Abstract
Previous studies showed that binding of the CBF/NF-Y (CBF) transcription factor to cellular promoters is essential for cell proliferation. This observation prompted us to investigate the function of CBF in relation to cell cycle progression and in cell-cycle-regulated transcription. In this study, we used a tetracycline-inducible adenoviral vector to express a truncated CBF-B subunit, Bdbd, lacking a transcription activation domain in various mammalian cell lines. The Bdbd polypeptide interacts with cellular CBF-A/CBF-C and binds to promoters containing CBF-binding sites. Interestingly, expression of Bdbd in various mammalian cells resulted in the inhibition of cell proliferation and specific cell cycle arrest at G2/M phase. Gene expression analysis demonstrated that the expression of Bdbd strongly suppressed cell cycle-dependent transcription activation of Cyclin B1, Aurora A and CDK1 genes, key regulators for cell cycle progression at G2/M phase. Chromatin immunoprecipitation analysis showed that Bdbd significantly inhibited binding of TATA-binding protein, TBP to both Cyclin B1 and Aurora A promoters, but did not inhibit binding of E2F3 activator to Cyclin B1 promoter. This study suggested that the activation domain of CBF-B plays an essential role in the transcription activation of Cyclin B1 and Aurora A genes at G2/M phase, thus regulating cell cycle progression at G2/M phase.
INTRODUCTION
In mammalian cells, transcription of several genes including Cyclin B1, CDK1 (also known as CDC2) and Aurora A is activated at G2/M phase of the cell cycle. The proteins encoded by these genes play crucial roles in progression through mitosis. Inhibition of the activity of any of these proteins often leads to arrest of cells at G2/M phase (1–4). Thus, coordinated transcription activation of these genes is believed to be essential for cell cycle progression at G2/M phase. Also, expression of both Cyclin B1 and Aurora A genes is increased in various human tumors (5,6).
Previous studies of Cyclin B1, CDK1, CDC25C and topoisomerase IIα promoters showed that binding of the CBF/NF-Y (CBF) transcription factor to these promoters plays a crucial role in transcription activation of them at G2/M phase (2,7–9). Comparative genomic analysis identified a conserved regulatory promoter module consisting of a CBF-binding site, a cell cycle-dependent element (CDE) and a cell cycle homology region (CHR). The proposed module is present in different human genes that are activated at G2/M phase. This suggested that CBF controls transcription of multiple genes at G2/M phase (10,11).
Mammalian CBF consists of three subunits, CBF-A (NF-YB), CBF-B (NF-YA) and CBF-C (NF-YC), which are all needed for DNA binding (12,13). CBF consists of two transcription activation domains: one each in CBF-B and CBF-C. Interestingly, the activity of CBF-B is regulated by cyclin-dependent kinase 2 (CDK2) phosphorylation. Mutation of CBF-B that inhibits CDK2-dependent phosphorylation has been shown to decrease DNA binding of CBF (14). This study suggested that phosphorylation of CBF-B plays a role in the transcription activation of genes at G2/M phase. The tumor suppressor protein p53 inhibits transcription activation of Cyclin B1, CDK1, securin and topoisomerase IIα promoters through CBF-binding sites. Recent studies showed that p53 inhibits CBF activity through inhibition of CDK2-dependent phosphorylation as well as through direct interaction with CBF (8,14–18). Altogether these studies indicated that CBF-binding sites in the G2/M specific promoters are needed for transcription activation as well as for transcription repression.
The function of CBF in the cellular transcription was studied by the expression of dominant-negative CBF-B mutants and also by conditional inactivation of the mouse CBF-B gene (19–21). When a dominant-negative CBF-B mutant that interacted with CBF-A/CBF-C but did not bind DNA was expressed in mouse fibroblasts, this resulted in the retardation of cell growth. Similarly, expression of a CBF-B mutant defective in CDK2-dependent phosphorylation resulted in inhibition of the proliferation of human colorectal cancer cells. Further analysis of the cells showed that the growth arrest occurred at both G1/S and G2/M. Inactivation of the CBF-B gene in mouse embryonic fibroblasts also resulted in complete inhibition of cell proliferation and growth arrest at various phases of the cell cycle. Taken together, these studies demonstrated that inhibition of DNA binding by CBF leads to growth arrest at multiple phases of the cell cycle.
Since previous studies dissected various domains of CBF involved in DNA binding and transcription activation, this prompted us to investigate whether specific domain of CBF could contribute to the regulation of cell cycle. To do this, as described herein, we expressed a truncated CBF-B, Bdbd, lacking a transcription activation domain but containing a DNA-binding domain in various human and mouse cells. Bdbd formed a CBF–DNA complex that lacked one transcription activation domain. Our results showed that expression of Bdbd in various cell lines resulted in cell cycle arrest specifically at G2/M phase. Gene expression analysis showed that Bdbd inhibited transcription activation of Cyclin B1, Aurora A, and CDK1 genes, which all are required for cell cycle progression at G2/M phase. Chromatin immunoprecipitation analysis demonstrated that Bdbd primarily inhibited recruitment of TATA-binding protein to both cellular Cyclin B1 and Aurora A promoters. Together, our study provided evidence that the CBF-B activation domain controls cell cycle progression at G2/M phase, and plays an essential role in transcription activation of Cyclin B1 and Aurora A genes, two key regulators for G2/M progression.
MATERIALS AND METHODS
Cell lines
The HeLa tet-off, Saos-2 tet-off and U2 OS tet-on cell lines were purchased from BD Biosciences Clontech and maintained as instructed. The NIH 3T3 tet-off cell line was established and maintained as reported previously (19). HeLa and Saos-2 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 100 μg/ml G418. For NIH 3T3 cells, calf serum instead of FBS and 200 μg/ml G418 were used. U2 OS cells were maintained in a culture medium containing 90% McCoy's 5A medium, 10% FBS, 4 mM l-glutamine, 200 μg/ml G418, 4.5 g/l glucose, 10 mM HEPES, 1 mM sodium pyruvate and 0.2 U/ml bovine insulin. The human embryonic kidney 293 IQ cell line was purchased from Microbix Biosystems, and was used for the development and amplification of recombinant adenoviruses. The cells were maintained in the same medium where HeLa cells were maintained in with the exception of 150 μg/ml hygromycin instead of 100 μg/ml G418. All the cells were maintained in an incubator with 5% CO2.
Recombinant adenoviral expression vector and virus
The recombinant adenoviral expression vector was constructed by using the Adeno-X Tet-Off Expression Systems (BD Biosciences Clontech). First, EcoRV/XbaI-restricted gene fragments of FLAG-tagged CBF-B (Bwt) CBF-Bmut (Bmut), and CBF-Bdbd (Bdbd) were generated from their parental plasmids (pTRE-flagB, pTRE-flagBmut and pTRE-flagBdbd, respectively) (9,19). Each gene was cloned into the shuttle vector pTRE-Shuttle2. A Tet-responsive expression cassette was excised from the recombinant shuttle vector by PI-SceI/I-CeuI digestion and then cloned into the replication-incompetent (E1/E3-deleted) Adeno-X viral DNA to form a recombinant adenoviral expression vector. Two such vectors, Ad-Bwt and Ad-Bdbd, were generated, transformed and amplified in HB101. To make the recombinant adenoviruses, each expression vector was linearized by PacI digestion and transfected into E1-complementing 293 IQ cells. Amplification was achieved by sequential infection into the same cell line. The titer of the final virus stocks at a level of 8 × 107 pfu/ml was determined by following the instructions in the manual of BD Biosciences Clontech. To express the interested genes, the recombinant adenoviruses were used to infect the tet-on or tet-off cells with or without the addition of Tc or doxycycline. Unless otherwise indicated, an MOI (multiplicity of infection) of 1:3 (three plaque forming units of virus per cell, an equivalent of 25 μl stock) was used for experiments in this study.
