Abstract
Phage display technology involves the display of proteins or peptides, as coat protein fusions, on the surface of a phage or phagemid particles. Using standard technology, helper phage are essential for the replication and assembly of phagemid particles, during library production and biopanning. We have eliminated the need to add helper phage by using 'bacterial packaging cell lines' that provide the same functions. These cell lines contain M13-based helper plasmids that express phage packaging proteins which assemble phagemid particles as efficiently as helper phage, but without helper phage contamination. This results in genetically pure phagemid particle preparations. Furthermore, by using constructs differing in the form of gene 3 that they contain, we have shown that the display, from a single library, can be modulated between monovalent (phagemid-like) and multivalent display (phage-like) without any further engineering. These packaging cells eliminate the use of helper phage from phagemid-based selection protocols; reducing the amount of technical preparation, facilitating automation, optimizing selections by matching display levels to diversity, and effectively using the packaged phagemid particles as means to transfer genetic information at an efficiency approaching 100%.
INTRODUCTION
Phage display is a widely used method to select antibody fragments (1–9), peptides (10–14) and other scaffolds (15–20) from large libraries, as well as to increase the affinity of antibodies for their antigens (21–24) and other proteins for their receptors (25). Polypeptides are displayed as phage coat protein fusions and the corresponding gene is contained within the particle. It is usually mediated by the fusion of the displayed polypeptide to a coat protein, and encapsulating the gene encoding the fusion protein within the phage particle. This coupling of phenotype and genotype ensures that selection of the displayed protein simultaneously selects the encoding gene, allowing further selection rounds and the eventual isolation of a population highly enriched in phage displaying polypeptides of interest. Vectors based on filamentous Ff phage are the most popular, and of the five coat proteins, the gene 3 protein (g3p) is most commonly used for display.
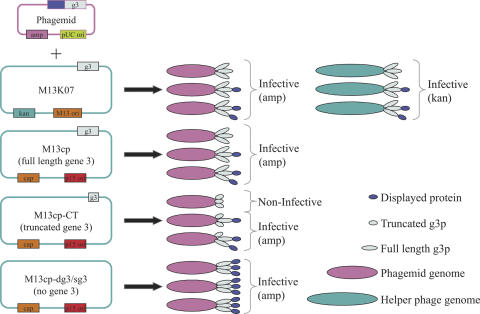
There are two broad categories of vectors used for phage display: phage and phagemid. When proteins are displayed using phage vectors, which are not of the 33 or 88 kind, the bacteria produce phage particles that all display recombinant protein. The gene encoding the recombinant displayed protein is included in the phage genome and as a result the phage population produced by a single clone is phenotypically and genotypically homogenous, excluding the effects of proteolysis. In contrast, phagemid make recombinant displayed protein, but require the additional proteins provided by helper phage to create phage particles that display recombinant protein. Helper phage are essential for phagemid systems as they supply all the other proteins required to make functional phage. Helper phage (26,27) are normal Ff phages with a number of modifications: they contain an additional origin of replication, they usually carry antibiotic resistance genes and their packaging signal is severely disabled.
When a bacterium is infected with helper phage, the disabled packaging signal does not prevent the production of phage particles. However, when a bacterium is infected with both phagemid and helper phage, the phagemid DNA (containing an optimal packaging signal) is packaged in preference. As a result, phagemid preparations are both phenotypically and genotypically heterogeneous (Figure 1): the display protein may be either wild type (derived from the helper phage) or recombinant (derived from the phagemid), and the packaged genome may be either phage or phagemid. In theory, the disabled packaging signal should significantly reduce the number of helper phage particles in any phagemid preparation. However, the number of helper phage can sometimes equal, or exceed, the number of phagemid particles, which can significantly compromise subsequent selections.
Figure 1.
The expected phage particle genotypes and phenotypes when using standard phagemid and helper phage (M13K07) as well as the four helper plasmid constructs described here (M13cp, M13cp-CT, M13cp-dg3 and M13cp-sg3).
The different antibiotic resistance genes carried by the helper phage and phagemid genomes allows the selection of bacteria that contain both, resulting in the recovery of functional phagemid particles displaying the recombinant protein of interest.
Practically, phage and phagemid libraries have a number of differences. At the DNA level (preparing DNA, cloning, transfection efficiency) it is easier to work with phagemids than phage. As a result, phagemid libraries can be made far larger than phage libraries. It is also easier to produce soluble proteins using phagemids by the insertion of an amber stop codon between the displayed protein and g3p (28). Although soluble protein could theoretically be made in phage libraries using a similar genetic arrangement, the low copy number of the vector and the weakness of the g3p promoter and ribosome-binding site, results in levels of soluble protein that are too low for most practical purposes, requiring subsequent recloning into expression vectors (29). Another advantage of phagemids concerns the relative resistance to deletions of extraneous genetic material. Filamentous phage vectors, in general, have a tendency to delete unnecessary DNA, due to the selective growth advantage that a smaller phage genome has over a larger one. Phagemids suffer far less from such deletions and as a result are more genetically stable.
Phage libraries have considerable operational advantages. First, they do not require the use of helper phage for phage production. As a result, the additional technical procedures associated with helper phage infection, such as monitoring the absorbance of bacterial cultures, are omitted from protocols. To amplify phage libraries it is sufficient to grow bacteria containing phage genomes and phage particles are produced. This makes phage far easier to use in selections, particularly in high-throughput selections where antibodies are selected against up to 96 targets simultaneously (30,31).
Second, each phage particle in a phage library displays up to five copies of the displayed protein (using a g3p display system), whereas only 1–10% of phage particles in a phagemid library display a single copy of the displayed protein (32). As a result, a greater number of binders in a library are recovered, and therefore antibodies tend to be more diverse. However, this is counterbalanced by a lower average affinity (29): phagemid display, by virtue of the display of single proteins, results in the selection of fewer unique binders, which tend to have higher affinities (29). For similar reasons, affinity maturation (21–23) can only be carried out using phagemid vectors.
Recently, a number of groups (27,33–38) have developed alternative helper phage systems, compared in Table 1, designed to improve display in either of two ways: by increasing the display level, so that it resembles the display obtained with phage vectors (27,34–36), or by reducing the background from non-displaying phage by rendering them non-infective (37,38). However, none of these has been addressed the need for helper phage itself.
