can be isolated when nitrogenase is acid-quenched during N2 reduction (23).
We recently reported a strategy for trapping states with bound species when N2, a diazene, or hydrazine, are used as substrate (26). Key to trapping certain of these species in high occupancy was the use of MoFe proteins that contain amino acid substitutions, separately, or in combination, that compromise proton delivery to nitrogenous substrates (α-195Gln) or expand accessibility to the substrate reduction site (α-70Ala). EPR and ENDOR analysis of these trapped states revealed that each of them represents either the initial substrate or a reduced species bound to FeMo cofactor and indicated that at least two, and possibly three, distinct states could be trapped (26). We have suggested that these species could be analogous to different stages along the N2 reduction pathway. Here, we describe a multinuclear ENDOR analysis of a species that was trapped during nitrogenase catalysis when 15N- and 13C- or 2H-labeled methyldiazene isotopomers were used as substrate. These studies expand our initial insight into the nature of the methyldiazene-bound state and present powerful constraints on possible mechanisms of its reduction by the α-195Gln MoFe protein.
Results
Trapping Nitrogenous Species Bound to FeMo Cofactor.
We have sought to gain insight into the nitrogenase mechanism by using freeze-quench experiments to trap nitrogenous species bound at the active site when N2 and other nitrogenous compounds that could be considered as intermediates or analogs in the reduction of N2 are used as substrates (25, 26). An important aspect in the development of this strategy was the prediction that restricting proton delivery to substrates could result in the accumulation of turnover states with enzyme-bound semireduced species. This prediction was supported by freeze-quench experiments where an S = ½ state was trapped when hydrazine was used as a substrate for the α-195Gln-substituted MoFe protein (25). The α-195Gln-substituted MoFe protein appears to be specifically compromised in its ability to effectively deliver protons to nitrogenous substrates (27, 28). The accumulation of this state could be further increased (25) by placing another substitution (α-70Ala) within the MoFe protein that lowers constraints on the accessibility of substrates to the active site (Fig. 1). Our next goal was to trap a state earlier along the N2 reduction pathway by using a less-reduced nitrogenous compound as substrate. In the present work we elected to use methyldiazene as substrate because it is asymmetrical and such asymmetry could be exploited to enable discrimination of binding modes by using specifically labeled 15N or 13C isotopomers in freeze-quench ENDOR experiments.
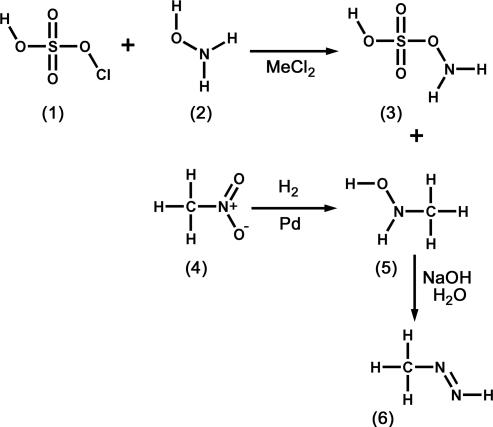
The synthesis and properties of methyldiazene were described some years ago (29). Of importance for the current studies, methyldiazene can be synthesized (Scheme 1) from readily available starting materials with appropriate 14/15N or 12/13C labels to produce the requisite isotopomers. The short half-life of methyldiazene in solution necessitated special handling procedures that typically involved synthesis of methyldiazene immediately before it was needed, stabilization by freezing, and rapid transfer of the gas directly to the nitrogenase assay vessel.
Scheme 1.
Methyldiazene synthetic scheme. Hydroxylamine-O-sulfonic acid (3) was synthesized as a white precipitate after reaction of chlorosulfonic acid (1) and hydroxylamine (2, where the N atom was either 15N or 14N). N-methylhydroxylamine (5) was synthesized by the reduction of nitromethane (4, where the N atom was either 14N or 15N and the C atom was either 12C or 13C). Methyldiazene (6) was prepared by the base-catalyzed reaction of hydroxylamine-O-sulfonic acid (3) with N-methylhydrolyamine (5), and the gas was isolated by freezing. See Materials and Methods for details.
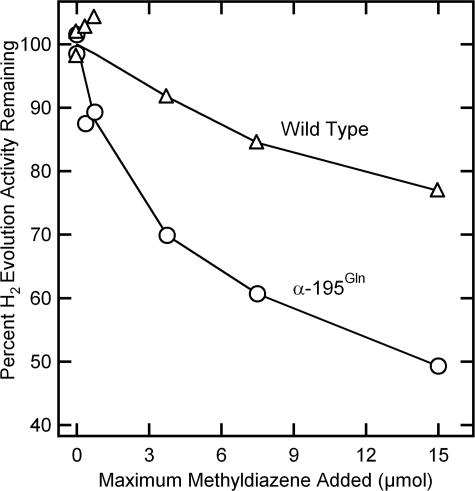
The instability of methyldiazene and accumulation of its nonenzymatic degradation products precludes a direct demonstration that methyldiazene is an effective nitrogenase substrate. Interaction of methyldiazene with the nitrogenase active site was therefore established by its capacity to inhibit nitrogenase-catalyzed H2 evolution (30). In the absence of other substrates, nitrogenase reduces protons yielding H2. The presence of substrates such as N2 or acetylene diverts electron flow away from proton reduction, resulting in lower H2 evolution rates. When an increasing quantity of methyldiazene is added to WT nitrogenase under proton reduction conditions, a progressive inhibition of the H2 evolution rate is observed (Fig. 2). Methyldiazene was found to be an even stronger inhibitor of proton reduction catalyzed by the α-195Gln-substituted MoFe protein, providing support for the idea that restriction of proton delivery results in a stronger association of methyldiazene, or one of its reduction products, with the substituted MoFe protein.
