Abstract
Although Sgf73p, a yeast homologue of human Sca7p, has recently been implicated as a new component of Spt-Ada-Gcn5-acetyltransferase (SAGA), its association with SAGA and functional role in regulation of transcription remain unknown in vivo. Here, using a chromatin immunoprecipitation (ChIP) assay, we show in vivo that, like SAGA, Sgf73p is recruited to the upstream activating sequence (UAS) of a SAGA-dependent gene, GAL1, in an activator-dependent manner. Further, Sgf73p is required for recruitment of SAGA to the GAL1 UAS, and facilitates formation of the preinitiation complex (PIC) assembly at the GAL1 promoter. When PIC is not formed in Δsgf73, histone H3 is not evicted from the GAL1 promoter. Interestingly, PIC formation at GAL1 is not regulated by histone H3 acetylation or histone acetyltransferase (HAT) activity of SAGA. Similarly, Sgf73p facilitates PIC formation at another SAGA-dependent gene, ADH1, independent of histone H3 acetylation or HAT. In contrast, Sgf73p stimulates PIC formation at PHO84 (a SAGA-dependent gene), in a HAT-dependent-manner. Collectively, our data reveal that Sgf73p is required for SAGA recruitment, and stimulates PIC formation either in a HAT-dependent or -independent manner, providing significant information on how Sgf73p and possibly human Sca7p function physiologically.
INTRODUCTION
Spt-Ada-Gcn5-acetyltransferase (SAGA), a large multi-protein complex with Gcn5p-HAT (histone acetyltransferase) activity, is required for the normal transcription of ∼10% of yeast genes (1). The role of SAGA in transcriptional activation has been studied extensively at GAL1 (2–8). At the GAL1 promoter, the activator Gal4p first recruits SAGA to the upstream activating sequence (UAS), and then the UAS-bound SAGA facilitates recruitment of TATA-box binding protein (TBP) to the core promoter, thereby stimulating formation of preinitiation complex (PIC) and hence transcription (5,7,8).
Recently, Sgf73p has been biochemically identified as a new component of SAGA (9,10). Sgf73p is a yeast homologue of the human Spinocerebellar ataxia type 7 (SCA7) gene product, ataxin-7 or Sca7p. It is one of the eight known autosomal dominant neurodegenerative disorders which are caused by CAG (that encodes glutamine, Q) nucleotide repeat expansions from <35 (non-pathogenic) to an extreme of 300 (pathogenic) copies (11–15). However, it is not known how CAG nucleotide repeat expansion within ataxin-7 causes neurodegenerative diseases. Recent biochemical studies (16,17) have demonstrated that, like Sgf73p, wild-type ataxin-7 is a component of mammalian SPT3-TAFII31-GCN5L acetylase (STAGA) transcription coactivator complex (18,19), a human homologue of yeast SAGA complex, and it regulates HAT activity of STAGA and its interaction with activators. The poly(Q)-expanded ataxin-7 gets incorporated into STAGA as well as SAGA (10,16), but inhibits Gcn5p-HAT activities of both STAGA and SAGA (10,16), and thus alters gene expression, providing significant information on the molecular basis of poly(Q) disorders.
Although previous biochemical studies (10,16,17) have defined the functional roles of Sgf73p and ataxin-7 in maintaining the overall structural integrities of SAGA and STAGA, respectively, and their HAT activities, the molecular role of Sgf73p or ataxin-7 in transcriptional regulation remains mostly unknown in vivo. Here, using a formaldehyde-based in vivo crosslinking and chromatin immunoprecipitation (ChIP) assay, we have analyzed the role of Sgf73p in recruitment of SAGA and formation of the PIC assembly (and hence transcription) at the SAGA-dependent promoter in Saccharomyces cerevisiae. Further, we have determined the role of histone H3 acetylation or the HAT activity of SAGA in regulation of the PIC formation at the SAGA-dependent promoters in vivo. Our results reveal that Sgf73p is required for SAGA recruitment and stimulates formation of the PIC assembly at the SAGA-dependent promoters. Consistently, transcription of the SAGA-regulated genes is significantly impaired in Δsgf73. Interestingly, PIC formation at the SAGA-regulated genes is differentially regulated by histone H3 acetylation or the HAT activity of SAGA in vivo.