Table 1.
Different published helper phages
| Helper phage name | Mutation/mechanism | aHelper phage titers/ml | Display levels | a,bRescued phagemid titers/ml | Use in display | Helper phage propagation | Reference |
|---|---|---|---|---|---|---|---|
| M13K07 | 1011c | Low | 2 × 1010−12 | Standard infection | Growth | (67) | |
| KM13 | Trypsin site in g3p, elution with trypsin | 1011 | Low | 2 × 1010−12 | Standard infection | Growth | (38) |
| g3 deletion | 105−6 | High | ∼1010 | Standard infection | g3 plasmid expression under lac promoter | (68) | |
| M13MDD3.2 | g3 deletion | 2 × 109 | NT | 109 | Standard infection | g3p plasmid expression | (33) |
| R408d3 | g3 deletiond | 1010 | NT | NT | Standard infection | g3p plasmid expression under pspA promoter | (27) |
| Hyperphage | g3 deletion (8-406) | 109 | High | 109−10 | Standard infection | g3p integrated into E.coli genome | (35) |
| CT helper phage | g3 N1 & N2 domains deletede | 3 × 1011 | Low | 1011 total 5 × 108 infective | Standard infection | g3p plasmid expression | (37) |
| Ex-phage | Amber stop codon 5′ g3 | 1012−13f | High | 1010−11 | Non-suppressor strain | Suppressor strain | (36) |
| Phaberge | Amber stop codon 3′ g3 | 1011 | High | 109−10 | Non-suppressor strain | Suppressor strain | (34) |
The mechanisms of action, as well as the reported titers of both packaged phage and helper phage are given. NT-not tested.
aTiters are unconcentrated supernatant titers—i.e. not PEG precipitated.
bRefers to the number of infectious phagemid particles, independent of whether they carry antibody or no.
cData from our laboratory.
dSome reversion due to packaging of plasmid expressing p3 observed-probably also occurs in other plasmid expression systems, but not examined.
eRescued phagemid are two populations, displaying and infective, non-displaying and non-infective.
fStandard M13K07 titers obtained in this laboratory tend to be 10–100-fold higher than obtained elsewhere.
Initial experiments to increase display involved the creation of g3p deleted helper phage, packaged in bacterial strains expressing gene 3 in trans. These allowed higher display levels, but suffered from the problem that when the g3p was derived from plasmids, although not when integrated into the Eschericia coli chromosome (35), such plasmids could also be packaged at low levels (27). Helper phage titers also tend to be very low. More recently, conditional g3p deletions have been created by the introduction of suppressible stop codons in g3 (34,36), allowing production of helper phage in suppressor strains, and the packaging of phagemids in non-suppressor strains, where the helper phage is unable to make its own g3p.
Two approaches have been used to reduce background, in both of which only phagemid particles containing the recombinant g3p are infectious. These involve either the deletion of the portions of g3p required for infection (37), or the incorporation of a trypsin site within the g3p of the helper phage, and using trypsin-treated phage for infection (38). Both of these systems result in a lower background during selection by eliminating those phagemid particles which contain only helper phage derived g3p.
Although these systems may overcome some of the disadvantages of helper phage, they do not avoid one of the main problems associated with the use of helper phage: the need to make helper phage and add it to growing bacterial cultures at relatively restricted phases of the growth cycle. In this paper we describe a series of constructs which eliminate the need for helper phage altogether, creating a system in which bacterial packaging cell lines replace the use of helper phage, making the generation of pure phagemid particles as straightforward as using a phage-based system. Furthermore, by using different packaging bacteria the phagemid particles produced are either monovalent or multivalent. The concept is illustrated in Figure 1.
MATERIALS AND METHODS
Cloning
M13c was created by amplifying M13mp19 with two outward facing primers, M13 MluI (5′-ttgatgacgcgtcctattggttaaaaaatgagctg) and M13 MluI (3′-ttgatgacgcgtccgaaatcggcaaaatcc) using a high-fidelity proof reading polymerase which amplified the whole plasmid and put MluI sites at the junctions between M13 and lac Z. This large PCR product was digested with MluI and the chloramphenicol resistance gene from pBSL121 (39) cloned in after cutting with MluI. This produced an M13 phage which confers chloramphenicol resistance. M13cp was created by amplifying M13c with the two outwards primers, gene 4 3′ (ccacacctgcagcgcttaatgcgccgctacagggcgcgtact) and CATgene 5′ (tgatttctgcagacgcgtgtccgaatttctgccattcatcc). These primers amplify the whole plasmid without the M13 origin and place PstI sites at the ends. The p15a origin was amplified from pMPM-K3 (39) with 5′ P15 ori (taacgctgcagagaacatggcttcatgtgg) and 3′ P15 ori (actgttctgcagagcagacagttttattgttc). This yielded an 875 bp fragment containing the P15 origin of replication flanked by PstI sites which could be cloned into the large M13c PCR fragment using PstI. M13cp-CT was created by amplifying M13cp with two outward primers: g3sig 3′ (acaactttcggatccttcagcggagtgagaatag) and g3D3 5′ (ggtggctctggatccggtgattttgattatgaaaag). The resultant fragment was cleaved with BamHI and religated. These primers are located within g3, and after amplification, the leader of g3 is joined directly to the C-terminal 151 amino acids of g3p, which comprise the C-terminal domain. M13cp-dg3 was made similarly, using gene 6 5′ (ccatatgaattctctattgattgtgacaaaataaacttattcc) and gene 8 3′ (gaaaggaacaactaaaggaattccgaataataattttttcac) to amplify M13cp. These amplify the whole plasmid without gene 3, and can be ligated using EcoRI. However, the PCR product does include the terminator (T0.25) found at the end of gene 8 and the C-terminal portion of gene 3, which is thought to contain the p6 promoter. M13cp-sg3 was also made by amplifying the whole of M13cp using g3 stop BclIS (gaaagt tga tca gca taa ccc cat aca tga aat tca ttt act aac gtc) and g3 stop BclAS (ttt tgc tga tca act ttc aac agt tca agc gga gtg aga ata g), cutting with BclI and religating. This inserted four stop codons (underlined in the primers) in the first 32 codons of g3. The junctions of all constructs were confirmed by sequencing.