Synchronization, BrdU labeling and cell cycle analysis
HeLa cell synchronization at G1/S phase was achieved by using a double thymidine block (22). Briefly, the cells were incubated with 2 mM thymidine for 18 h for the first block, released for 9 h after the thymidine was washed out, and then incubated with thymidine at the same concentration for 14 h for the second block. The cells were released by washing out the thymidine and analyzed for cell cycle progression by flow cytometry as described previously (19). For S phase analysis, cells were labeled with BrdU by using a FLUOS Kit (Roche Applied Science) as described in the manufacturer's protocol. Flow cytometry analyses were performed with a FACS Calibur (BD Biosciences) using the Cell Quest Pro software program.
Detection of CBF-B polypeptides in cell extracts and after immunoprecipitation
To detect adenoviral mediated CBF-B expression, HeLa cells at 22 h in six-well plates after adenovirus infection were washed twice with PBS, and were lysed in 80 μl of 1× SDS gel loading buffer (50 mM Tris–HC1, pH 6.8, 2% SDS, 10% glycerol and 0.1% bromophenol blue). About 15 μg of each cell extracts were separated using SDS–PAGE and then analyzed by western blotting using an anti-CBF-B monoclonal antibody (Santa Cruz Biotech).
To determine the association of cellular CBF-A/CBF-C heterodimer with Bdbd, whole cell extracts of Ad-Bdbd infected HeLa cells were prepared by lysing the cells in a buffer containing 150 mM NaCl, 1% NP-40 and 50 mM Tris–HCl, pH 8.0. The cell lysates were incubated with anti-CBF-A antibody or IgG as a control at 4°C for 1.5 h, and then precipitated with protein A–Sepharose 6B resin (Amersham Biosciences). The resin bound proteins were eluted with SDS gel loading buffer, and were separated by SDS–PAGE followed by western blotting using an anti-CBF-B monoclonal antibody.
Plasmids and transfection
A DNA fragment between −177 and +268 of human Aurora A gene was isolated by genomic PCR and was cloned between the SacI and XhoI sites of the pGL3-basic vector to generate wild-type Aurora A promoter–reporter construct (−177/+268). Single nucleotide mutation in each CCAAT motif of the wild-type construct was introduced by two-step PCR method (9) to generate Mt I, Mt II, and Mt I and II constructs, as indicated in Figure 8D. These constructs were transfected into mouse fibroblast NIH3T3 cells by using the LipofectAMINE reagent (Invitrogen), and expression of the luciferase gene was measured as described previously (9).
DNA-binding assay
Cultured cells in 60 mm plates were first washed twice with PBS and then lysed in 400 μl of buffer consisting of 150 mM NaCl, 50 mM Tris–HC1, pH 8.0 and 1% NP-40. Lysis was performed for 30 min with occasional shaking at 4°C. The cellular extract was obtained by high-speed centrifugation of the lysates in a microcentrifuge. In each DNA-binding assay reaction, 3 μl of the extract was used. The DNA-binding assay method was described previously (23).
RNA isolation and northern blot analysis
Total RNA was isolated from 2 × 106 cultured cells by using an RNeasy Mini Kit (Qiagen). RNA (10 μg) from each sample was used for northern blot analysis as described previously (19). After the analysis, the signal intensity of the radioactive bands in the blot was quantified by using a Phosphorimager in combination with the use of an image analysis software ImageQuant 5.2 (Molecular Dynamics). For each band, the mean intensity value and the standard deviation for three independent experiments were calculated. The Cyclin E1 (207–697 nt), CDK1 (216–652 nt), Cyclin D1 (311–731 nt) and FOXM1 (1206–1770 nt) cDNA probes were obtained by PCR amplification from cDNA clones, which were purchased from either Harvard Institute of Proteomics (Cyclin E1 and CDK1) or American Tissue Culture Center (Cyclin D1). The cDNA clone for FOXM1 was a gift from Dr R.H. Costa (24). The Cyclin B1 (800–1036 nt) and the thymidine kinase I, TK1 (570–979 nt) cDNA probes were generated by RT–PCR amplification using total RNAs of HeLa cells. The probe for Aurora kinase A (186–1038 nt) was isolated by HindIII/EcoRI digestion from a GFP-Aurora A construct (3). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was isolated by EcoRI and HindIII digestion of pTRI-GAPDH plasmid (Ambion).
Immunofluorescence analysis
HeLa cells grown on cover slips were fixed with 4% formaldehyde in PEM buffer (80 mM potassium–PIPES, pH 6.8, 5 mM EGTA, pH 7.0 and 2 mM MgCl2) for 30 min on ice; permeabilized with 0.5% Triton X-100 in the PEM buffer for 30 min at room temperature. After incubation for another 30 min with a blocking solution (1% BSA, 20 mM Tris–HCl, pH 7.4, 137 mM NaCl and 0.1% Tween-20), the cells were incubated for 1 h each with the primary antibody anti-phosphoserine 10-histone H3 (Upstate) and an Alexa Fluor 488-conjugated secondary antibody (Molecular Probes). The DNA of the cells was counterstained with DAPI (20 μg/ml) in the blocking solution. Finally, a ProLong Antifade kit (Molecular Probes) was used to complete the mounting of the cover slips. The staining was visualized under a Leica DMR microscope and the fluorescent images were acquired by using the MetaMorph software program (Universal Imaging).
Chromatin immunoprecipitation analysis
The chromatin immunoprecipitation (ChIP) was performed as described previously (25). Briefly, ∼107 HeLa cells were incubated with formaldehyde for 10 min to cross-link the chromatin DNA with bound proteins. Cross-linking was stopped by the addition of glycine to a 125 mM final concentration. The cells were then washed, lysed and sonicated to produce an average chromatin DNA fragment length of 0.5–1 kb. The lysis buffer consists of 5mM PIPES, pH 8.0, 85 mM KCl, 0.5% Igepal CA-630, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 mM PMSF. The cell lysate containing chromatin DNAs was precleared with 20 μl of packed protein A/G–agarose at 4°C for 2 h. Then, the chromatin DNAs were incubated and precipitated with each of polyclonal antibodies against E2F3, E2F4 and TBP (Santa Cruz Biotech), CBF-A (25) and rabbit IgG as a control. The immunoprecipitation buffer consists of 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris–HCl, pH 8.1, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 mM PMSF. In the end, the precipitated chromatin DNAs were extracted after reverse cross-linking and were used in either regular or real-time PCRs to detect the promoter DNA regions of specific genes. Ten percent of chromatin DNA before each immunoprecipitation was utilized to isolate DNA by reverse cross-linking and extraction, which was subsequently used as a source for input DNA in each PCR. The PCR primers for each specific promoter are listed in Supplementary Table S1.
Real-time PCRs with a SYBR Green approach were performed to quantify ChIP DNAs corresponding to the promoters of Cyclin B1, Aurora A, and TK1 genes. Primers for these PCRs were designed by using Primer Express 2.0 (Applied Biosystems). The PCRs were set-up in optical 96-well plates and performed in 20 μl of mixtures containing 1× SYBR Green Mix (Applied Biosystems), 1/100 fraction of the ChIP-enriched DNA and 100 nM primers. A series of dilutions of input DNAs were used in PCRs to determine a standard curve for each individual pair of primers. PCRs using ChIP DNAs precipitated with rabbit IgG were run for the purpose of background subtraction. The plates were read in an ABI Prism 7900HT real-time PCR instrument. The amount of ChIP DNAs, as immunoprecipitated by antibodies against different transcription factors, was calculated by normalizing to the values of input material in the linear range of PCR cycles. For each antibody, three independent ChIP assay were performed to isolate ChIP DNAs, which were then quantified using real-time PCR.