Fig. 2.
Methyldiazene inhibition of proton reduction activity. The percentage of maximal H2 evolution activity (nmol H2/min per mg of MoFe protein) remaining in the presence of increasing methyldiazene is shown for the WT (△) and α-195Gln (○) MoFe proteins. The maximum quantity of methyldiazene added to a 9-ml vial assuming 100% yield is shown.
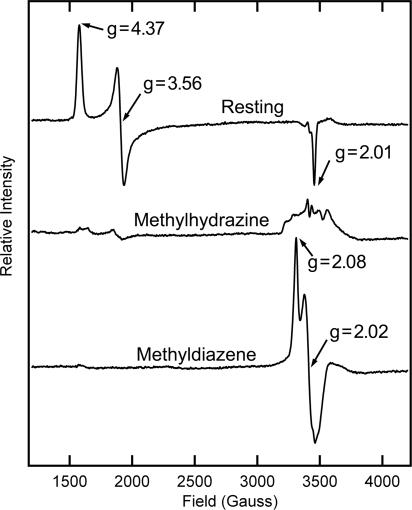
This result suggested it might be possible to trap an intermediate state during turnover of methyldiazene. The addition of methyldiazene to the resting-state α-195Gln-substituted MoFe protein does not perturb the distinctive resting-state EPR signal of S = 3/2 FeMo cofactor, with g = [4.37, 3.56, 2.01] (Fig. 3). When the α-195Gln-substituted MoFe protein is freeze-trapped during turnover without added substrate under argon, the FeMo cofactor EPR spectrum is greatly decreased in intensity (31), indicating reduction to an EPR-silent state (data not shown). When methyldiazene was included in the turnover mixture and the sample was freeze-quenched, an EPR spectrum was observed corresponding to an S = ½ spin state. At X band the spectrum appears roughly axial, with g‖ = 2.08, g⊥ = 2.02 (Fig. 3); spectra collected at 35 GHz (data not shown) revealed a rhombic signal with g(m) = [2.083, 2.021, 1.993]. This spectrum was not observed when methyldiazene was allowed to decompose before addition to nitrogenase. The methyldiazene-dependent spectrum is similar but not identical to the S = ½ signals of states trapped with other substrates, such as N2 (26), hydrazine (25), propargyl alcohol (12), acetylene (13), and carbon disulfide (32). Like these other states, this state represents a majority species: conversion of the resting-state FeMo cofactor to this state is over >50% based on spin integration of the EPR signal. Because the α-195Gln-substituted MoFe protein is compromised in proton delivery (27), the trapped species most likely represents either bound methyldiazene or an early stage in the reduction of methyldiazene. A methyldiazene-dependent EPR signal was not detected when the WT MoFe protein was used. Such binding would be analogous to a middle stage in N2 reduction, which leads us to use the shorthand m(CH3 N
N NH) to designate the state. It should be emphasized that whether or not m(CH3
NH) to designate the state. It should be emphasized that whether or not m(CH3 N
N NH) represents a faithful mimic of an enzyme-bound semireduced N2 species is not yet established, although our experimental strategy and interpretations are framed around this possibility.
NH) represents a faithful mimic of an enzyme-bound semireduced N2 species is not yet established, although our experimental strategy and interpretations are framed around this possibility.
Fig. 3.
EPR spectra are shown for the resting state (Top) and turnover trapped states with methylhydrazine (Middle) or methyldiazene (Bottom) present. EPR conditions are described in Materials and Methods. Calculated g values are noted for some inflections.
To test the possibility that m(CH3 N
N NH) represents a nonenzymatic breakdown product of methyldiazene, the α-195Gln MoFe protein was freeze-trapped during turnover in the presence of ammonia, methyl amine, or N2. These potential breakdown products did not generate an EPR spectrum like that observed when methyldiazene was used as substrate. Nitrogenase trapped during turnover in the presence of another possible breakdown product, methylhydrazine, did reveal a minor EPR signal in the g ≈ 2 region, but that signal was not similar to the one elicited by methyldiazene (Fig. 3).
NH) represents a nonenzymatic breakdown product of methyldiazene, the α-195Gln MoFe protein was freeze-trapped during turnover in the presence of ammonia, methyl amine, or N2. These potential breakdown products did not generate an EPR spectrum like that observed when methyldiazene was used as substrate. Nitrogenase trapped during turnover in the presence of another possible breakdown product, methylhydrazine, did reveal a minor EPR signal in the g ≈ 2 region, but that signal was not similar to the one elicited by methyldiazene (Fig. 3).
The microwave power dependence of the EPR signal associated with m(CH3 N
N NH) was also established (see Fig. 6, which is published as supporting information on the PNAS web site). The results reveal the expected difference of the microwave power saturation for an S = ½ state from that of the S = 3/2 resting-state FeMo cofactor. The microwave power dependence for the m(CH3
NH) was also established (see Fig. 6, which is published as supporting information on the PNAS web site). The results reveal the expected difference of the microwave power saturation for an S = ½ state from that of the S = 3/2 resting-state FeMo cofactor. The microwave power dependence for the m(CH3 N
N NH)-associated EPR signal is significantly different from that reported for the trapped state formed during hydrazine turnover (25). This latter species is denoted l(N2H4) because it is suggested to represent a late stage in N2 reduction. The pH dependence of the EPR signal intensity for m(CH3
NH)-associated EPR signal is significantly different from that reported for the trapped state formed during hydrazine turnover (25). This latter species is denoted l(N2H4) because it is suggested to represent a late stage in N2 reduction. The pH dependence of the EPR signal intensity for m(CH3 N
N NH) is also distinctly different from the pH dependence of the EPR signal intensity for the l(N2H4) trapped state (see Fig. 7, which is published as supporting information on the PNAS web site). Such features that distinguish m(CH3
NH) is also distinctly different from the pH dependence of the EPR signal intensity for the l(N2H4) trapped state (see Fig. 7, which is published as supporting information on the PNAS web site). Such features that distinguish m(CH3 N
N NH) and l(N2H4) provides further evidence that the two states are different.