Phagemid production
Transformation Single colonies were picked from each helper cell construct (M13cp, M13cp-CT, M13cp-dg3 and M13cp-sg3), made chemically competent and transformed with pDan5-scFv DNA. After growth on 2XTY-ampicillin-chloramphenicol agar plates, colonies from each transformation were picked and grown overnight in 2XTY-ampicillin-chloramphenicol media at 30°C at 250 r.p.m.
Infection and direct growth Bacteria infected as described above at an absorbance of OD600 of 0.5, were grown overnight at 30°C, 250 r.p.m., without retention on ice. DH5αF cells containing no helper plasmid were also infected with M13K07 helper phage for an additional 30 min at 37°C without shaking before dilution and overnight growth.
Infection and growth from single colony Single bacterial colonies obtained following a procedure similar to that described for titration above were picked and grown overnight in 2XTY-ampicillin-chloramphenicol at 30°C, 250 r.p.m.
For each phage production protocol, after overnight growth phagemids were collected from the supernatant by centrifugation (4000 r.p.m., 30 min), and the levels of phage production determined by titration in DH5αF cells plated on both 2XTY-ampicillin and 2XTY-chloramphenicol plates.
Determination of infectability
A single colony starter culture from DH5αF alone, or containing one of the four M13 helper plasmid constructs, was grown overnight at 37°C, 250 r.p.m., in 2XTY-chloramphenicol in the case of the helper plasmids and 2XTY alone for DH5αFT. The following day each culture was diluted and re-grown at 37°C, 250 r.p.m., to an absorbance OD600 of 0.5. As culture growth rates were variable, each culture was stored on ice after reaching OD600 0.5, and for a further 30 min after all cultures had reached this absorbance. The samples were then returned to the 37°C incubator to warm for 30 min. Each sample was infected with D1.3 phagemid made from the M13cp transformation (and so genetically homogenous) at a multiplicity of infection of 1:1 and left to infect for 30 min at 37°C without shaking. Aliquots of infected bacteria were removed from each sample and titered to determine the ability of the bacteria to support phage infection.
ELISA
Phage ELISAs were carried out essentially as described previously (40), with phage either used directly after growth (Figure 4), or purified by PEG precipitation, titered and then diluted to appropriate titers (Figure 5). Antigens were adsorbed to Nunc immunosorb microtiter plates (Nunc) overnight at 4°C in PBS at 1–10 μg/ml and blocked with 1% BSA/PBS. Phage were added in blocking buffer, incubated for 60–90 min, washed in PBS and PBS-T (PBS/0.1% Tween-20), incubated with horseradish peroxidase labeled anti-M13 phage monoclonal antibody (Pharmacia) for 60 min diluted in blocking buffer, washed in PBS and PBS-T, followed by treatment with 1-Step Turbo TMB-ELISA substrate (Pierce). The reaction was terminated with 1 N sulfuric acid, and read using absorbance at 405 nm.
Western blotting
PEG precipitated phage particles, corresponding to 500 μl of culture supernatant, were reduced with sample buffer [7.5% β-mercaptoethanol, 75 mM Tris (pH 6.8), 2% SDS, 15% glycerol and 0.002% Bromophenol Blue] and heat-treated at 100°C for 10 min. Phage proteins were resolved by SDS–PAGE (NuPAGE™ 4–12% Bis–Tris; Invitrogen), in MES buffer, against a protein standard. Separated proteins were then transferred to 0.2 μM nitrocellulose membrane (Protran BA83; Schleicher & Schuell Bioscience #10401396) using the XCell II™ Blot Module (Invitrogen), according to the manufacturer's instructions. The membrane was blocked (1% Skim Milk Powder + 1% BSA in PBS) overnight at 4°C and subsequently probed with either Anti-SV5 mouse monoclonal or Anti-M13pIII mouse monoclonal antibody (1/1000 dilution; NEB #E8033S). The membrane was washed (2 × 5 min) with PBS-T before the addition of secondary Anti-Mouse IgG-Alkaline Phosphatase (AP) conjugate (1/1000; Dakocytomation #D0486). Following incubation, the membrane was washed once with PBS-T, PBS-LT (0.01% Tween-20) and PBS. Displayed proteins were detected following treatment with AP substrate (NBT/BCIP; Pierce #34042).
RESULTS
Plasmid design
In initial experiments we attempted to integrate portions of M13 into the E.coli genome by Tn5 transposition (41). However, although we could obtain clones that contained all the M13 protein-coding regions none was able to package phagemid DNA into phagemid particles. As an alternative, we turned to the use of plasmids (see Materials and Methods for construction details). These 'helper plasmids' were all based on M13mp19 (26). In the starting construct, the M13 origin was removed and replaced with the p15a origin, which provides ∼15 copies per cell and allows it to co-exist with ColE1-based origins (42), including pUC derivatives such as our phage display vector, pDAN5 (3). Next, the polylinker and lac gene of M13mp19, located in the M13 intergenic region and known to be non-disruptive to M13 function (43), were replaced by the chloramphenicol resistance gene (39) to create M13cp. M13cp, which contains full-length g3p, was further modified to create three derivative plasmids with changes made only to g3p. The first modification created a truncated g3p construct (M13cp-CT) which removed the N-terminal two domains of g3p, leaving only the C-terminal phage assembly domain connected to the leader sequence. The following two constructs inactivated g3p, either by complete deletion (M13cp-dg3) or by the insertion of four non-suppressible stop codons within the first 32 codons of g3 (M13cp-sg3). The latter strategy was similar to that used to make Ex-phage (36) and Phaberge (34). The expected phenotype of the phagemid particles produced by these different helper plasmids is illustrated in Figure 1.