MATERIALS AND METHODS
Plasmids
Plasmid SGP4 was generated by cloning a DNA fragment containing three Gal4p-binding sites into the low copy number plasmid pRS416 (5). The plasmid pFA6a-13Myc-KanMX6 (20) was used for genomic myc epitope-tagging of the proteins of interest. The Plasmid pRS416 (21) was used in the PCR-based gene disruption.
Yeast strains and media
Yeast strain harboring null mutation in GCN5 (FY1370), and its isogenic wild-type equivalent FY1369 were obtained from Fred Winston (Harvard Medical School, Boston, MA) (2). Multiple myc-epitope tags were added at the original chromosomal locus of SGF73 in W303a to generate ASY3 (Sgf73p-myc, Kan). The plasmid SGP4, carrying three Gal4p-binding sites, was transformed into ASY3 to generate ASY6. The endogenous SGF73 gene of W303a was disrupted using the PCR-based gene knock-out method (22) to generate ASY9 (Δsgf73::URA3). Multiple myc-epitope tags were added at the original chromosomal locus of SPT20 in W303a and ASY9 to generate ASY10 (Spt20p-myc, Kan) and ASY13 (Spt20p-myc, Kan; Δsgf73), respectively.
For the studies at the GAL1 promoter in Δsgf73 and Δgcn5 mutant strains, and their isogenic wild-type equivalents, cells were first grown in YPR (yeast extract containing peptone plus 2% raffinose) to an optical density at 600 nm (OD600) of 0.9 and then transferred to YPG (yeast extract–peptone plus 2% galactose) for 90 min at 30°C prior to formaldehyde crosslinking. Minimal media containing either 2% galactose or raffinose were used for the strain (ASY6) with reporter plasmid, SGP4. The yeast strains were grown in YPD (yeast extract containing peptone plus 2% dextrose) to an OD600 of 1.0 at 30°C for the studies at the ADH1, PHO84 and RPS5 promoters.
ChIP assay
The ChIP assay was performed as described previously (23,24). Briefly, yeast cells were treated with 1% formaldehyde, collected and resuspended in lysis buffer. Following sonication (that generates the average size of ∼400 bp of sheared DNA), cell lysates (400 μl lysate from 50 ml of yeast culture) were precleared by centrifugation, and then 100 μl lysate was used for each immunoprecipitation (IP). Immunoprecipitated (IP) protein–DNA complexes were treated with proteinase K, the crosslinks were reversed, and the DNA was purified. IP DNA was dissolved in 20 μl TE 8.0 [10 mM Tris–HCl (pH 8.0) and 1 mM EDTA], and 1 μl of IP DNA was analyzed by PCR. The PCR (a total of 23 cycles) contained [α-32P]dATP (2.5 μCi for each 25 μl reaction) and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, ‘input’ DNA was isolated from 5 μl lysate without going through the IP step and suspended in 100 μl TE 8.0. To compare the PCR signal arising from the IP DNA with the input DNA, 1 μl of input DNA was used in PCR analysis.
For analysis of histone H3 levels at the promoters, we modified the above ChIP protocol as follows. Eight hundred microlitre lysate was prepared from 100 ml of yeast culture. Four hundred microlitre lysate was used for each IP (using 5 μl of anti-histone H3 antibody from Abcam and 100 μl of protein A/G plus agarose beads from Santa Cruz Biotechnology, Inc.), and IP DNA sample was dissolved in 10 μl TE 8.0 of which 1 μl was used in PCR analysis. In parallel, the PCR for ‘input’ DNA was performed using 1 μl DNA that was prepared by dissolving purified DNA from 5 μl lysate in 1000 μl TE 8.0.