NH) and l(N2H4) provides further evidence that the two states are different.
ENDOR Studies of m(HN N
N CH3).
CH3).
To further characterize m(HN N
N CH3) and any species bound to FeMo cofactor, methyldiazene was synthesized with 15N in either the terminal position or the internal position (Scheme 1). As expected, the EPR signal of m(H15N
CH3) and any species bound to FeMo cofactor, methyldiazene was synthesized with 15N in either the terminal position or the internal position (Scheme 1). As expected, the EPR signal of m(H15N N
N CH3) was indistinguishable from that observed when 14N-methyldiazene was used. It was shown (26) that the 15N Mims pulsed ENDOR spectra collected at g1 = 2.08 for m(15NH
CH3) was indistinguishable from that observed when 14N-methyldiazene was used. It was shown (26) that the 15N Mims pulsed ENDOR spectra collected at g1 = 2.08 for m(15NH N
N CH3) contains a single 15N doublet that is centered at the 15N Larmor frequency and has a hyperfine splitting, A(g1) = 1.5 MHz. In a g1 spectrum, such a 15N doublet represents a single type of 15N of the nitrogenous moiety bound to the FeMo cofactor. A spectrum taken at g2 = 2.03 (Fig. 4) also exhibits a single doublet, although at this g value the individual peaks have structure and breadth associated with hyperfine anisotropy. These signals are absent in spectra of samples prepared with 14N-labeled methyldiazene, which demonstrates that m(HN
CH3) contains a single 15N doublet that is centered at the 15N Larmor frequency and has a hyperfine splitting, A(g1) = 1.5 MHz. In a g1 spectrum, such a 15N doublet represents a single type of 15N of the nitrogenous moiety bound to the FeMo cofactor. A spectrum taken at g2 = 2.03 (Fig. 4) also exhibits a single doublet, although at this g value the individual peaks have structure and breadth associated with hyperfine anisotropy. These signals are absent in spectra of samples prepared with 14N-labeled methyldiazene, which demonstrates that m(HN N
N CH3) contains a species bound to FeMo cofactor through a nitrogen that originates as the terminal N of methyldiazene.
CH3) contains a species bound to FeMo cofactor through a nitrogen that originates as the terminal N of methyldiazene.
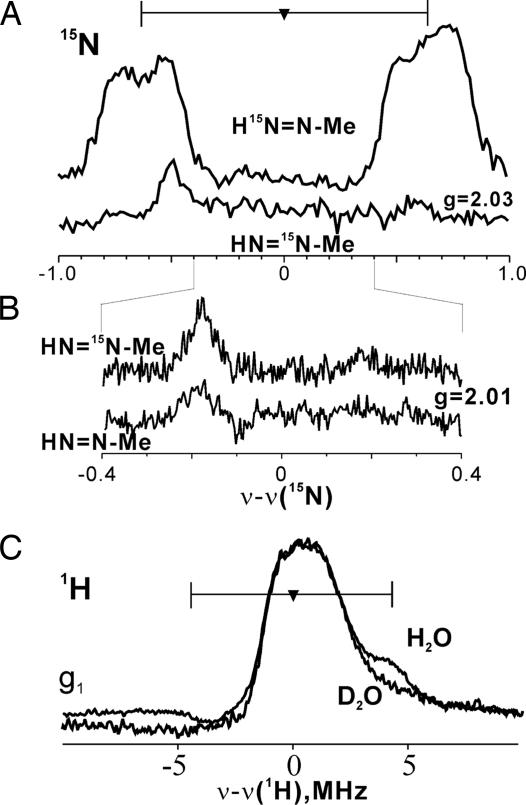
Fig. 4.
35-GHz ENDOR spectra. (A) 15N Mims ENDOR spectra of m(CH3 N
N NH). For H15N
NH). For H15N N
N CH3, ν = 34.808 GHz, g = 2.03, π/2 = 52 ns, τ = 400 ns, T = 25.352 μs, radio frequency (RF) = 20 μs, eight scans, 30 shots per point, 20-ms shot repetition time, 2 K. For HN
CH3, ν = 34.808 GHz, g = 2.03, π/2 = 52 ns, τ = 400 ns, T = 25.352 μs, radio frequency (RF) = 20 μs, eight scans, 30 shots per point, 20-ms shot repetition time, 2 K. For HN 15N
15N CH3, ν = 34.784 GHz, and other conditions are the same as for H15N
CH3, ν = 34.784 GHz, and other conditions are the same as for H15N N
N CH3. Spectra are normalized for comparison. (B) As in A, but narrower scan and g = 2.01, τ = 552 ns, and 15 scans. Backgrounds have been corrected by simple subtractions as needed. (C) Continuous wave (CW) 1H-ENDOR of m(CH3
CH3. Spectra are normalized for comparison. (B) As in A, but narrower scan and g = 2.01, τ = 552 ns, and 15 scans. Backgrounds have been corrected by simple subtractions as needed. (C) Continuous wave (CW) 1H-ENDOR of m(CH3 N
N NH) in H2O and D2O. Conditions were: microwave frequency of 35.057–35.171 GHz, modulation amplitude = 4 G, RF sweep speed = 1 MHz/s, bandwidth of RF broadened to 100 kHz, 2 K. Methyldiazene is abbreviated as HN
NH) in H2O and D2O. Conditions were: microwave frequency of 35.057–35.171 GHz, modulation amplitude = 4 G, RF sweep speed = 1 MHz/s, bandwidth of RF broadened to 100 kHz, 2 K. Methyldiazene is abbreviated as HN N
N Me.