Phagemid particle production
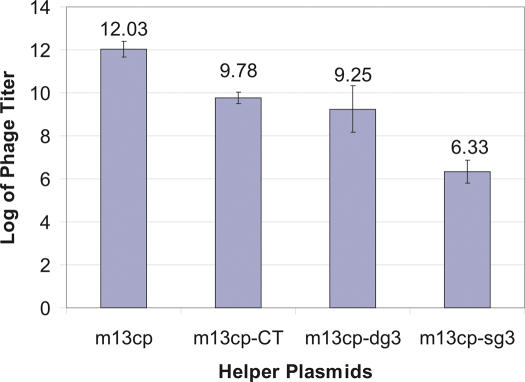
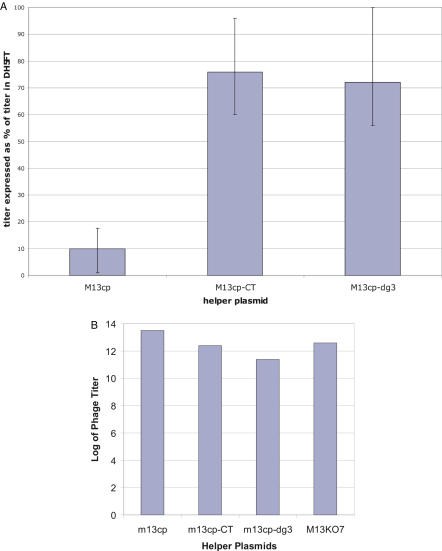
A first experiment was carried out to determine whether these constructs were capable of transforming bacteria into filamentous phage packaging cell lines. DH5αF bacteria containing each of the helper plasmids were made competent and transformed with our standard phagemid display vector pDAN5-D1.3 (3). Each transformation was plated on agar media containing ampicillin and chloramphenicol. Following overnight growth single colonies were picked and further grown in liquid media containing ampicillin and chloramphenicol. Figure 2 shows that helper plasmids were able to supply all the necessary phage proteins to produce phagemid particles carrying ampicillin resistance. M13cp gave ampicillin titers of 1011−12 ml−1, M13cp-CT and M13cp-dg3 gave titers ∼50–100-fold less, whereas M13cp-sg3 gave titers of only 106 phagemid particles/ml. Given the very low titers obtained with M13cp-sg3, no further work regarding this construct is discussed. Phagemid particles produced in this experiment were also tested for their resistance to kanamycin (carried by standard helper phage, M13K07) and chloramphenicol (carried by the helper plasmids). We were unable to obtain colonies with resistance to either, indicating that the helper plasmids were unable to package themselves, and that the packaged phagemid particles were genetically homogenous. Furthermore, the titers obtained with M13cp were equal to those produced by standard helper phage (M13K07) infection (Table 2). The next set of experiments served two purposes. First, and most important, was to determine whether the presence of helper plasmids within bacteria reduced their ability to be infected (plating efficiency). This is important if selection outputs are to be infected directly into such cells. The second, was to determine whether after infection and plating bacteria containing helper plasmids could produce phagemid particles at levels similar to those derived from direct transformation. Genetically homogenous phagemid particles prepared using the M13cp helper plasmid were infected into DH5αF bacteria, or DH5αF bacteria containing each of the three helper plasmids (M13cp, M13cp-CT and M13cp-dg3). The results in Figure 3A, expressed as a percentage of the plating efficiency in DH5αF, show that although phagemid particles could infect DH5αF containing M13cp-CT or M13cp-dg3 at levels comparable to unmodified DH5αF under these experimental conditions, their ability to infect bacteria containing M13cp was reduced by at least 10-fold. The reduced plating efficiency, when M13cp is present, relative to DH5αF alone, could not be overcome by increasing the number of bacteria available for infection, modifying antibiotic concentrations, changing the OD at which infection occurred, or the temperature at which bacteria were grown after infection (data not shown). This suggests that the problem is not a reduction in the number of bacteria displaying pili and so competent for infection, but a failure of propagation within the bacteria after interaction with the pilus has occurred.
Figure 2.
Titer of pDAN5-D1.3 phagemid produced by different packaging bacteria after transformation. Bacteria containing the different helper plasmids were transfected with pDAN5-D1.3 phagemid DNA, plated on ampicillin chloramphenicol plates, single colonies picked and grown overnight. The titers for each packaging cell line is shown in figures above the error bar.
Table 2.
The properties, and potential uses, of the different helper plasmids created here are indicated
| Helper plasmid | Form g3p | Phage production titer | Infectability of bacteria bearing plasmid | Notes | Potential use |
|---|---|---|---|---|---|
| M13cp | Full length | Equal M13K07 | 10% DH5αFT | Most similar to phage produced using standard helper phage | Transfer genetic material. Monovalent phage display. |
| M13cp-CT | Truncated | 5−20% M13K07 | Equal DH5αFT | Behaves more like g3p deletion. Only phagemid with recombinant g3p are infectious | Multivalent phage display |
| M13cp-dg3 | Absent | 0.1−10% M13K07 | Equal DH5αFT | All phage produced have recombinant g3p | Multivalent phage display |
Figure 3.
(A) DH5αFT bacteria containing each of the helper plasmids were infected with a genetically homogenous preparation of D1.3 phagemid (previously prepared using M13cp) and the titers obtained are expressed as the mean percentage of the titers obtained using DH5αFT. Error bars indicate the range of results obtained in three independent experiments. (B) Titer of pDAN5-D1.3 phagemid produced by different packaging bacteria after infection. Genetically homogenous pDAN5-D1.3 phagemid particles (previously prepared using M13cp) were infected into bacteria and plated onto ampicillin chloramphenicol plates. A single colony was picked, allowed to grow overnight and the titers determined. The results shown here are from a single experiment.
Bacterial colonies obtained after infection and plating were further grown in liquid media to determine the titer of the phagemid particles produced. Figure 3B shows that the relative order of phagemid particle production was consistent with the results obtained by transformation (M13cp>M13cp-CT>M13cp-dg3), although the phagemid particle titers produced by cells containing the helper plasmids were 10-fold higher.