Serial dilutions of the input and IP DNAs were used to asess the linear range of PCR amplification as described previously (6). The PCR data presented in this paper are within the linear range of PCR analysis. All the ChIP experiments were repeated at least three times and consistent results were obtained. Primer-pairs used for PCR analysis were as follows: GAL1(UAS): 5′-CGCTTAACTGCTCATTGCTATATTG-3′, 5′-TTGTTCGGAGCAGTGCGGCGC-3′; GAL1(Core): 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′, 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; ADH1(Core): 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′, 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′; PHO84(Core): 5′-GATCCACTTACTATTGTGGCTCGT-3′, 5′-GTTTGTTGTGTGCCCTGGTGATCT-3′; RPS5(Core): 5′-GGCCAACTTCTACGCTCACGTTAG-3′, 5′-CGGTGTCAGACATCTTTGGAATGGTC-3′; GAL4 [open reading frame (ORF)]: 5′-CTTGTTCAATGCAGTCCTAGTACCC-3′, 5′-CACAAGTCTGGATTTTAAAAGTGGCC-3′.
Primers flanking Gal4p-binding sites in the plasmid SGP4 are 5′-GGTGGCGGCCGCTCTAGAACTAGT-3′ and 5′-TTGACCGTAATGGGATAGGTCACG-3′.
Autoradiograms were scanned and quantitated by the National Institutes of Health image 1.62 program. The ratio of IP DNA over the input was presented as %IP. For mutant strains, DNA IP relative to wild-type (WT) was presented as %WT.
Primer extension analysis
Primer extension analysis was performed as described previously (5–7,23,25,26). Briefly, total RNA was prepared from 10 ml of yeast culture. Fifteen micrograms of total RNA was used in the primer extension analysis using 32P-labeled primer and AMV reverse transcriptase (Promega). The primers used for analysis of GAL1, ADH1, PHO84 and RPS5 mRNAs were as follows: GAL1: 5′-CCTTGACGTTACCTTGACGTTAAAGTATAGAGG-3′; ADH1: 5′-TATCCTTGTGTTCCAATTTACCGTGG-3′; PHO84: 5′-GAAGACTTCTTTCAGCAACATG-3′; RPS5: 5′-GACTGGGGTGAATTCTTCAACAACTTC-3′.
RESULTS
Sgf73p is an integral component of SAGA in vivo
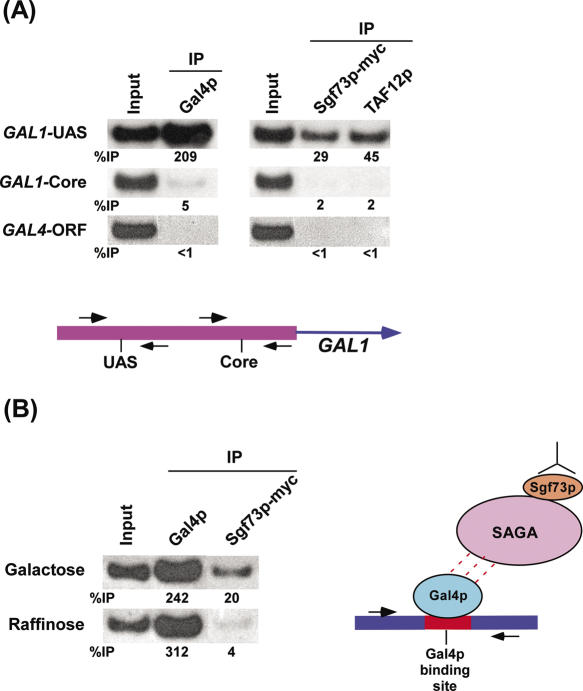
Yeast SAGA is a large multi-protein complex that contains Ada1p, Ada2p, Ada3p, Ada5p/Spt20p, Spt3p, Spt7p, Spt8p, Gcn5p, Tra1p, TAF5p, TAF6p, TAF9p, TAF10p, TAF12p, Ubp8p, Chd1 and Sgf11 (9,27–35). Recently, Sgf73p has been biochemically implicated as a new component of SAGA (10). To confirm whether Sgf73p is a new component of SAGA in vivo, we analyzed recruitment of Sgf73p to the UAS of a well-characterized SAGA-dependent gene, GAL1, using a ChIP assay. We used either a c-myc mouse monoclonal antibody and yeast strains expressing a C-terminal myc epitope-tagged Sgf73p or polyclonal antibody to SAGA TAF (e.g. TAF12p). Two sets of promoter-specific primer-pairs were used that, as shown below in Figure 1A, could distinguish binding to the GAL1 UAS or core promoter. If Sgf73p is an integral component of the SAGA complex, it would be recruited to the GAL1 UAS, but not core promoter, since previous ChIP studies (5) have demonstrated recruitment of SAGA to the GAL1 UAS.