Me.
The 15N-ENDOR spectrum associated with m(HN 15N
15N CH3) at g = 2.03 (Fig. 4) shows a background feature presumed to be from enzyme 14N, but the 15N doublet seen with the m(H15N
CH3) at g = 2.03 (Fig. 4) shows a background feature presumed to be from enzyme 14N, but the 15N doublet seen with the m(H15N N
N CH3) state is gone; neither is an 15N doublet seen in broader scans at this and other g values. Narrowing the scan at this and other g values shows only weak features that persist in the natural-abundance sample; for example, a narrow g = 2.01 spectrum shows a single weak feature in this range in both 15N- and 14N-labeled trapped states (Fig. 4). Thus, whereas a signal from what originates as the terminal 15N is readily observed, no signal associated with the 15N of HN
CH3) state is gone; neither is an 15N doublet seen in broader scans at this and other g values. Narrowing the scan at this and other g values shows only weak features that persist in the natural-abundance sample; for example, a narrow g = 2.01 spectrum shows a single weak feature in this range in both 15N- and 14N-labeled trapped states (Fig. 4). Thus, whereas a signal from what originates as the terminal 15N is readily observed, no signal associated with the 15N of HN 15N
15N CH3 is detected. HN
CH3 is detected. HN N
N 13CH3 and HN
13CH3 and HN N
N CD3 also were prepared (4 in Scheme 1) and used in freeze-quench experiments. No differences were observed between the 13C or 2H ENDOR spectra of the trapped states when labeled or unlabeled methyldiazene were used as substrate (data not shown). These measurements indicate that the central N from methyldiazene is not bound to a metal ion of FeMo cofactor, or at least not to one with a significant electron-spin projection. In part because of interference from the background 14N signals, determination of whether the methyldiazene framework is intact or not will require further study and comparison to data from model compounds.
CD3 also were prepared (4 in Scheme 1) and used in freeze-quench experiments. No differences were observed between the 13C or 2H ENDOR spectra of the trapped states when labeled or unlabeled methyldiazene were used as substrate (data not shown). These measurements indicate that the central N from methyldiazene is not bound to a metal ion of FeMo cofactor, or at least not to one with a significant electron-spin projection. In part because of interference from the background 14N signals, determination of whether the methyldiazene framework is intact or not will require further study and comparison to data from model compounds.
1H ENDOR spectra of the m(NH N
N CH3) state in H2O and D2O buffers taken at g1 = 2.08 (Fig. 4) show an unresolved, nonexchangeable (unchanged in D2O buffer) central peak at the proton Larmor frequency primarily from the “matrix” protons of nearby residues. As indicated in Fig. 4, the spectra further show a signal from an exchangeable proton(s) with A(g1) ≈9 MHz. This coupling is too small to be associated with the H+/H− bound to FeMo cofactor studied previously (17). It is similar to those of the protons of an alkene bound to FeMo cofactor during alkyne reduction (12–14). As a result, this proton can be assigned to an [-NHx] moiety that is bound directly to the FeMo cofactor. This assignment of the proton signal, rather than to an exchangeable proton associated with an H-bonding amino acid residue, is supported by comparison with the state that forms upon turnover with N2 itself (26). That state also displays a 15N ENDOR signal when formed with 15N2 but does not exhibit a resolved signal from an exchangeable proton. With this assignment, m(NH
CH3) state in H2O and D2O buffers taken at g1 = 2.08 (Fig. 4) show an unresolved, nonexchangeable (unchanged in D2O buffer) central peak at the proton Larmor frequency primarily from the “matrix” protons of nearby residues. As indicated in Fig. 4, the spectra further show a signal from an exchangeable proton(s) with A(g1) ≈9 MHz. This coupling is too small to be associated with the H+/H− bound to FeMo cofactor studied previously (17). It is similar to those of the protons of an alkene bound to FeMo cofactor during alkyne reduction (12–14). As a result, this proton can be assigned to an [-NHx] moiety that is bound directly to the FeMo cofactor. This assignment of the proton signal, rather than to an exchangeable proton associated with an H-bonding amino acid residue, is supported by comparison with the state that forms upon turnover with N2 itself (26). That state also displays a 15N ENDOR signal when formed with 15N2 but does not exhibit a resolved signal from an exchangeable proton. With this assignment, m(NH N
N CH3) cannot contain bound, deprotonated methyldiazene or the bound [
CH3) cannot contain bound, deprotonated methyldiazene or the bound [ N
N NHCH3] isomer (Fig. 5). 1H couplings to a distal [-NH], if present, would be too small to resolve.
NHCH3] isomer (Fig. 5). 1H couplings to a distal [-NH], if present, would be too small to resolve.