Assessing display levels
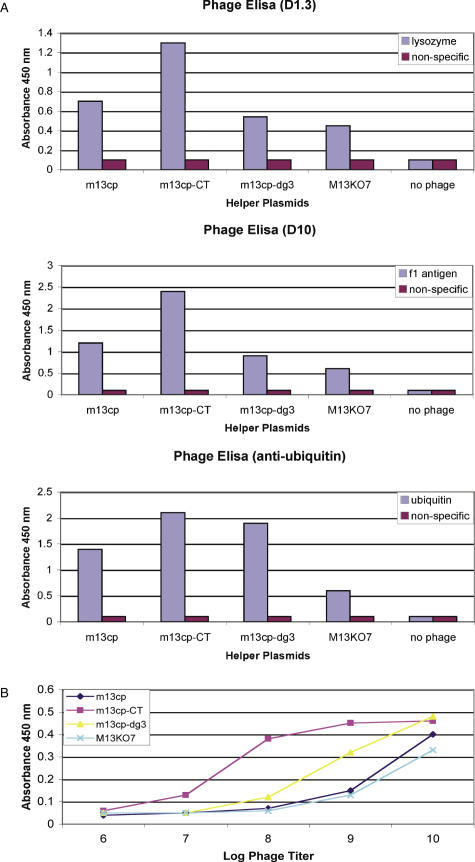
The final set of experiments were conducted to determine the display levels of phagemid particles produced by each helper plasmid, using phage enzyme-linked immunosorbent assay (ELISA) and western blot. The most stringent assessment of functional display is to carry out phage ELISAs, since only well displayed and correctly folded scFvs will give positive signals. In order to provide practical comparisons, phage ELISAs were carried out for three different scFvs using growth supernatants directly after growth in the three different packaging cells, without purification or concentration by PEG precipitation. This mimics the true experimental use of such a system in selection and screening. Figure 4A shows that M13cp-CT provides the highest ELISA signal in all cases. Although there was some variation, M13K07, M13cp and M13cp-dg3 all gave approximately similar signals.
Figure 4.
(A) Phage ELISA signals obtained with phagemid particles from three different scFvs (D1.3, recognizing lysozyme; anti-ubiquitin; and F10, recognizing the Y. pestis f1 antigen) prepared using each of the different helper plasmids or M13K07. M13K07 alone indicates the use of helper phage without rescued phagemid, as a negative control. An anti-M13 phage monoclonal labeled with horseradish peroxidase (Pharmacia) was used as the secondary antibody. The results shown are representative of typical experiments which have been repeated on separate occasions at least three times. (B) Correlation of phage ELISA signals with phagemid particle numbers. D1.3 phagemid particles, recognizing lysozyme, were prepared using each of the different packaging cells, as well as with M13K07 under standard protocols. After the titers of the different phagemid particles were determined, samples of equal titer were prepared and ELISA signals determined for a dilution series as shown in the figure.
In order to determine the relative display levels, pDAN5-D1.3 phagemid particles prepared using the different helper plasmids were also assessed in terms of the ELISA signals obtained at identical phage titers. The results in Figure 4B show that M13cp-CT produces by far the most functionally active phage at the lowest phage titer, followed by M13cp-dg3. To obtain ELISA signals comparable to M13cp-CT, ∼100-fold more M13K07-derived phagemid particles are required.
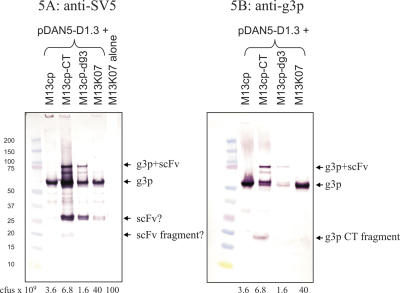
D1.3 phagemid particles were further examined by two western blots: the first using an anti-g3p antibody and the second using the SV5 anti-tag antibody (44). In these experiments phagemid were not normalized for titer, but culture volume, giving an indication of how much recombinant protein will be displayed under standard use, with the actual numbers of phagemids loaded indicated below (Figure 5). The SV5 antibody recognizes the tag placed between the displayed protein and g3p and will only recognize recombinant g3p from the display vector. This gives an assessment of the total amount of incorporated recombinant g3p (equivalent to the sum of the g3p+scFv and g3p bands derived from the proteolytic degradation of g3p+scFv) as well as that portion which is full length and displaying scFv (the g3p+scFv band). The results (Figure 5) show again that M13cp-CT gives the highest display level, as shown by the intensity of the g3p+scFv band, even though the number of phagemid particles loaded is 5-fold less than those produced using M13K07. Consistent with the ELISA results, M13cp-dg3 also gave good display, and full-length display was barely detectable with either M13cp or M13K07 packaged phagemid particles. The second western blot was carried out using an anti-g3p monoclonal (New England Biolabs) which recognizes the C-terminal portion of g3p. This will reveal the presence of all g3p, whether derived from phagemid or helper plasmid, including the truncated g3p (as a band of 19 kDa). This western shows that a large fraction of the g3p incorporated using M13cp-CT is derived from the recombinant phagemid g3p (compare the intensity of the g3p-CT fragment—derived from the helper plasmid—with that of the g3p and g3p+scFv bands). A slightly larger band was occasionally seen with M13cp-CT and M13cp-dg3 prepared phagemid (Figure 5A). Given the exquisite specificity of this antibody (the helper phage alone shows no bands), we believe this to be an scFv fragment containing an SV5 epitope. Although the amount of total recombinant phagemid g3p incorporated in M13cp-dg3, M13cp and M13K07 appears to be similar, functional display (g3p+scFv band) is only seen for M13cp-dg3. This suggests that proteolysis is greater in phagemid prepared using M13cp and M13K07.
Figure 5.
Western blots of pDAN5-D1.3 phagemid packaged using M13K07 and the different helper plasmids. (A) The blot was probed with SV5 which recognizes the tag between g3p and D1.3. The lane showing M13K07 alone was taken from a different position in the same gel. (B) An anti-g3p monoclonal (New England Biolabs) which recognizes a linear epitope in the C-terminal domain of g3p, was used.
DISCUSSION
In this paper we describe a novel method to eliminate the use of helper phage from phagemid preparation, allowing the production of phagemid by simple bacterial growth. This has been carried out by the creation of bacterial packaging cell lines, containing helper plasmids, that are able to provide all the proteins required for phagemid packaging.
These helper plasmids are based on M13mp19, with the phage packaging signal/origin replaced by p15a, and the addition of a chloramphenicol resistance gene. The absence of packaging signals in these helper plasmids prevents their DNA from being packaged. This provides a number of significant advantages over the use of both standard (26,27) and modified helper phages (27,33–37). Perhaps most important is the purity of the phagemids produced: we have been unable to detect contamination of phagemids prepared using these helper plasmids with any helper phage genomes whatsoever. For phage display, this avoids the occasional problem of helper phage overgrowth, which can result in failed selections. For other applications, the genetic purity of the phagemid particles, combined with their extremely high titers, makes this a powerful method to transfer genetic material between bacteria. This is likely to be particularly useful in the generation of antibody diversity by recombination (3,45), and in genetic selection protocols carried out in bacteria (46–50), in which it will be possible to replace library transfection by infection. As infection is far more efficient than transfection, this will be especially applicable to library-versus-library (45,51) selection protocols, where increased efficiency of entry by infection will result in higher sampling of library diversity (52).