Figure 1.
Recruitment of Sgf73p to the GAL1 UAS or minimal Gal4p binding site. (A) Recruitment of Sgf73p to the UAS, but not core promoter, of the GAL1 gene. Yeast strain expressing myc-epitope tagged Sgf73p was grown at 30°C in 1% yeast extract containing 2% peptone plus 2% galactose (YPG) up to an OD600 of 1.0 prior to formaldehyde-based in vivo crosslinking. The chromatin (IP) was performed as previously described (23,24). Primer-pair located at the UAS or core promoter of the GAL1 gene (see Materials and Methods section) was used for PCR analysis of the (IP) DNA samples. A PCR fragment corresponding to the GAL4 ORF was used as a control for background binding. (IP) was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz Biotechnology, Inc.) or a polyclonal antibody against TAF12p (from Michael R. Green, University of Massachusetts Medical School). A mouse monoclonal antibody against the DNA binding domain of Gal4p (RK5C1; Santa Cruz Biotechnology, Inc.) was used. The primer-pairs are indicated on the left. The ratio of immunoprecipitate over the input is indicated below each band as %IP. (B) Recruitment of Sgf73p to the minimal Gal4p-binding sites. The plasmid containing minimal Gal4p-binding sites was transformed into the yeast strain expressing myc-epitope tagged Sgf73p. Yeast strain thus generated, was grown at 30°C in 1% yeast extract containing 2% peptone plus 2% galactose or raffinose. The ChIP analysis was performed as described in (A). The primers used for the PCR analysis are adjacent to the Gal4p-binding sites in the plasmid (see Materials and Methods section). The growth media are indicated on the left.
Figure 1A shows that under condition in which Gal4p activation domain was active (galactose-containing growth medium), the TAF12p subunit of SAGA as well as Sgf73p were present at the GAL1 UAS. Similarly, Gal4p was recruited to the GAL1 UAS (Figure 1A). Significantly, these same proteins were not associated with the GAL1 core promoter as well as an irrelevant DNA sequence (GAL4 ORF) (Figure 1A). These observations supported the possibility that Sgf73p is a component of SAGA in vivo. However, because these experiments were performed with the intact GAL1 promoter, it remained possible that other transcription components, such as general transcription factors (GTFs) played a role in recruitment of Sgf73p to the UAS. To address this issue, we asked whether Sgf73p could be recruited by Gal4p to a plasmid bearing only Gal4p-binding sites, but not other promoter elements. Figure 1B shows that Sgf73p was recruited to a plasmid bearing Gal4p-binding sites in galactose-containing growth medium in which Gal4p activation domain was active. Significantly, Sgf73p was not associated with the Gal4p-binding sites when Gal4p activation domain was inactive (raffinose-containing growth medium) even though the level of Gal4p recruitment was almost equal to that in galactose-containing growth medium (Figure 1B). Further, our previous studies (5) have demonstrated that, like Sgf73p, other SAGA components, such as Spt3p and Spt20p, were also recruited to the minimal Gal4p-binding sites in the plasmid in galactose-containing growth medium. Significantly, the GTFs, such as TBP, TFIIB, Mediator and RNA polymerase II, were not recruited to the minimal Gal4p binding sites in the plasmid (5). Taken together, our data provide evidence that Sgf73p is a new component of the SAGA complex in vivo, consistent with previous biochemical data (9,10).