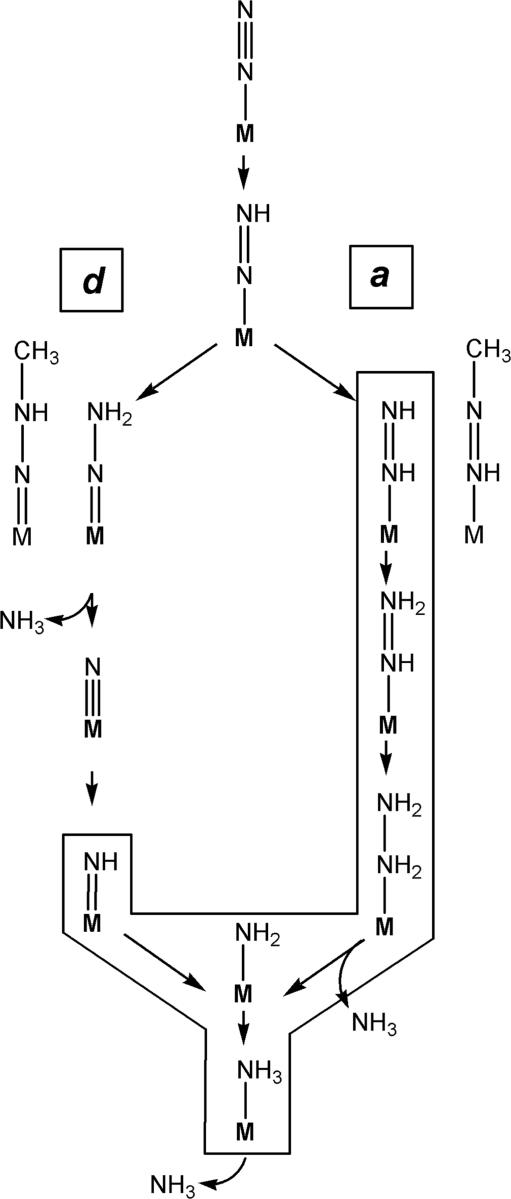
Fig. 5.
Alternating and distal N2 reduction mechanisms at a metal center (M). Essential intermediates are shown for N2 reduction mechanisms with distal proton addition (d-mechanism) (Left) and alternating proton addition (a-mechanism) (Right), bound end-on to a metal center (M). Small straight arrows represent the addition of H+/e− to substrate. Methyldiazene is placed where it might enter into each mechanism. Only the boxed intermediates are consistent with the ENDOR data for the methyldiazene-derived species trapped on the FeMo cofactor.
Discussion
The biochemical, EPR, 14,15N-, 13C-, and 1H-ENDOR studies reported here have established: (i) An S = ½ state denoted m(CH3N NH) is trapped during the reduction of methyldiazene by the α-195Gln-substituted MoFe protein. (ii) m(CH3N
NH) is trapped during the reduction of methyldiazene by the α-195Gln-substituted MoFe protein. (ii) m(CH3N NH) contains a methyldiazene-derived species bound to the FeMo cofactor. (iii) This species binds through an [-NHx] fragment whose N derives from the methyldiazene terminal N. (iv) Interaction with the internal N (and methyl) is weak or absent, suggesting that the internal N does not bind to FeMo cofactor. These results provide powerful constraints on possible mechanisms for reduction of methyldiazene by the α-195Gln MoFe protein and allow steps toward experimental discrimination between the two principal mechanistic views of N2 reduction.
NH) contains a methyldiazene-derived species bound to the FeMo cofactor. (iii) This species binds through an [-NHx] fragment whose N derives from the methyldiazene terminal N. (iv) Interaction with the internal N (and methyl) is weak or absent, suggesting that the internal N does not bind to FeMo cofactor. These results provide powerful constraints on possible mechanisms for reduction of methyldiazene by the α-195Gln MoFe protein and allow steps toward experimental discrimination between the two principal mechanistic views of N2 reduction.
In the Chatt-Schrock mechanism (22) for N2 reduction by Mo complexes, H atoms are sequentially added to the distal N of N2 before N-N bond cleavage. The metal-bound nitride formed by N-N bond cleavage is subsequently reduced to yield the second NH3 (Fig. 5). This mechanism is denoted as the d-mechanism to indicate the exclusive protonation of the distal N before N-N bond cleavage. For the d-mechanism to operate in the reduction of methyldiazene, the [-NHx] fragment observed to be bound to FeMo cofactor could be formed only after reduction by ≥3 electrons and only after N-N bond cleavage. Thus, the appearance of bound [-NHx] is compatible with the d-mechanism only if m(CH3N NH) represents a late stage in the reduction process. In contrast are mechanisms where H atoms are added alternately to the distal and proximal N before N-N cleavage (denoted as the a-mechanism in Fig. 5) (33–37). An [-NHx] fragment would be bound at every stage of methyldiazene reduction by an a-mechanism (Fig. 5). These results for m(CH3
NH) represents a late stage in the reduction process. In contrast are mechanisms where H atoms are added alternately to the distal and proximal N before N-N cleavage (denoted as the a-mechanism in Fig. 5) (33–37). An [-NHx] fragment would be bound at every stage of methyldiazene reduction by an a-mechanism (Fig. 5). These results for m(CH3 N
N NH) do not rule out the d-mechanism, but make the a-mechanism more attractive because the α-195Gln-substituted MoFe protein is specifically compromised in its capacity for delivery of protons to nitrogenase substrates, making it unlikely that [-NHx] could represent a species that is trapped at a late stage of diazene reduction. The findings that N2H4 is both a substrate (24, 25) and minor product (23) during N2 reduction, and that an intermediate can be trapped during N2H4 reduction, lend powerful support to the a-mechanism: at no stage in the d-mechanism does N2H4 appear.
NH) do not rule out the d-mechanism, but make the a-mechanism more attractive because the α-195Gln-substituted MoFe protein is specifically compromised in its capacity for delivery of protons to nitrogenase substrates, making it unlikely that [-NHx] could represent a species that is trapped at a late stage of diazene reduction. The findings that N2H4 is both a substrate (24, 25) and minor product (23) during N2 reduction, and that an intermediate can be trapped during N2H4 reduction, lend powerful support to the a-mechanism: at no stage in the d-mechanism does N2H4 appear.