RESULTS
Expression of a full-length CBF-B, Bwt, and a truncated CBF-B, Bdbd, under the control of a tetracycline-inducible adenoviral vector
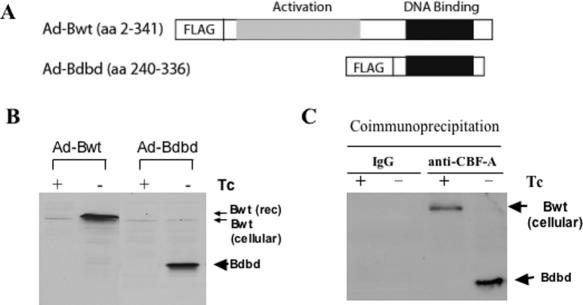
We used a tetracycline (Tc)-regulated adenoviral vector to express each of the full-length CBF-B (Bwt) and truncated CBF-B (Bdbd) polypeptides in mammalian cells. Bdbd consists of 96 C-terminal amino acids containing a DNA-binding domain. Each of the CBF-B polypeptides was constructed as a fusion protein with the FLAG epitope (Figure 1A). Recombinant adenoviruses Ad-Bwt and Ad-Bdbd were used to infect HeLa cells expressing a tet-off activator, which induces the expression of CBF-B polypeptide in the absence but not the presence of Tc. Expression of both CBF-B polypeptides was detected by using western blot with an anti-CBF-B antibody. Both Bwt and Bdbd were expressed at a comparable level in HeLa cells in the absence but not the presence of Tc (Figure 1B). The cellular or endogenous CBF-B was also detected, as represented by the faint band just below that for Bwt. This experiment showed that each of Bwt and Bdbd polypeptides was overexpressed ∼10 times more than that of cellular CBF-B.
Figure 1.
Expression of full-length and truncated CBF-B in HeLa cells and association of the truncated CBF-B with cellular CBF-A. (A) A full-length (amino acids 2–341) CBF-B (Bwt) containing both transcription activation and DNA-binding domains, and a truncated version (amino acids 240–336) of CBF-B (Bdbd) lacking the activation domain were cloned in fusion with FLAG into a Tc-induced adenoviral expression system. (B) After infection with the recombinant adenovirus, HeLa cells in the presence or absence of Tc were lysed to prepare whole cell extracts, which were analyzed by western blotting with an anti-CBF-B antibody to detect Bwt (41 kDa) and Bdbd (14 kDa). (C) Ad-Bdbd infected HeLa cells in the presence or absence of Tc were lysed to prepare whole cell extracts and an anti-CBF-A antibody or IgG was used to carry out an immunoprecipitation experiment. The immunoprecipitated complex was then subjected to western blotting with an anti-CBF-B antibody, which detected co-immunoprecipitated Bdbd in the absence of Tc, but cellular CBF-B in the presence of Tc.
Previously, we demonstrated that Bwt was associated with cellular CBF-A/CBF-C complex and formed a DNA–protein complex with CCAAT motif (26). To determine the interaction of Bdbd with cellular CBF-A/CBF-C complex, cellular extracts prepared from HeLa cells infected with Ad-Bdbd in the presence and absence of Tc were immunoprecipitated with anti-CBF-A antibody or rabbit IgG as a control. The immunoprecipitated complex was analyzed by western blotting using an anti-CBF-B antibody. This showed that Bdbd was precipitated with anti-CBF-A antibody in cell extracts in the absence of Tc (Figure 1C). In contrast, cellular CBF-B was precipitated with anti-CBF-A antibody in cell extracts in the presence of Tc. This indicated that similar to cellular CBF-B, Bdbd was associated with cellular CBF-A/CBF-C complex. Since no cellular CBF-B was detectable in the immunoprecipitate containing Bdbd, it was suggested that cellular CBF-A/CBF-C complex was exclusively associated with Bdbd in HeLa cells expressing Bdbd. This was possibly due to complete displacement of cellular CBF-B from CBF-A/CBF-C complex by Bdbd.
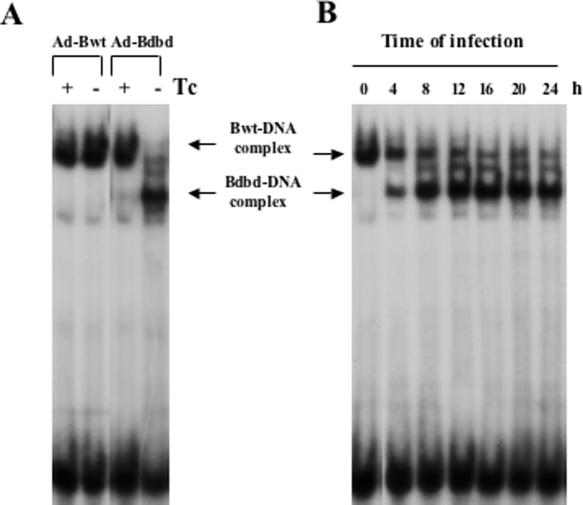
The HeLa cell extracts were also monitored for CBF-binding activity. This showed that the expression of Bwt in the absence of Tc did not significantly change the cellular CBF-binding activity (Figure 2A). However, expression of Bdbd in the absence of Tc changed the mobility of the cellular CBF–DNA complex by forming a faster moving DNA–protein complex. An anti-FLAG antibody supershifted the faster moving complex, indicating that the complex contained Bdbd (data not shown). We interpreted the change in mobility as being a result of the smaller size of Bdbd, which displaced the larger size cellular CBF-B from CBF complex. Consistent with the co-immunoprecipitation experiment, there was an indication of complete displacement of CBF-B by Bdbd in HeLa cells expressing Bdbd (MOI 1:3). DNA-binding analysis of cell extracts prepared at various time points after Ad-Bdbd infection (MOI 1:1.5) showed that the Bdbd complex began to form 4 h after infection and reached its maximum amount at 12–16 h of infection (Figure 2B).
Figure 2.
DNA-binding activity of CBF in HeLa cells expressing Bwt or Bdbd. (A) Whole cell extracts from the recombinant adenovirus-infected HeLa cells (MOI 1:3) in the presence or absence of Tc were used for the DNA-binding assay. Expression of Bwt (−Tc) slightly increased DNA-binding activity of CBF. Expression of Bdbd (−Tc) competed out the endogenous CBF-B (Bwt) complex completely and formed a CBF complex containing Bdbd. (B) The adenovirus Ad-Bdbd infected HeLa cells (MOI 1:1.5) in the absence of Tc were collected at various time points after the infection and used in a DNA-binding assay to monitor formation of the subshifted Bdbd–DNA complex. The results showed that the maximum amount of the Bdbd–DNA complex was formed from 12 to 16 h after the infection with disappearance of the cellular CBF–DNA (Bwt–DNA) complex.
Expression of Bdbd inhibits cell cycle progression
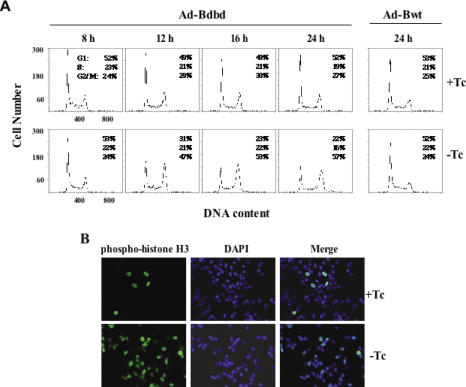
Infection of HeLa cells with Ad-Bwt did not inhibit cell proliferation in the absence of Tc (Supplementary Table S2). In contrast, infection of HeLa cells with Ad-Bdbd greatly inhibited cell proliferation in the absence of Tc. This indicated that expression of Bdbd inhibited cell proliferation. To determine the cell cycle distribution profile of Bdbd-expressing cells, the HeLa cells were analyzed by using flow cytometry at various time points after adenoviral infection (Figure 3A). Expression of Bdbd resulted in a time-dependent increase in the percentage of cells at G2/M phase. Specifically, the data showed that the cell cycle distribution of the HeLa cells after 24 h of infection with Ad-Bdbd in the presence of Tc was 52% at G1, 19% at S and 27% at G2/M. In contrast, the cell cycle distribution in the absence of Tc was 22% at G1, 16% at S and 57% at G2/M. As a control, HeLa cells infected with Ad-Bwt had a similar cell cycle distribution between in the presence and absence of Tc. Interestingly, dose-dependent effect of Bdbd was evident. HeLa cells infected with increasing MOIs of Ad-Bdbd that resulted in gradual increase of Bdbd level, also resulted in gradual increase in the percentage of cells arrested at G2/M phase (Supplementary Figure S1A–C). DNA-binding assay of CBF at various Bdbd concentrations showed that the cellular CBF (Bwt) was partially displaced by Bdbd at MOIs of 0.75 or 1.5. Thus, accumulation of cells at G2/M phase was observed even when the cellular CBF was partially displaced by Bdbd. In contrast, expression of Bwt had no effect on cell cycle distribution (Supplementary Figure S1D). As another control, a dominant-negative mutant of CBF-B (Bmut) that inhibited DNA binding of CBF was also expressed in HeLa cells using the adenoviral vector. This showed that the expression of Bmut in the absence of tetracycline strongly inhibited DNA binding of CBF and also resulted in retardation of cell growth (Supplementary Figure S2A–C), similarly as observed in mouse fibroblast cells in our previous publication (19). Flow cytometry analysis of Ad-Bmut infected cells showed that no cell cycle distribution was changed with and without expression of Bmut (Supplementary Figure S2D). This suggests that Bmut inhibited HeLa cell proliferation through growth arrest at multiple phases of cell cycle, similarly as we have reported its effect on mouse fibroblast cells. This experiment supports the specific role of Bdbd at G2/M phase of HeLa cells.