Sgf73p regulates the integrity of SAGA in vivo
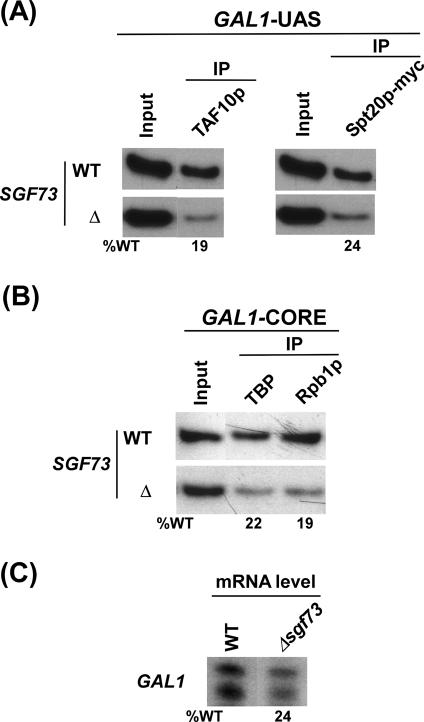
To understand the role of the newly identified Sgf73p component of SAGA in transcriptional activation in greater detail, it is necessary to determine its contribution in maintaining the integrity of SAGA. Recently, McMahon et al. (10) have demonstrated biochemically that Sgf73p is required to maintain SAGA integrity. Similarly, if Sgf73p maintains SAGA integrity in vivo, Spt20p and other SAGA components will not be recruited to the GAL1 UAS in the Δsgf73 strain. To test this idea, we analyzed recruitment of SAGA to the GAL1 UAS in the SGF73 deletion mutant. Figure 2A shows that the deletion of SGF73 significantly lowered recruitment of the Spt20p and TAF10p components of SAGA to the GAL1 UAS. Similarly, our previous studies (5,6) have also demonstrated that recruitment of SAGA was lost in the SPT20 [that integrates SAGA (3)] deletion mutant strain. On the other hand, recruitment of the SAGA components such as TAF10p, TAF12p and Spt3p was not altered in the deletion mutant of GCN5 (5) that is dispensable for the integrity of SAGA (3). Since Spt20p maintains the integrity of SAGA (3,5) and its recruitment was significantly reduced in the absence of Sgf73p (Figure 2A), thus Sgf73p, like Spt20p, seems to regulate the structural integrity of the SAGA complex in vivo, consistent with previous biochemical data (10). However, our ChIP data also raised the possibility that Sgf73p might be required for recruitment of Spt20p (and hence SAGA) to the GAL1 UAS by its interaction with the activator, Gal4p. In fact, our previous studies (7) have demonstrated that SAGA is recruited to the GAL1 UAS by interaction of its Tra1 subunit with Gal4p in vivo. Thus, it is more likely that Sgf73p is required to maintain the integrity of SAGA in vivo, consistent with previous biochemical studies (10).
Figure 2.