Additional ENDOR/electron spin-echo envelope modulation studies should disclose more detail about the m(CH3 N
N NH) state and thus tighten the above constraints. In particular, while the a- and d-mechanisms involve diazene isomers when N2 is doubly reduced, they incorporate different bound species at reduction levels three and four: the d-mechanism has bound
NH) state and thus tighten the above constraints. In particular, while the a- and d-mechanisms involve diazene isomers when N2 is doubly reduced, they incorporate different bound species at reduction levels three and four: the d-mechanism has bound  N and
N and  NH, whereas the a-mechanism has bound hydrazide and hydrazine species. We anticipate that ENDOR studies of the intermediates trapped with nitrogenous substrates will be able to distinguish among these species. As another ultimately testable difference between the a- and d-mechanisms, each predicts that the first NH3 is released at different stages of substrate reduction (Fig. 5). Provided that m(CH3N
NH, whereas the a-mechanism has bound hydrazide and hydrazine species. We anticipate that ENDOR studies of the intermediates trapped with nitrogenous substrates will be able to distinguish among these species. As another ultimately testable difference between the a- and d-mechanisms, each predicts that the first NH3 is released at different stages of substrate reduction (Fig. 5). Provided that m(CH3N NH) indeed is a state before N-N bond cleavage, then the present results suggest a fundamental mechanistic difference between reduction of nitrogenous and alkyne substrates. The alkyne substrate-derived intermediates are bound side-on (for two intermediates) (13, 15), whereas the nitrogenous substrate methyldiazene is bound end-on. In any case, mechanistic difference between alkyne and nitrogenous substrates is underscored by the differential effects on substrate reduction elicited by the α-195Gln substitution within the MoFe protein, where alkyne reduction is unperturbed by this amino acid substitution (27).
NH) indeed is a state before N-N bond cleavage, then the present results suggest a fundamental mechanistic difference between reduction of nitrogenous and alkyne substrates. The alkyne substrate-derived intermediates are bound side-on (for two intermediates) (13, 15), whereas the nitrogenous substrate methyldiazene is bound end-on. In any case, mechanistic difference between alkyne and nitrogenous substrates is underscored by the differential effects on substrate reduction elicited by the α-195Gln substitution within the MoFe protein, where alkyne reduction is unperturbed by this amino acid substitution (27).
Materials and Methods
Materials and Proteins.
All reagents were obtained from Sigma-Aldrich and used as supplied unless stated otherwise. 15N-labeled hydroxylamine and 15N-labeled nitromethane were obtained from Cambridge Isotope Laboratories (Andover, MA), and 13C- and 2H-labeled nitromethane were obtained from Sigma-Aldrich. Azotobacter vinelandii strains DJ995 (WT MoFe protein), DJ1310 (α-70Ala MoFe protein), DJ997 (α-195Gln MoFe protein), and DJ1316 (α-195Gln/α-70Ala MoFe protein) were constructed and nitrogenase proteins were expressed as described (25). The MoFe protein from each strain was purified by using a metal affinity chromatography system (38). All proteins used were >95% pure as judged by SDS/PAGE analysis using Coomassie blue staining. Manipulation of proteins was done in septum-sealed serum vials under an argon atmosphere. All transfer of gases and liquids was done with gas-tight syringes.
N-Methylhydroxylamine Synthesis.
The synthesis of isotopically labeled N-methylhydroxylamine (5 in Scheme 1) was accomplished following a modification of a procedure from Ackermann et al. (29) by reduction of nitromethane (4 in Scheme 1) using palladium at room temperature to form the oxalate salt. Briefly, 20 mg of palladium catalyst (5% on barium sulfate) was added to a 50-ml Erlenmeyer flask. To this, 80 mg of oxalic acid and 1 ml of distilled water were added. The flask was then capped and connected with tubing to a second sealed flask (500-ml minimum volume). This second flask was in turn connected with a second piece of tubing to a 500-ml graduated cylinder filled with water. The entire system was flushed with hydrogen gas delivered into the first (50 ml) flask for 5 min, resulting in the catalyst turning black. Finally, 110 μl of 14N- or 15N-, 13C-, or 2H-labeled nitromethane (4 in Scheme 1) was added via a gas-tight syringe to the first flask containing the catalyst. As hydrogen gas was consumed during the reaction, water from the graduated cylinder was drawn into the second flask, maintaining 1 atm (1 atm = 101.3 kPa) of hydrogen in the first flask. The reaction was run ≈12 h (until hydrogen was no longer consumed). The reaction solution was filtered through a 0.2-μm syringe filter to remove any remaining catalyst, and the product N-methylhydroxylamine (5 in Scheme 1) was recrystallized in absolute ethanol, yielding the oxalate salt upon drying under vacuum. This salt could be stored for months at room temperature in the presence of a desiccant.
Hydroxylamine-O-Sulfonic Acid Synthesis.
The synthesis of hydroxylamine-O-sulfonic acid (3 in Scheme 1) was accomplished by a variation of a published protocol (26, 39). 14N- or 15N-labeled hydroxylamine (50 mg; 2 in Scheme 1) was added to a 50-ml Erlenmeyer flask that was then capped with a rubber septum connected to a condenser tube. Methylene chloride (5 ml) was added through the condenser tube, and the reaction was brought to 50°C. Chlorosulfonic acid (100 μl; 1 in Scheme 1) was added to the reaction, and the mixture was allowed to react at 50°C for ≈5 min, resulting in a white flaky crystalline precipitate. The flask was then returned to 25°C, and the methylene chloride solvent was removed with a syringe, leaving the white precipitate. The precipitate was then washed twice with methylene chloride at room temperature (≈2 ml each wash), followed by several washes with diethyl ether at room temperature (≈2 ml each wash) to remove excess acid and water. The resulting white crystals were dried under vacuum for several minutes and used immediately in the synthesis of methyldiazene as described below.