Figure 3.
Cell-cycle analysis of HeLa cells expressing Bdbd or Bwt. (A) Flow cytometric analysis of HeLa cells at different time points after infection with Ad-Bdbd or Ad-Bwt in the presence or absence of Tc. Fluorescence of propidium iodide-stained nuclei was used to measure DNA content. Expression of Bdbd in the absence of Tc led to the accumulation of cells at G2/M phase in a time-dependent manner. (B) Analysis of HeLa cells after 20 h of Ad-Bdbd infection with an anti-phosphoserine 10-histone H3 antibody, which was detected by the Alexa 488 labeled secondary antibody. The nuclei were stained with DAPI. The merge shows an overlay of green and blue fluorescence data.
We analyzed HeLa cells by performing bromodeoxyuridine (BrdU) labeling to determine S phase at various time points after adenoviral infection (Supplementary Table S3). At 12 and 16 h of Ad-Bdbd infection, a significant percentage of BrdU-labeled cells were detected both in the presence and absence of Tc. However, at 24 h of Ad-Bdbd infection, a small percentage of BrdU-labeled cells were present in the absence of Tc. This indicated that the expression of Bdbd did not initially inhibit the HeLa cells at S phase. The loss of HeLa cells at S phase at a later time point (24 h) was likely caused by cell cycle arrest at G2/M phase.
The HeLa cells were also analyzed by immunostaining with an antibody against phosphoserine 10-histone H3, a marker for G2 phase and mitotic cells (27). At 20 h of infection with Ad-Bdbd, only ∼5% of the cells in the presence of Tc were stained with the antibody, whereas >50% of the cells in the absence of Tc were stained with the antibody (Figure 3B). This supported the earlier observation that the expression of Bdbd resulted in the arrest of cells at G2/M phase.
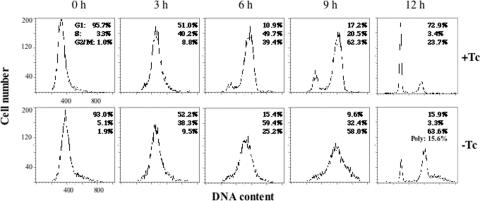
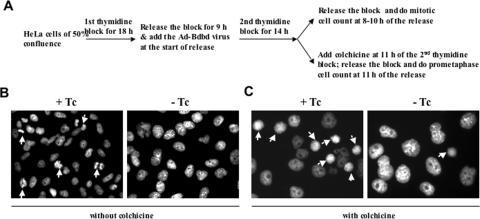
To further analyze the effect of Bdbd expression on progression of the cell cycle, HeLa cells were first synchronized by using a double thymidine block and then analyzed by using flow cytometry (22). The cells were infected with Ad-Bdbd when the first release of the thymidine block started, and then analyzed by flow cytometry at various time points after releasing the second block (Figure 4). Most of the cells in the presence or absence of Tc were at G1 phase at 0 h of release. At 3 h of release, a large percentage of the cells were at S phase, and at 6 and 9 h of release, the percentage of cells at G2/M phase was increased both in the presence and absence of Tc. At 12 h, most of the cells in the presence of Tc completed mitosis and were at G1 phase; in contrast, most of the cells in the absence of Tc were arrested at G2/M phase along with some cells displaying polyploidy. In comparison, analysis of Ad-Bwt infected HeLa cells in the presence or absence of Tc showed similar cell cycle progression pattern as that for Ad-Bdbd infected cells in the presence of Tc (data not shown). These results indicated that the expression of Bdbd specifically inhibited the progression of normal cell cycle at G2/M phase. We counted the mitotic cells by performing 4′,6′-diamidin-2-phenylindone (DAPI) staining at various time points from 8 to 10 h of release (Figure 5A). This showed that at any time point during this time range, <1% of the Bdbd-expressing cells were mitotic compared with 8% of the non-Bdbd-expressing cells (Figure 5B). We also analyzed HeLa cells after incubating them with colchicine to arrest the cells at prometaphase during the thymidine block and release experiment (28) (Figure 5A). In this experiment, the cells were stained with DAPI at 11 h of release from the second thymidine block. About 40% of the cells without Bdbd expression were at prometaphase (Figure 5B). In contrast, only 5% of the cells with Bdbd expression were at prometaphase. This suggested that the expression of Bdbd inhibits the entry of the cells into mitosis and arrests cells at G2 phase.
Figure 4.
Progression of the cell cycle after the expression of Bdbd in synchronized HeLa cells at G1/S phase. HeLa cells were synchronized by using a double thymidine block and infected with Ad-Bdbd in the presence or absence of Tc. After release of the block, the cells were collected at different time points and then analyzed by using flow cytometry, similarly as described in Figure 3A.
Figure 5.
Effects of Bdbd expression on mitosis of HeLa cells. (A) Experimental design for mitotic cell count or prometaphase cell count after Bdbd expression. Mitotic cell count (B) and prometaphase cell count (C) were performed after DAPI staining of Ad-Bdbd infected HeLa cells in the presence or absence of Tc. The results showed that the expression of Bdbd significantly inhibited both mitotic and prometaphase cell counts. Arrows indicate mitotic or prometaphase cells.
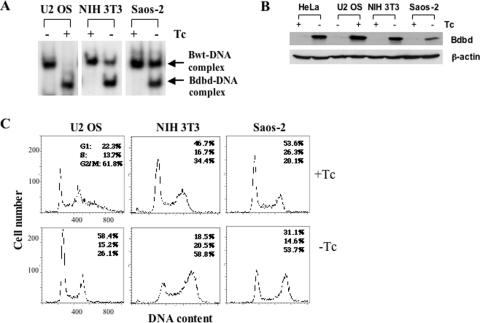
To determine whether the expression of Bdbd has a similar effect in cells other than HeLa cells, we expressed Bdbd in two human osteosarcoma cell lines, Saos-2 and U2 OS, and the mouse fibroblast line NIH 3T3 (Figure 6). Saos-2 and NIH 3T3 constitutively expressed the tet-off activator. Thus, as in HeLa cells, Bdbd was expressed in the absence but not in the presence of Tc in these cell lines. In contrast, U2 OS constitutively expressed the tet-on activator, which induced Bdbd expression in the presence but not in the absence of Tc. We infected each of these cell lines with Ad-Bdbd and grew them in the presence and absence of Tc. At 48 h after infection, cell extracts were prepared to determine the level of CBF-binding activity. We found a Bdbd-specific DNA–protein complex was observed in all of the cell extracts only when Bdbd expression was induced (Figure 6A). In U2 OS cells in the presence of Tc, only Bdbd-specific DNA–protein complex was observed, indicating that Bdbd completely displaced cellular CBF-B. In both NIH 3T3 and Saos-2 cells in the absence of Tc, a Bdbd–DNA complex was formed along with a residual cellular Bwt–DNA complex. The results indicated that Bdbd did not completely displace cellular CBF-B in these two cell lines.
Figure 6.