Although the helper plasmids were designed to lack the M13 packaging signal, the complete absence of packaging into phage particles is at first somewhat surprising, considering that many other plasmids, without M13 origins, do show low levels of packaging (27). This is usually attributed to the presence of cryptic packaging signals in plasmids which can be inefficiently recognized by the M13 packaging machinery. It is likely that evolution has selected against the presence of other putative packaging sequences in the M13 genome to ensure that only the correct single site is used, so guaranteeing correct orientation within the phage particle (53). As a result elimination provides no alternative signals in the phage, and it is clear that neither the p15a origin nor the added chloramphenicol resistance gene are able to provide alternative signals.
The helper plasmids differ in the form of g3 they contain; the g3p in M13cp is full length, that in M13cp-CT is truncated. M13cp-CT contains the portion of g3p responsible for phage assembly and release (54,55), but lacks the domains involved in bacterial toxicity (56,57), phage infection (58–60) and the inhibition of infection by bacteria carrying g3p (27,61,62). M13cp-dg3 contains no g3, by virtue of genetic deletion, and gave lower titers than M13cp or M13cp-CT, but ELISA and western signals intermediate between the two.
When phagemid are infected into bacteria containing M13cp the plating efficiencies obtained are consistently lower than those obtained in bacteria containing the other helper plasmids, or no plasmids at all. As this cannot be overcome by increa-sing the number of bacteria, the problem is not a reduction in infectivity (e.g. by a reduction in the number of bacteria displaying pili), as might be expected, but a failure of these phagemids to establish themselves within the bacteria (e.g. an inability to replicate). This could occur at a number of different levels, including sequestration of the TolA co-receptor by the helper plasmid g3p (58,60), preventing productive phagemid particle entry into the bacteria, or a failure of phagemid DNA replication or survival after entry into the cytoplasm.
In phagemid particles packaged using M13cp most of the g3p appears to be derived from the helper plasmid, and very little from the display vector. As a result, low levels of monovalent display occur (Figure 5). However, in the case of phagemid particles made using M13cp-CT, a large proportion of the incorporated g3p in phagemid is derived from the display vector (compare the intensity of the g3p-CT fragment in Figure 5B with that of the g3p and g3p+scFv bands), and of this almost 50% is full length (compare g3p+scFv with g3p in the anti-g3p blot), leading to the very high multivalent display levels seen. This is also reflected in the ELISA results shown in Figure 4B: in order to mimic the signal obtained with phagemid particles produced using M13cp-CT, 100-fold more phagmid particles produced using M13K07 or M13cp are required. This suggests that there is a preference for the full-length g3p provided by the display vector over the truncated form provided by the helper plasmid during phage assembly. Although the C-terminal domain is known to be sufficient to allow phage assembly to occur (63), this result suggests that additional portions of g3p facilitate the process, leading to increased incorporation levels when full-length g3p is used.
M13cp and M13cp-CT stand out as being the most useful helper plasmids, each with potentially different applications. For phage display, it is likely that a selection approach combining both, in which phagemid packaged by M13cp-CT, providing multivalent display able to capture full diversity, is used in early rounds of selection, and M13cp, packaging monovalent phagemids, is used in later rounds to select higher affinity clones, will be the most useful. Given the lower plating efficiency of M13cp, it should only be used once selected phagemids are well represented (e.g. after the second round), in order to avoid loss of diversity.
M13cp could also be very useful for transferring genetic material between bacteria when maximum diversity must be maintained (3,45), for making phagemid particles for other purposes, and also in phage display when display proteins other than p3 are used. Surprisingly, M13cp-dg3 does not appear to have any advantages over M13cp-CT, giving lower titers, and lower ELISA signals at similar titers, and at the moment it is difficult to find a use for this construct.
At a practical level, these helper plasmids are easier to use than helper phage-based systems. Bacteria containing the helper plasmid can be either infected or transformed with phagemid and then grown up in selective media without the need to further monitor bacterial OD. In fact, we have found that it is sufficient to add phagemid particles to a freshly diluted overnight culture of M13-cap-p15-CT bacteria and grow: bacteria become infected and phage production follows. Similarly, bacterial colonies can be scraped after overnight growth, and phagemid particles can be harvested from the supernatant after centrifugation. Although these helper plasmids will simplify the use of phage display under standard selection conditions, it is in high-throughput antibody selection projects (31,64–66), where minimal oversight is desired, and it is impractical to closely monitor bacterial OD in order to add helper phage, that they will prove particularly useful.