Methyldiazene Synthesis.
Methyldiazene (6 in Scheme 1) was synthesized by using a modification of a method described (40). In a sealed flask (115 ml) under vacuum, 60 mg of hydroxylamine-O-sulfonic acid (3 in Scheme 1) was dissolved in 1.0 ml of water. To this was added a solution (70 mg dissolved in 1.0 ml of water) of N-methylhydroxylamine hydrochloride (Sigma) or the oxalate salt synthesized as described above (5 in Scheme 1) followed by the addition of 1.0 ml of 5 M NaOH. Methyldiazene was evolved as a gas from the solution over the course of several minutes and isolated by freezing in a custom-made gas trap (with a gas sampling port) immersed in liquid nitrogen. The trap containing the frozen methyldiazene was isolated from the rest of the system and quickly warmed to room temperature. Argon was then added to achieve 1 atm pressure. Aliquots of this methyldiazene/argon mixture were withdrawn by syringe and used immediately.
Nitrogenase Activity Assays.
Substrate reduction reactions for nitrogenase proteins were determined as described at pH 7.2 for 10 min at 30°C (41). MoFe protein was added (100 μg) followed by Fe protein (500 μg) to initiate the reaction. Reactions were quenched by the addition of 300 μl of 400 mM EDTA. Hydrogen was quantified by gas chromatography (41). Methyldiazene inhibition of proton reduction was determined as described above except that the pH was adjusted to 7.0.
X-Band EPR Sample Preparation and Analysis.
EPR samples were prepared in a solution containing a MgATP regeneration system as described (25). MoFe protein was added to a final concentration of ≈75 μM (20 mg/ml). Methyldiazene was generated in the same vial containing the protein to maximize the concentration of the substrate. Turnover conditions were initiated by the addition of 50 μM Fe protein. EPR samples under resting conditions were prepared as described above, except that Fe protein was not included. All X-band EPR samples were flash-frozen in liquid nitrogen in 4-mm calibrated quartz EPR tubes. For the pH profile, the buffer was composed of 50 mM Mes, 50 mM 3-[Tris(hydroxymethl)methylamino]-1-propane sulfonic acid, and 50 mM Mops, and the pH was adjusted by the addition of HCl or NaOH. X-band EPR spectra were recorded with a Bruker ESP-300 E spectrometer with an ER 4116 dual-mode X-band cavity equipped with an Oxford Instruments ESR-900 helium flow cryostat. Spectra were obtained at a microwave frequency of ≈9.65 GHz. Initial spectra were obtained at a microwave power of 1.0 mW, a modulation frequency of 1.26 mT, and a temperature of 8 K and were the sum of five scans. The microwave power dependence on EPR signal intensity was determined at 4.7 K with microwave powers ranging from 5 μW to 1 mW. Spin integration of EPR signals was accomplished by comparison to the integrated area of the resting-state FeMo cofactor signal in the MoFe protein.
35-GHz EPR/ENDOR Spectroscopy.
Q-band samples were prepared as described above, except that the MoFe protein concentration was brought to ≈200 μM, and the reactions were initiated with a 100 μM concentration of Fe protein. Continuous wave and Mims pulsed 35-GHz ENDOR spectra were taken as described (15).
Supplementary Material
Acknowledgments
We thank Martin Ackerman for technical advice on the synthesis of methyldiazene and Mike Yurth for assistance with graphics. This work was supported by National Institutes of Health Grants R01-GM59087 (to L.C.S. and D.R.D.) and HL13531 (to B.M.H.), National Science Foundation Grant MCB-0316038 (to B.M.H.), and U.S. Department of Agriculture Postdoctoral Fellowship 2004-35318-14905 (to B.M.B.).