Expression of Bdbd in various cell lines caused arrest at G2/M phase. (A) DNA binding of CBF after the expression of Bdbd in various cell lines. Whole cell extracts were prepared at 48 h after infection with Ad-Bdbd and used in a DNA-binding assay. (B) Flow cytometric analysis of various cell lines at 48 h after infection with Ad-Bdbd in the presence or absence of Tc. Expression of Bdbd was induced in the absence of Tc in NIH 3T3 and Saos-2 cells, whereas it was induced in the presence of doxycycline in U2 OS cells. (C) The MOIs used were 1:3 for U2 OS cells, 1:6 for NIH 3T3 cells and 1:9 for Saos-2 cells.
To determine the cause for partial displacement of CBF complex by Bdbd, we compared expression of Bdbd polypeptide in HeLa, U2 OS, NIH3T3 and Saos-2 by western blot (Figure 6B). This showed that a low quantity of Bdbd was expressed in Saos-2 cells compared with the other cell lines. Quantification of the western blot showed that Bdbd was expressed 2.3-, 3- and 3.7-fold more in NIH3T3, HeLa and U2 OS cells, respectively, in comparison with its expression in Saos-2 cells. This suggests that the partial displacement of CBF complex in Saos-2 cells is due to low quantity of Bdbd expression. The partial displacement of CBF complex in NIH3T3 cells could also be due to lower level of Bdbd in comparison to HeLa and U2 OS cells. However, we cannot rule out a possibility that other parameters such as secondary modifications might play a role in the interactions among CBF subunits and caused variation at the level of displacement by Bdbd in different cell lines. Despite this difference in displacement of cellular CBF-B, analyses of these three cell lines by using flow cytometry showed that the expression of Bdbd resulted in an increase in the percentage of cells at G2/M phase in each cell line (Figure 6C). Consistent with this observation, expression of Bdbd also inhibited proliferation of each cell line (Supplementary Table S2). As a control, expression of Bwt had no effect on cell cycle distribution in any of the cell lines (Supplementary Figure S3). Thus, we concluded that expression of Bdbd inhibits the proliferation of these cells and arrests them at G2/M phase.
Expression of Bdbd suppresses transcription activation at G2/M phase
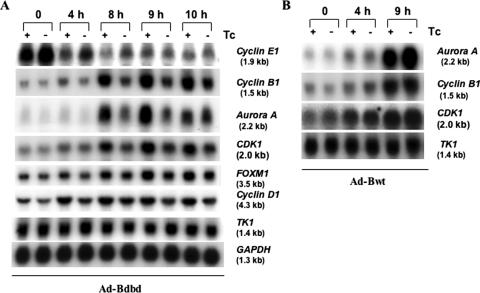
As multiple CBF-binding sites are present in the promoters of several genes that are activated at G2/M phase [(7–11); Supplementary Table S4], we examined the expression of some of these genes during cell cycle progression after release of the double thymidine block (Figure 7). Northern blot analysis showed that the expression of Cyclin B1 and Aurora A is activated at 8 h of release, consistent with previously published observations (3,4). Interestingly, expression of Bdbd strongly inhibited the transcription activation of Aurora A and Cyclin B1 at 8 and 9 h of release of the second thymidine block (Figure 7A). Expression of Bdbd also reduced the activation of CDK1, and only slightly reduced the expression of FOXM1 and Cyclin D1, but did not alter the expression of TK1 and GAPDH. Quantification of the northern blot showed that Bdbd caused the fold inhibition of gene expression at 9 h time point approximately 3.1, 2.6, 2.1, 1.5 and 1.5 for Aurora A, Cyclin B1, CDK1, FOXM1 and Cyclin D1, respectively (Table 1). As a control, expression of Bwt in the absence of Tc had little effect on gene expression (Figure 7B). Quantification data for the northern blot at 9 h time point showed that the expression of Bwt resulted in 1.26 ± 0.16 fold increase of Aurora A but no change of Cyclin B1, CDK1 and TK1 genes expression. Taken together these results demonstrated that the expression of Bdbd-inhibited transcription activation of Cyclin B1, Aurora A and CDK1 at late S and G2/M phases. As the cell cycle arrest occurred specifically at G2/M phase, we did not extensively examine the impact of Bdbd on the expression of G1/S genes. However, we did analyze the expression of one G1/S gene Cyclin E1 after Bdbd expression, with a result showing no inhibition of its expression at G1/S by Bdbd (Figure 7A).
Figure 7.
Analysis of the expression of various genes during cell cycle progression in HeLa cells after the expression of Bdbd or Bwt. The HeLa cells were synchronized by using a double thymidine block, infected with Ad-Bdbd or Ad-Bwt, and then released from the block as described in Figure 4. The cells were collected at different time points after the release and used to isolate total RNAs, which were analyzed by using northern blot analysis. Each lane contains 10 μg of RNA. Analysis was carried out for RNA samples prepared from HeLa cells infected with Ad-Bdbd (A) or with Ad-Bwt (B) in the presence or absence of Tc. This experiment was repeated three times specifically at 0 h, 8 h and 9 h time points.
Table 1.
Quantification of the mRNA levels at 9 h time point of Figure 7A
| Gene | Relative band signal intensity of detected genes (×105) | Fold inhibition | |
|---|---|---|---|
| +Tc | −Tc | ||
| Cyclin B1 | 45.39 ± 3.78 | 17.75 ± 1.19 | 2.56 |
| Aurora A | 7.28 ± 0.59 | 2.33 ± 0.18 | 3.12 |
| CDC2 | 4.45 ± 0.36 | 2.14 ± 0.15 | 2.08 |
| FOXM1 | 1.90 ± 0.17 | 1.24 ± 0.08 | 1.53 |
| Cyclin D1 | 7.70 ± 0.46 | 5.25 ± 0.28 | 1.47 |
| TK1 | 33.25 ± 2.30 | 32.96 ± 0.72 | 1.01 |
The band signal intensity representing the mRNA levels at 9 h time point in Figure 7A was quantified by using a Phosphoimager. Data collected from three independent experiments were normalized with the value for GAPDH and used for calculation of the mean and standard deviation. Fold inhibition of gene expression was calculated as the ratio of the mean intensity value for without Bdbd expression (+Tc) versus with Bdbd expression (−Tc).
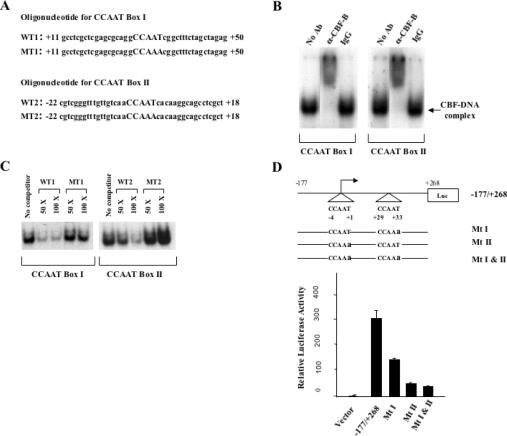
Although the role of CBF binding in transcription of Cyclin B1 and CDK1 promoters was studied in previous reports (8), the function of CBF in transcription of the Aurora A promoter is not known. The human Aurora A promoter contains two CCAAT motifs, one of which is located on the start of transcription, but it does not contain a TATA box (29). Double-stranded oligonucleotides corresponding to each CCAAT motifs were used in DNA-binding assay with HeLa cell nuclear extracts (Figure 8A). This showed that a DNA–protein complex formed with each labeled oligonucleotide was supershifted with an anti-CBF-A antibody (Figure 8B). Each DNA–protein complex was competed by excess wild-type oligonucleotide, but not by excess of mutant oligonucleotide containing mutation of CCAAT to CCAAA, indicating that both of the CCAAT motifs of Aurora A promoter specifically interact with CBF (Figure 8C). A promoter fragment containing −177 to +268 bp of human Aurora A gene was used to study the role of CBF-binding sites in transcription activity. The wild-type promoter DNA and three mutant promoter DNAs containing mutations in the CBF-binding sites were constructed in a luciferase reporter gene vector (Figure 8D). Activity of eachreporter construct was analyzed by transient transfection in NIH 3T3 cells. This showed that mutations of distal (Mt I) and proximal (Mt II) CCAAT motifs resulted in 2-and 6-fold reduction of promoter activity, respectively. Mutations of both CCAAT motifs (Mt I and Mt II) reduced promoter activity by 7-fold. This indicated that CBF binding to the Aurora A promoter is required for its high level promoter activity.