Acknowledgments
This work was funded by a DOE GTL grant awarded to A.B. Funding to pay the Open Access publication charges for this article was provided by the DOE GTL grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vaughan T.J., Williams A.J., Pritchard K., Osbourn J.K., Pope A.R., Earnshaw J.C., McCafferty J., Hodits R.A., Wilton J., Johnson K.S. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 2.Marks J.D., Hoogenboom H.R., Bonnert T.P., McCafferty J., Griffiths A.D., Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 3.Sblattero D., Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 4.de Haard H.J., van Neer N., Reurs A., Hufton S.E., Roovers R.C., Henderikx P., de Bruine A.P., Arends J.W., Hoogenboom H.R. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 5.Sheets M.D., Amersdorfer P., Finnern R., Sargent P., Lindqvist E., Schier R., Hemmingsen G., Wong C., Gerhart J.C., Marks J.D. Efficient construction of a large nonimmune phage antibody library; the production of panels of high affinity human single-chain antibodies to protein antigens. Proc. Natl Acad. Sci. USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nissim A., Hoogenboom H.R., Tomlinson I.M., Flynn G., Midgley C., Lane D., Winter G. Antibody fragments from a 'single pot' phage display library as immunochemical reagents. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths A.D., Williams S.C., Hartley O., Tomlinson I.M., Waterhouse P., Crosby W.L., Kontermann R.E., Jones P.T., Low N.M., Alison T.J., et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Kruif J., Boel E., Logtenberg T. Selection and application of human single chain Fv antibody fragments from a semi-synthetic phage antibody display library with designed CDR3 regions. J. Mol. Biol. 1995;248:97–105. doi: 10.1006/jmbi.1995.0204. [DOI] [PubMed] [Google Scholar]
- 9.Knappik A., Ge L., Honegger A., Pack P., Fischer M., Wellnhofer G., Hoess A., Wolle J., Pluckthun A., Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 10.Cwirla S.E., Peters E.A., Barrett R.W., Dower W.J. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl Acad. Sci. USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott J.K., Smith G.P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 12.Kay B.K., Adey N.B., He Y.S., Manfredi J.P., Mataragnon A.H., Fowlkes D.M. An M13 phage library displaying random 38-amino-acid peptides as a source of novel sequences with affinity to selected targets. Gene. 1993;128:59–65. doi: 10.1016/0378-1119(93)90153-t. [DOI] [PubMed] [Google Scholar]
- 13.Cortese R., Monaci P., Nicosia A., Luzzago A., Felici F., Galfre G., Pessi A., Tramontano A., Sollazzo M. Identification of biologically active peptides using random libraries displayed on phage. Curr. Opin. Biotechnol. 1995;6:73–80. doi: 10.1016/0958-1669(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 14.Healy J.M., Murayama O., Maeda T., Yoshino K., Sekiguchi K., Kikuchi M. Peptide ligands for integrin alpha v beta 3 selected from random phage display libraries. Biochemistry. 1995;34:3948–3955. doi: 10.1021/bi00012a012. [DOI] [PubMed] [Google Scholar]
- 15.Beste G., Schmidt F.S., Stibora T., Skerra A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc. Natl Acad. Sci. USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nord K., Gunneriusson E., Ringdahl J., Stahl S., Uhlen M., Nygren P.A. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 17.Koide A., Bailey C.W., Huang X., Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 18.Wang C.I., Yang Q., Craik C.S. Isolation of a high affinity inhibitor of urokinase-type plasminogen activator by phage display of ecotin. J. Biol. Chem. 1995;270:12250–12256. doi: 10.1074/jbc.270.20.12250. [DOI] [PubMed] [Google Scholar]
- 19.Jespers L.S., Messens J.H., De Keyser A., Eeckhout D., Van Den Brande I., Gansemans Y.G., Lauwereys M.J., Vlasuk G.P., Stanssens P.E. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (NY) 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 20.Rebar E.J., Pabo C.O. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 21.Schier R., Bye J., Apell G., McCall A., Adams G.P., Malmqvist M., Weiner L.M., Marks J.D. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J. Mol. Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 22.Schier R., McCall A., Adams G.P., Marshall K.W., Merritt H., Yim M., Crawford R.S., Weiner L.M., Marks C., Marks J.D. Isolation of picomolar affinity anti-c-erB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J. Mol. Biol. 1996;263:551–567. doi: 10.1006/jmbi.1996.0598. [DOI] [PubMed] [Google Scholar]
- 23.Yang W.P., Green K., Pinz-Sweeney S., Briones A.T., Burton D.R., Barbas C.F.R. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J., Pope T., Tung J.S., Chan C., Hollis G., Mark G., Johnson K.S. Affinity maturation of a high-affinity human monoclonal antibody against the third hypervariable loop of human immunodeficiency virus: use of phage display to improve affinity and broaden strain reactivity. J. Mol. Biol. 1996;256:77–88. doi: 10.1006/jmbi.1996.0069. [DOI] [PubMed] [Google Scholar]
- 25.Lowman H.B., Wells J.A. Affinity maturation of human growth hormone by monovalent phage display. J. Mol. Biol. 1993;234:564–578. doi: 10.1006/jmbi.1993.1612. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Rakonjac J., Jovanovic G., Model P. Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene. 1997;198:99–103. doi: 10.1016/s0378-1119(97)00298-9. [DOI] [PubMed] [Google Scholar]
- 28.Hoogenboom H.R., Griffiths A.D., Johnson K.S., Chiswell D.J., Hudson P., Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huie M.A., Cheung M.C., Muench M.O., Becerril B., Kan Y.W., Marks J.D. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc. Natl Acad. Sci. USA. 2001;98:2682–2687. doi: 10.1073/pnas.051631798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou J., Marzari R., Verzillo V., Ferrero F., Pak D., Sheng M., Yang C., Sblattero D., Bradbury A. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries. J. Immunol. Methods. 2001;253:233–242. doi: 10.1016/s0022-1759(01)00385-4. [DOI] [PubMed] [Google Scholar]
- 31.Hallborn J., Carlsson R. Automated screening procedure for high-throughput generation of antibody fragments. Biotechniques. 2002;(Suppl):30–37. [PubMed] [Google Scholar]
- 32.Clackson T., Wells J.A. In vitro selection from protein and peptide libraries. TIBTECH. 1994;12:173–184. doi: 10.1016/0167-7799(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 33.Duenas M., Borrebaeck C.A. Novel helper phage design: intergenic region affects the assembly of bacteriophages and the size of antibody libraries. FEMS Microbiol. Lett. 1995;125:317–321. doi: 10.1111/j.1574-6968.1995.tb07375.x. [DOI] [PubMed] [Google Scholar]
- 34.Soltes G., Barker H., Marmai K., Pun E., Yuen A., Wiersma E.J. A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions. J. Immunol. Methods. 2003;274:233–244. doi: 10.1016/s0022-1759(02)00294-6. [DOI] [PubMed] [Google Scholar]