Abbreviations
- ENDOR
electron nuclear double resonance
- N2
dinitrogen
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Smil V. Enriching the Earth: Fritz Haber, Carl Bosch, and the Transformation of World Food Production. Cambridge, MA: MIT Press; 2001. [Google Scholar]
- 2.Rees DC, Howard JB. Curr Opin Chem Biol. 2000;4:559–566. doi: 10.1016/s1367-5931(00)00132-0. [DOI] [PubMed] [Google Scholar]
- 3.Shah VK, Brill WJ. Proc Natl Acad Sci USA. 1977;74:3249–3253. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkes TR, McLean PA, Smith BE. Biochem J. 1984;217:317–321. doi: 10.1042/bj2170317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Rees DC. Science. 1992;257:1677–1682. doi: 10.1126/science.1529354. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Woo D, Rees DC. Biochemistry. 1993;32:7104–7115. doi: 10.1021/bi00079a006. [DOI] [PubMed] [Google Scholar]
- 7.Chan MK, Kim J, Rees DC. Science. 1993;260:792–794. doi: 10.1126/science.8484118. [DOI] [PubMed] [Google Scholar]
- 8.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 9.Mayer SM, Lawson DM, Gormal CA, Roe SM, Smith BE. J Mol Biol. 1999;292:871–891. doi: 10.1006/jmbi.1999.3107. [DOI] [PubMed] [Google Scholar]
- 10.Mayer SM, Gormal CA, Smith BE, Lawson DM. J Biol Chem. 2002;277:35263–35266. doi: 10.1074/jbc.M205888200. [DOI] [PubMed] [Google Scholar]
- 11.Seefeldt LC, Dance I, Dean DR. Biochemistry. 2004;43:1401–1409. doi: 10.1021/bi036038g. [DOI] [PubMed] [Google Scholar]
- 12.Benton PMC, Laryukhin M, Mayer SM, Hoffman BM, Dean DR, Seefeldt LC. Biochemistry. 2003;42:9102–9109. doi: 10.1021/bi034595x. [DOI] [PubMed] [Google Scholar]
- 13.Lee HI, Sørlie M, Christiansen J, Yang TC, Shao J, Dean DR, Hales BJ, Hoffman BM. J Am Chem Soc. 2005;127:15880–15890. doi: 10.1021/ja054078x. [DOI] [PubMed] [Google Scholar]
- 14.Lee HI, Sørlie M, Christiansen J, Song R, Dean DR, Hales BJ, Hoffman BM. J Am Chem Soc. 2000;122:5582–5587. [Google Scholar]
- 15.Lee HI, Igarashi RY, Laryukhin M, Doan PE, Dos Santos PC, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2004;126:9563–9569. doi: 10.1021/ja048714n. [DOI] [PubMed] [Google Scholar]
- 16.Frohnapfel DS, Templeton JL. Coord Chem Rev. 2000;207:199–235. [Google Scholar]
- 17.Igarashi RY, Laryukhin M, Dos Santos PC, Lee HI, Dean DR, Seefeldt LC, Hoffman BM. J Am Chem Soc. 2004;127:6231–6241. doi: 10.1021/ja043596p. [DOI] [PubMed] [Google Scholar]
- 18.Chatt J, Dilworth JR, Richards RL. Chem Rev. 1978;78:589–625. [Google Scholar]
- 19.Pickett CJ. J Biol Inorg Chem. 1996;1:601–606. [Google Scholar]
- 20.Yandulov DV, Schrock RR. Science. 2003;301:76–78. doi: 10.1126/science.1085326. [DOI] [PubMed] [Google Scholar]
- 21.Schrock RR. Chem Commun. 2003:2389–2391. doi: 10.1039/b307784p. [DOI] [PubMed] [Google Scholar]
- 22.Schrock RR. Philos Trans R Soc London A. 2005;363:959–969. doi: 10.1098/rsta.2004.1541. [DOI] [PubMed] [Google Scholar]
- 23.Thorneley RNF, Eady RR, Lowe DJ. Nature. 1978;272:557–558. [Google Scholar]
- 24.Davis LC. Arch Biochem Biophys. 1980;204:270–276. doi: 10.1016/0003-9861(80)90033-8. [DOI] [PubMed] [Google Scholar]
- 25.Barney BM, Laryukhin M, Igarashi RY, Lee HI, Dos Santos PC, Yang TC, Hoffman BM, Dean DR, Seefeldt LC. Biochemistry. 2005;44:8030–8037. doi: 10.1021/bi0504409. [DOI] [PubMed] [Google Scholar]
- 26.Barney BM, Yang TC, Igarashi RY, Dos Santos PC, Laryukhin M, Lee HI, Hoffman BM, Dean DR, Seefeldt LC. J Am Chem Soc. 2005;127:14960–14961. doi: 10.1021/ja0539342. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH, Newton WE, Dean DR. Biochemistry. 1995;34:2798–2808. doi: 10.1021/bi00009a008. [DOI] [PubMed] [Google Scholar]
- 28.Fisher K, Dilworth MJ, Newton WE. Biochemistry. 2000;39:15570–15577. doi: 10.1021/bi0017834. [DOI] [PubMed] [Google Scholar]
- 29.Ackermann MN, Ellenson JL, Robison DH. J Am Chem Soc. 1968;90:7173–7174. [Google Scholar]
- 30.Guth JH, Burris RH. Biochemistry. 1983;22:5111–5122. doi: 10.1021/bi00291a010. [DOI] [PubMed] [Google Scholar]
- 31.Orme-Johnson WH, Hamilton WD, Jones TL, Tso MYW, Burris RH, Shah VK, Brill WJ. Proc Natl Acad Sci USA. 1972;69:3142–3145. doi: 10.1073/pnas.69.11.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryle MJ, Lee HI, Seefeldt LC, Hoffman BM. Biochemistry. 2000;39:1114–1119. doi: 10.1021/bi9919100. [DOI] [PubMed] [Google Scholar]
- 33.Hinnemann B, Nørskov JK. J Am Chem Soc. 2004;126:3920–3927. doi: 10.1021/ja037792s. [DOI] [PubMed] [Google Scholar]
- 34.Rod TH, Hammer B, Nørskov JK. Phys Rev Lett. 1999;82:4054–4057. [Google Scholar]
- 35.Stavrev KK, Zerner MC. Int J Quantum Chem. 1998;70:1159–1168. [Google Scholar]
- 36.Dance I. Chem Commun. 1997:165–166. [Google Scholar]
- 37.Siegbahn PEM, Westerberg J, Svensson M, Crabtree RH. J Phys Chem B. 1998;102:1615–1623. [Google Scholar]
- 38.Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. Biochemistry. 1998;37:12611–12623. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- 39.Rathke MW, Millard AA. Org Synth. 1988;50:943–946. [Google Scholar]
- 40.Ackermann MN, Hallmark MR, Hammond SK, Roe AN. Inorg Chem. 1972;11:3076–3082. [Google Scholar]
- 41.Barney BM, Igarashi RY, Dos Santos PC, Dean DR, Seefeldt LC. J Biol Chem. 2004;279:53621–53624. doi: 10.1074/jbc.M410247200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.