Figure 8.
The activity of Aurora A promoter is dependent on CBF binding to the two CCAAT motifs located within the promoter. (A) Oligonucleotide sequences containing wild or mutant CCAAT box I or II. (B) DNA-binding assay for CBF binding to CCAAT box I or II. Radiolabeled oligonucleotide probes corresponding to WT1 or WT2 and HeLa cell nuclear extracts were used for the assay. The DNA–protein complexes for both probes were supershifted by an anti-CBF-B antibody, but not by IgG. (C) Competition of DNA–protein complexes by WT1 and WT2 but not by MT1 and MT2 unlabeled oligouncleotides of 50 or 100 times excess. (D) Wild-type CBF-binding sites in the Aurora A promoter is required for the promoter activity. Wild-type Aurora A promoter (−177/+268) and mutant promoters containing one or two mutated CCAAT boxes (from CCAAT to CCAAa) were constructed into a luciferase reporter vector. They were transiently transfected into NIH 3T3 fibroblast cells to measure relative luciferase activity. This was done for a minimum of three times and in duplication each time.
Inhibition of TBP recruitment to Cyclin B1 and Aurora A promoter by Bdbd
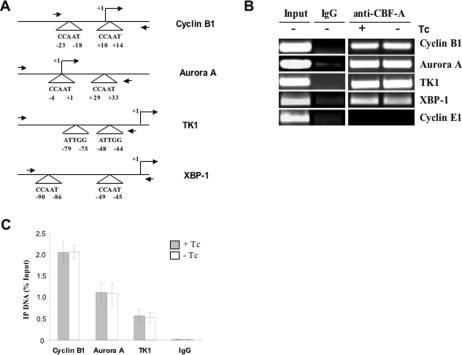
To determine whether CBF interacts with the cellular promoters of the studied genes in HeLa cells, we performed chromatin immunoprecipitation (ChIP) experiments with the anti-CBF-A antibody (Figure 9), which was used in earlier experiment. This showed that CBF interacted with the cellular promoters of both Cyclin B1 and Aurora A genes. We also performed the ChIP analysis at various time points after the release of the double thymidine block. The results showed that CBF interacted with the promoters at all of the time points (supplementary Figure S2), indicating that CBF constitutively occupied these promoters at all phases of the cell cycle, consistent with previous publications (30–32). To determine CBF occupancy after Bdbd expression, the ChIP analysis was performed after infection of HeLa cells with Ad-Bdbd in the presence and absence of Tc (Figure 9B and C). This showed that anti-CBF-A antibody pulled down equal amount of promoter DNAs with or without Tc. Since CBF-A was exclusively associated with Bdbd in cells expressing Bdbd, this indicated that CBF complex containing Bdbd binds to promoter DNAs equally to CBF complex containing full-length cellular CBF-B. As a control, Bdbd binds to the promoter of TK1 gene. However, gene expression of TK1 was not changed by Bdbd expression (Figure 7A). As another control, Bdbd also binds to the promoter of XBP-1 gene, which is usually activated by endoplasmic reticulum stress but expressed at a very low level in normal HeLa cells.
Figure 9.
Chromatin immunoprecipitation (ChIP) analysis of CBF binding to cellular promoters. (A) Schematic representation of Cyclin B1, Aurora A, TK1 and XBP-1 promoters highlighting the CCAAT motif positions in relation to the transcription start site. Two side arrows indicate primer positions for ChIP experiments. (B) The ChIP experiment was performed with a polyclonal anti-CBF-A antibody or rabbit IgG as a control using Ad-Bdbd infected HeLa cells in the presence or absence of Tc. For input, PCR amplification was done using sonicated chromatin DNA before immunoprecipitation. The promoter of Cyclin E1 gene, which does not contain CBF-binding sites, was PCR amplified as a control. (C) The amount of ChIP DNAs corresponding to the promoters of Cyclin B1, Aurora A and TK1 genes was quantified by real-time PCR with a SYBR Green approach. It is expressed as a percentage of the input DNAs used in immunoprecipitation. Data presented in the histogram is the mean for three independent experiments. Standard deviations are displayed as error bars.
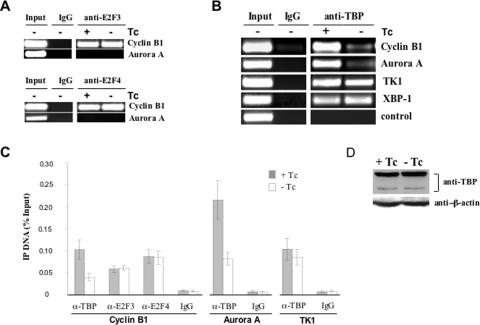
A recently published study demonstrated that a CBF-binding site in the human Cyclin B1 promoter plays a role in the recruitment of the E2F3 transcription factor during the cell cycle (31). Another recent published study also demonstrated that CBF binding to various promoters is needed for the recruitment of a general transcription factor and RNA polymerase II during inducible transcription (33). To test these two possibilities, we examined the binding of E2F3 and TATA-binding protein (TBP) to cellular promoters using ChIP method after the expression of Bdbd. To examine E2F3 binding, HeLa cells were synchronized by thymidine block and infected with Ad-Bdbd similarly as described in Figure 5A. The cells collected at 4 h of release from the second thymidine block were used for ChIP experiment. This showed that the expression of Bdbd, however, did not change binding of E2F3 to cellular Cyclin B1 promoter (Figure 10A). Similarly, binding of E2F4 to cellular Cyclin B1 promoter either did not change after the expression of Bdbd. This experiment also showed that neither E2F3 nor E2F4 interacted with the proximal promoter of cellular Aurora A gene.
Figure 10.
Chromatin immunoprecipitation (ChIP) analysis of E2F3, E2F4 and TBP binding to cellular promoters. (A) The ChIP experiment was performed with antibodies against E2F3 and E2F4 using Ad-Bdbd infected HeLa cells, which were synchronized and collected at 0 h of release similarly as described in Figure 4. (B) The ChIP experiment was performed with an anti-TBP antibody using Ad-Bdbd infected HeLa cells. A non-promoter genomic region at 3′ end of GRP78 gene was PCR amplified as a negative control. (C) The amount of ChIP DNAs was quantified and presented in the same way as for Figure 9C. (D) Analysis of TBP protein level in Ad-Bdbd infected HeLa cells in the presence or absence of Tc. Whole cell extracts were made and western blot analysis was performed with both anti-TBP and anti-β-actin antibodies. Detected amount of β-actin served as an internal control.
Binding of TBP to both Cyclin B1 and Aurora A promoters was examined by ChIP analysis in both synchronized and unsynchronized HeLa cells. This showed that TBP has equal occupancy to both Cyclin B1 and Aurora A promoters at various time points after the release of the thymidine block (Supplementary Figure S4). This indicated that similar to CBF, TBP constitutively occupied these promoters irrespective of transcription activation during the cell cycle. Interestingly, expression of Bdbd resulted in 2.3- and 2.6-fold reduction of TBP binding to Cyclin B1 and Aurora A promoters, respectively (Figure 10B and C). In contrast, TBP binding to TK1 promoter had slight reduction (1.18-fold) and its binding to XBP-1 promoter was not changed by the expression of Bdbd. As a control for this experiment, TBP only bound to the promoter region but not to a genomic region far from the start of transcription. As another control, expression of TBP was detected by western blot after Ad-Bdbd infection in the presence or absence of Tc. The results showed that the expression of Bdbd had no effect on the protein level of TBP (Figure 10D). Altogether, this study demonstrated that Bdbd significantly inhibited TBP occupancy to both Cyclin B1 and Aurora A promoters in HeLa cells.
DISCUSSION
Our study demonstrated that the expression of Bdbd containing a DNA-binding domain but lacking a transcription activation domain in various human and mouse cells inhibited cell growth and specifically arrested cells at G2/M phase of the cell cycle. The role of CBF in cell growth was previously demonstrated in various studies in which DNA binding of CBF was inhibited either by the expression of a dominant-negative CBF-B subunit or by inactivation of the CBF-B gene. Inhibition of DNA binding of CBF has been shown to arrest cell growth at all phases of the cell cycle (19–21). These studies indicated that CBF plays a role in the expression of genes required for cell cycle progression. However, the specific function of CBF in each phase of the cell cycle remains to be determined. In the present study, we unambiguously showed that the N-terminal activation domain of CBF-B is specifically required for cell cycle progression at G2/M phase.