- 35.Rondot S., Koch J., Breitling F., Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001;19:75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 36.Baek H., Suk K.H., Kim Y.H., Cha S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic Acids Res. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer R.A., Cox F., van der Horst M., van der Oudenrijn S., Res P.C., Bia J., Logtenberg T., de Kruif J. A novel helper phage that improves phage display selection efficiency by preventing the amplification of phages without recombinant protein. Nucleic Acids Res. 2003;31:e59. doi: 10.1093/nar/gng058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jestin J.L., Volioti G., Winter G. Improving the display of proteins on filamentous phage. Res. Microbiol. 2001;152:187–191. doi: 10.1016/s0923-2508(01)01191-3. [DOI] [PubMed] [Google Scholar]
- 39.Alexeyev M., Shokolenko I., Croughan T. Improved antibiotic-resistance gene cassettes and omega elements for Eschericia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 40.Marks J.D., Bradbury A. Selection of human antibodies from phage display libraries. Methods Mol. Biol. 2004;248:161–176. doi: 10.1385/1-59259-666-5:161. [DOI] [PubMed] [Google Scholar]
- 41.Goryshin I.Y., Jendrisak J., Hoffman L.M., Meis R., Reznikoff W.S. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 2000;18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 42.Chang A.C., Cohen S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messing J., Gronenborn B., Muller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc. Natl Acad. Sci. USA. 1977;74:3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanke T., Szawlowski P., Randall R.E. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 1992;73:653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 45.Sblattero D., Lou J., Marzari R., Bradbury A. In vivo recombination as a tool to generate molecular diversity in phage antibody libraries. J. Biotechnol. 2001;74:303–315. doi: 10.1016/s1389-0352(01)00022-8. [DOI] [PubMed] [Google Scholar]
- 46.Pelletier J.N., Arndt K.M., Pluckthun A., Michnick S.W. An in vivo library-versus-library selection of optimized protein–protein interactions. Nat. Biotechnol. 1999;17:683–690. doi: 10.1038/10897. [DOI] [PubMed] [Google Scholar]
- 47.Galarneau A., Primeau M., Trudeau L.E., Michnick S.W. β-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 48.Michnick S.W. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr. Opin. Struct. Biol. 2001;11:472–477. doi: 10.1016/s0959-440x(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 49.Michnick S.W., Remy I., Campbell-Valois F.X., Vallee-Belisle A., Pelletier J.N. Detection of protein–protein interactions by protein fragment complementation strategies. Methods Enzymol. 2000;328:208–230. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 50.Koch H., Grafe N., Schiess R., Pluckthun A. Direct selection of antibodies from complex libraries with the protein fragment complementation assay. J. Mol. Biol. 2006;357:427–441. doi: 10.1016/j.jmb.2005.12.043. [DOI] [PubMed] [Google Scholar]
- 51.Arndt K.M., Pelletier J.N., Muller K.M., Alber T., Michnick S.W., Pluckthun A. A heterodimeric coiled-coil peptide pair selected in vivo from a designed library-versus-library ensemble. J. Mol. Biol. 2000;295:627–639. doi: 10.1006/jmbi.1999.3352. [DOI] [PubMed] [Google Scholar]
- 52.Janssen D.B. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 2004;8:150–159. doi: 10.1016/j.cbpa.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 53.Marvin D.A. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 1998;8:150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]
- 54.Davis N.G., Model P. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell. 1985;41:607–614. doi: 10.1016/s0092-8674(85)80033-7. [DOI] [PubMed] [Google Scholar]
- 55.Davis N.G., Boeke J.D., Model P. Fine structure of a membrane anchor domain. J. Mol. Biol. 1985;181:111–121. doi: 10.1016/0022-2836(85)90329-8. [DOI] [PubMed] [Google Scholar]
- 56.Rampf B., Bross P., Vocke T., Rasched I. Release of periplasmic proteins induced in E.coli by expression of an N-terminal proximal segment of the phage fd gene 3 protein. FEBS Lett. 1991;280:27–31. doi: 10.1016/0014-5793(91)80196-a. [DOI] [PubMed] [Google Scholar]
- 57.Glaser-Wuttke G., Keppner J., Rasched I. Pore-forming properties of the adsorption protein of filamentous phage fd. Biochim. Biophys. Acta. 1989;985:239–247. doi: 10.1016/0005-2736(89)90408-2. [DOI] [PubMed] [Google Scholar]
- 58.Riechmann L., Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E.coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 59.Karlsson F., Borrebaeck C.A., Nilsson N., Malmborg-Hager A.C. The mechanism of bacterial infection by filamentous phages involves molecular interactions between TolA and phage protein 3 domains. J. Bacteriol. 2003;185:2628–2634. doi: 10.1128/JB.185.8.2628-2634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lubkowski J., Hennecke F., Pluckthun A., Wlodawer A. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Struct. Fold Des. 1999;7:711–722. doi: 10.1016/s0969-2126(99)80092-6. [DOI] [PubMed] [Google Scholar]
- 61.Boeke J.D., Model P., Zinder N.D. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 1982;186:185–192. doi: 10.1007/BF00331849. [DOI] [PubMed] [Google Scholar]
- 62.Stengele I., Bross P., Garces X., Giray J., Rasched I. Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J. Mol. Biol. 1990;212:143–149. doi: 10.1016/0022-2836(90)90311-9. [DOI] [PubMed] [Google Scholar]
- 63.Rakonjac J., Feng J., Model P. Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J. Mol. Biol. 1999;289:1253–1265. doi: 10.1006/jmbi.1999.2851. [DOI] [PubMed] [Google Scholar]
- 64.Bradbury A., Velappan N., Verzillo V., Ovecka M., Chasteen L., Sblattero D., Marzari R., Lou J., Siegel R., Pavlik P. Antibodies in proteomics I: generating antibodies. Trends Biotechnol. 2003;21:275–281. doi: 10.1016/S0167-7799(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 65.Bradbury A., Velappan N., Verzillo V., Ovecka M., Chasteen L., Sblattero D., Marzari R., Lou J., Siegel R., Pavlik P. Antibodies in proteomics II: screening, high-throughput characterization and downstream applications. Trends Biotechnol. 2003;21:312–317. doi: 10.1016/S0167-7799(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 66.Walter G., Konthur Z., Lehrach H. High-throughput screening of surface displayed gene products. Comb. Chem. High Throughput Screen. 2001;4:193–205. doi: 10.2174/1386207013331228. [DOI] [PubMed] [Google Scholar]
- 67.Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 68.Griffiths A.D., Malmqvist M., Marks J.D., Bye J.M., Embleton M.J., McCafferty J., Baier M., Holliger K.P., Gorick B.D., Hughes-Jones N.C., et al. Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 1993;12:725–734. doi: 10.1002/j.1460-2075.1993.tb05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]