We believe that the specific effect of Bdbd in cell cycle progression at G2/M results from the inhibition of CBF-dependent transcription activation of some essential G2/M regulatory genes. Our results showed that Bdbd was associated with cellular CBF-A/CBF-C and formed a DNA–protein complex similar to the complex formed with full-length CBF-B. The difference between the two DNA–protein complexes is that the one formed with full-length CBF-B contains two activation domains provided by CBF-B and CBF-C, whereas the one formed with Bdbd contains only one transcription activation domain provided by CBF-C. Our previous studies using an in vitro transcription system showed that Bdbd, together with full-length CBF-A/CBF-C activated transcription of various promoters, but the level of activation was almost half of that by the full-length CBF-B subunit (23). This indicated that the activation domain of CBF-B contributes half of the CBF-dependent activation in vitro. From this observation one would expect that Bdbd should inhibit expression of any cellular genes, which promoters contain CBF-binding site. In contrast, our results showed that Bdbd inhibited expression of Cyclin B1 and Aurora A but not TK1, although the promoters of all three genes interacted with CBF which was not altered by Bdbd expression as shown by ChIP analysis. This suggests that the activation domain of CBF-B plays a specific role in transcription activation of Cyclin B1 and Aurora A genes.
The gene expression analysis we performed during progression of the cell cycle showed that Bdbd primarily inhibited transcription activation of Cyclin B1, Aurora A and CDK1, which occurred from late S to G2/M phase of the cell cycle in HeLa cells. This also showed that Bdbd modestly inhibited activation of FOXM1 and Cyclin D1, but did not inhibit the expression of Cyclin E1 and TK1 (Figure 7A). Recent studies have shown that the inhibition of expression of either Cyclin B1 or Aurora A leads to cell cycle arrest at G2/M phase. The protein products of these genes play important roles in entry into mitosis and chromosome segregation (3,4). Similarly, inhibition of FOXM1 expression also resulted in cell cycle arrest at G2/M phase (34,35). Several studies demonstrated that the inhibition of DNA binding of CBF resulted in decreased expression of Cyclin A, CDC25C and topoisomerase IIα, whose expression is also activated at late S and G2/M phase (19,21), suggesting a possibility that Bdbd could also inhibit the expression of these additional genes in HeLa cells. Altogether, this suggests that the G2/M arrest by Bdbd is likely due to accumulative effect of the inhibition of multiple genes expression required for G2/M progression. Our gene expression analysis showed that Bdbd did not inhibit the expression of Cyclin E1, which is activated at G1/S phase. Our analysis, however, did not extensively examine the expression of genes at G1/S phase of the cell cycle. Since Bdbd did not inhibit cell cycle progression at G1/S phase, we speculate that Bdbd did not play a significant role in the expression of G1/S phase genes. In addition, previous study showed that inactivation of CDK1 in human cells resulted in initial accumulation at G2/M phase followed by increase of polyploidy without entering into mitosis (36). This study interpreted that the polyploidy without mitosis was developed due to re-replication of chromosome. Since Bdbd also reduced the expression of CDK1, which could partly explain the development of polyploidy in Bdbd expressing cells.
Analysis of Aurora A promoter showed that CBF binding to Aurora A promoter is needed for Aurora A promoter activity. Furthermore, ChIP analysis demonstrated that CBF constitutively occupied cellular Aurora A promoter at various stages of the cell cycle, indicating that CBF directly regulates transcription of Aurora A gene. Similarly, CBF also constitutively occupied cellular Cyclin B1 promoter, indicating that the activation of Cyclin B1 and Aurora A genes at G2/M is not due to increase in binding of CBF to these promoters.
A recently published promoter analysis demonstrated that CBF-binding site in CDK1 and Cyclin B1 promoters plays a role in the recruitment of the E2F3 transcription factor specifically at G2/M phase of the cell cycle (31). Although the function of the E2F activators (E2F1, E2F2 and E2F3) has been known to be a specific activator for S phase-specific genes, the function of these activators in transcription at G2/M phase has also been revealed in several publications (30,31,37). Our ChIP analysis showed that E2F3 interacts with cellular Cyclin B1 but not with Aurora A promoter. Currently it is not known whether E2F3 binds to Aurora A promoter. It is possible that E2F3-binding site is located far from CBF-binding sites in the Aurora A promoter; thus, it could not be detected by our ChIP analysis detecting only proximal promoter region. Nonetheless, we compared E2F3 binding to Cyclin B1 promoter with or without Bdbd expression. This showed that Bdbd did not change E2F3 binding to Cyclin B1 promoter. In contrast, Bdbd inhibited TBP binding to Cyclin B1 promoter. Similarly, Bdbd also inhibited TBP binding to Aurora A promoter. As a control, Bdbd caused slight inhibition of TBP binding to TK1 promoter and no inhibition to XBP-1 promoter, even though CBF also binds to both promoters. This indicated that Bdbd primarily inhibited TBP binding to Cyclin B1 and Aurora A promoters. Since Bdbd did not inhibit DNA binding of CBF, it is possible that only DNA binding of CBF is sufficient for E2F3 recruitment to Cyclin B1 promoter, whereas both DNA binding and activation domains of CBF are required for TBP binding to Cyclin B1 and Aurora A promoters.
In this regard, a recent publication demonstrated that CBF binding to an osteoclast differentiation factor (ODF) promoter is essential for activation of the ODF gene in the presence of vitamin D3, parathyroid hormone and prostaglandin E (33). Interestingly, this publication showed that CBF is also needed for the recruitment of TBP and RNA polymerase II to the cellular ODF promoter. Thus, the authors of this publication hypothesized that CBF regulates inducible transcription through recruitment of general transcription factor and RNA polymerase II. Similarly, CBF binding to a human γ-globin promoter is also required for the recruitment of TBP and RNA polymerase II to the promoter in adult erythroblasts but not in embryonic erythroid cells (38). This suggested a possibility that CBF controls inducible or developmentally specific transcription of various genes through recruitment of TBP and other general transcription factors. Based on our present study, we propose that the activation domain of CBF-B controls such recruitment of TBP and possibly other general transcription factors to the cellular promoters. However, we do not consider this function as a general property of CBF, since Bdbd significantly inhibited TBP recruitment to Cyclin B1 and Aurora A promoters but not to TK1 and XBP-1 promoters which also interacted with CBF.
To test whether CBF directly interacts with TBP, we had analyzed a purified CBF-B complex from HeLa cells using immunoaffinity method (26). Our previous analysis of CBF-B complex by mass spectrometry showed that CBF-B associated with CBF-A, CBF-C, and P32 but no TBP. Now we also analyzed the purified CBF-B complex by western blot method using anti-TBP antibody. This also showed that no TBP was associated with the CBF-B complex (data not shown). These results indicated that TBP does not directly interact with CBF in HeLa cells. Our study is also in agreement with a previous publication that showed CBF does not interact with TBP in a human γ-globin promoter, although CBF is required for TBP recruitment to the promoter (38). We speculate that the activation domain of CBF regulates the recruitment of TBP through an indirect mechanism, possibly required another factor that binds to Cyclin B1 and Aurora A promoters.
As mentioned in Introduction, comparative genomic analysis showed that a conserved promoter element consisting of CBF-binding site and two other elements, cell cycle-dependent element (CDE) and cell cycle homology region (CHR) is found in several human promoters that are specifically activated at G2/M phase of cell cycle (31). However, nuclear protein(s) that bind to CDE or CHR have not been identified. We speculate that CBF could cooperate with CDE- or CHR-binding protein(s) to recruit TBP and to activate transcription at G2/M phase. Future study of CDE- and CHR-binding protein(s) could elucidate specific role of the CBF activation domain in transcription activation at G2/M phase.
It is also possible that this specific function of CBF is dependent on promoter architecture. Both Cyclin B1 and Aurora A promoters contain CBF-binding sites, which are located very close to the start of transcription. However, none of these two promoters contains putative TATA box, the typical TBP-binding site. This suggests that TBP indirectly binds to Cyclin B1 and Aurora A promoters possibly through interaction with TBP-associated factors, TAFs, which are known to interact directly with TATA-less promoters (39). We speculate that due to close proximity of CBF binding to the start of transcription, the activation domain of CBF-B could regulate such indirect binding of TBP to the promoter DNAs. In this regard, it has been reported that CBF interacts with TBP and several TAFs, components of TFIID complex in vitro (40), suggesting that the activation domain of CBF-B could directly interact with TFIID complex and thus stabilize its binding to the cellular Cyclin B1 and Aurora A promoters.
In summary, our study demonstrated that the activation domain of CBF-B is specifically needed for the cell cycle progression at G2/M phase, and it plays a vital role in the cell cycle-dependent transcription activation of Cyclin B1 and Aurora A genes, which are very essential for cell cycle progression at G2/M phase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Henry Adams for assisting with the immunofluorescence studies and Wendy Schober-Ditmore with technical assistance in flow cytometry. We also thank Donald R. Norwood for editing this manuscript. This work was supported partly by National Institutes of Health Grants RO1 AR46264 (to S.N.M.) and RO1 CA89716 (to S.S.), and a Living Legend Allocation for Molecular Genetics and Developmental Biology Priority Program, a MRP project, and an Institutional Research Grant from The University of Texas M. D. Anderson Cancer Center (to S.N.M.). The DNA sequencing and flow cytometry facilities at The University of Texas M. D. Anderson Cancer Center are supported by a National Cancer Institute Grant CA 16672. Funding to pay the Open Access publication charges for this article was provided by an Institutional Research Grant from The University of Texas M. D. Anderson Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doree M., Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J. Cell. Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- 2.Fung T.K., Poon RY. A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 2005;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Katayama H., Sasai K., Kawai H., Yuan Z.M., Bondaruk J., Suzuki F., Fujii S., Arlinghaus R.B., Czerniak B.A., Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nature Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J.P., Yan R.L., Kramer A., Eckerdt F., Roller M., Kaufmann M., Strebhardt K. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23:5843–5852. doi: 10.1038/sj.onc.1207757. [DOI] [PubMed] [Google Scholar]
- 5.Katyama H., Brinkley W.R., Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 6.Whitfield M.L., George L.K., Grant G.D., Perou C.M. Common markers of proliferation. Nature Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 7.Zwicker J., Lucibello F.C., Wolfraim L.A., Gross C., Truss M., England K., Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcription respression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manni I., Mazzaro G., Gurtner A., Mantovani R., Haugwitz U., Krause K., Engeland K., Sacchi A., Soddu S., Piaggio G. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 2001;276:5570–5576. doi: 10.1074/jbc.M006052200. [DOI] [PubMed] [Google Scholar]
- 9.Hu Q., Bhattacharya C., Maity S.N. CCAAT binding factor (CBF) binding mediates cell cycle activation of topoisomerase IIalpha. Conventional CBF activation domains are not required. J. Biol. Chem. 2002;277:37191–37200. doi: 10.1074/jbc.M205985200. [DOI] [PubMed] [Google Scholar]
- 10.Linhart C., Elkon R., Shiloh Y., Shamir R. Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle. 2005;4:1788–1797. doi: 10.4161/cc.4.12.2173. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z., Shendure J., Church G.M. Discovering functional transcription-factor combinations in the human cell cycle. Genome Res. 2005;15:848–855. doi: 10.1101/gr.3394405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maity S.N., de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 14.Yun J., Chae H.D., Choi T.S., Kim E.H., Bang Y.J., Chung J., Choi K.S., Mantovani R., Shin D.Y. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J. Biol. Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q., Zambetti G.P., Suttle D.P. Inhibition of DNA topoisomerase II alpha gene expression by the p53 tumor suppressor. Mol. Cell. Biol. 1997;17:389–397. doi: 10.1128/mcb.17.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun J., Chae H.D., Choy H.E., Chung J., Yoo H.S., Han M.H., Shin D.Y. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y., Mehta K.R., Choi A.P., Scolavino S., Zhang X. DNA damage-induced inhibition of securin expression is mediated by p53. J. Biol. Chem. 2003;278:462–470. doi: 10.1074/jbc.M203793200. [DOI] [PubMed] [Google Scholar]
- 18.Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Q., Maity S.N. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J. Biol. Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya A., Deng J.M., Zhang Z., Behringer R., de Crombrugghe B., Maity S.N. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 21.Chae H.D., Yun J., Bang Y.J., Shin D.Y. Cdk2-dependent phosphorylation of the NF-Y transcription factor is essential for the expression of the cell cycle-regulatory genes and cell cycle G1/S and G2/M transitions. Oncogene. 2004;23:4084–4088. doi: 10.1038/sj.onc.1207482. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield M.L., Zheng L.X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coustry F., Hu Q., De Crombrugghe B., Maity S.N. CBF/NF-Y functions both in nucleosomal disruption and transcription activation of the chromatin-assembled topoisomerase IIalpha promoter. Transcription activation by CBF/NF-Y in chromatin is dependent on the promoter structure. J. Biol. Chem. 2001;276:40621–40630. doi: 10.1074/jbc.M106918200. [DOI] [PubMed] [Google Scholar]
- 24.Kalinichenko V.V., Major M.L., Wang X., Petrovic V., Kuechle J., Yoder H.M., Dennewitz M.B., Shin B., Datta A., Raychaudhuri P., et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Y., Jahroudi N. The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J. Biol. Chem. 2003;278:8385–8394. doi: 10.1074/jbc.M213156200. [DOI] [PubMed] [Google Scholar]
- 26.Chattopadhyay C., Hawke D., Kobayashi R., Maity S.N. Human p32, interacts with B subunit of the CCAAT-binding factor, CBF/NF-Y, and inhibits CBF-mediated transcription activation in vitro. Nucleic Acids Res. 2004;32:3632–3641. doi: 10.1093/nar/gkh692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 28.Sirri V., Roussel P., Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 2000;148:250–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka M., Ueda A., Kanamori H., Ideguchi H., Yang J., Kitajima S., Ishigatsubo Y. Cell-cycle-dependent regulation of human aurora A transcription is mediated by periodic repression of E4TF1. J. Biol. Chem. 2002;277:10719–10726. doi: 10.1074/jbc.M108252200. [DOI] [PubMed] [Google Scholar]
- 30.Tommasi S., Pfeifer G.P. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W., Giangrande P.H., Nevins J.R. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciortino S., Gurtner A., Manni I., Fontemaggi G., Dey A., Sacchi A., Ozato K., Piaggio G. The cyclin B1 gene is actively transcribed during mitosis in HeLa cells. EMBO Rep. 2001;2:1018–1023. doi: 10.1093/embo-reports/kve223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabe Y., Yamada J., Uga H., Yamaguchi Y., Wada T., Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell. Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laoukili J., Kooistra M.R., Bras A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 35.Wang I.C., Chen Y.J., Hughes D., Petrovic V., Major M.L., Park H.J., Tan Y., Ackerson T., Costa R.H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itzhaki J.E., Gilbert C.S., Porter A.C.G. Construction by gene targeting in human cells of a ‘conditional’ CDC2 mutant that replicates its DNA. Nature Genet. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- 37.Cam H., Dynlacht B.D. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell. 2003;3:311–316. doi: 10.1016/s1535-6108(03)00080-1. [DOI] [PubMed] [Google Scholar]
- 38.Fang X., Han H., Stamatoyannopoulos G., Li Q. Developmentally specific role of the CCAAT box in regulation of human gamma-globin gene expression. J. Biol. Chem. 2004;279:5444–5449. doi: 10.1074/jbc.M306241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke T.W., Kadonaga J.T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frontini M., Imbriano C., di Silvio A., Bell B., Bogni A., Romier C., Moras D., Tora L., Davidson I., Mantovani R. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 2002;277:5841–5848. doi: 10.1074/jbc.M103651200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.