Requirement of Sgf73p for recruitment of SAGA and formation of the PIC assembly at GAL1. (A) Sgf73p is required for recruitment of SAGA to the GAL1 UAS. The SGF73 deletion mutant and its isogenic wild-type equivalent were first grown in raffinose-containing (YPR) and then shifted to galactose-containing (YPG) growth media 90 min before treatment with formaldehyde. (IPs) were performed as described in Figure 1A. A polyclonal antibody against the TAF10p (from Michael R. Green, University of Massachusetts Medical School) was used. Primer-pair located at the GAL1 UAS was used for PCR analysis of the IP DNA samples. The percentage of DNA immunoprecipitated relative to wild-type (%WT) is indicated below each band of the mutant strains. (B) Sgf73p is required for formation of the PIC assembly at the GAL1 core promoter. Yeast strains were grown, crosslinked and immunoprecipitated as in panel A. Primer-pair located at the GAL1 core promoter was used for PCR analysis of the immunoprecipitated DNA samples. IPs were performed using polyclonal antibodies against TBP (obtained from Michael R. Green, University of Massachusetts Medical School) and the mouse monoclonal antibody 8WG16 (Covance) against the carboxy terminal domain of the RNA polymerase II large subunit (Rpb1p). (C) Transcription. Total cellular RNA was prepared from the wild-type or Δsgf73, and mRNA level from the GAL1 gene was quantitated by primer-extension. The percentage transcription relative to wild-type is indicated below.
Sgf73p facilitates formation of the PIC assembly at the core promoters of the SAGA-dependent genes in vivo
To better understand the role of Sgf73p in formation of the PIC assembly and hence transcriptional activation, we analyzed its contribution in recruitment of TBP and RNA polymerase II (two important components of the PIC assembly) to the GAL core promoter. Figure 2B shows that recruitment of TBP and the largest subunit (Rpb1p) of RNA polymerase II to the GAL1 core promoter was dramatically reduced in the SGF73 deletion mutant strain. Consistently, transcription of GAL1 was significantly impaired in Δsgf73 (Figure 2C). Thus, the Sgf73p component of SAGA regulates GAL1 transcription by facilitating formation of PIC at the core promoter. However, formation of the PIC assembly at the GAL1 core promoter was not completely impaired in Δsgf73. On the other hand, our previous studies (5,6) have demonstrated that PIC formation at the GAL1 core promoter was completely impaired in the absence of Spt20p which maintains SAGA integrity. Such a discrepancy in PIC formation in the absence of Sgf73p or Spt20p is due to the fact that the loss of Spt20p leads to complete impairment of SAGA recruitment to the GAL1 UAS (5,6) while SAGA recruitment was not completely lost in Δsgf73 (Figure 2A). Consistently, SAGA has been shown biochemically to be completely disintegrated in the absence of Spt20p but not Sgf73p (3,10).
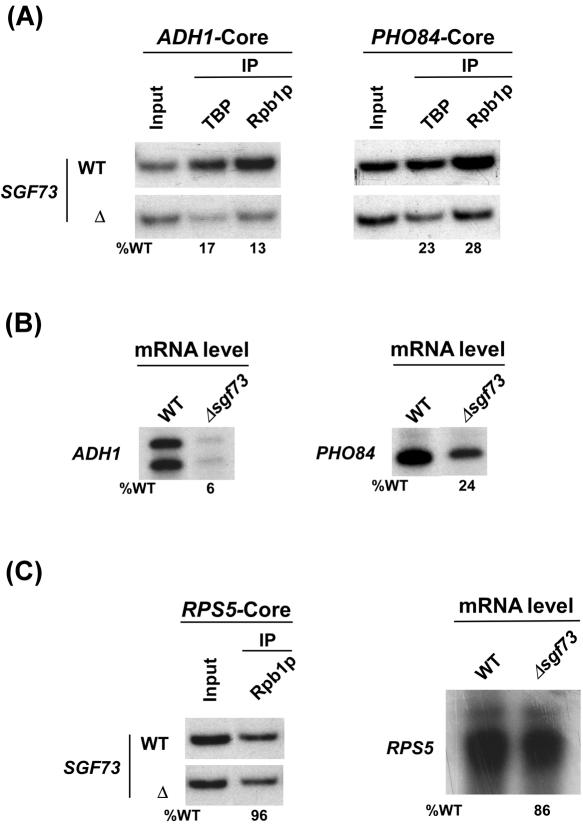
Since Sgf73p regulates recruitment of SAGA, the formation of the PIC assembly at the SAGA-dependent promoters in general would be altered in Δsgf73. To test this idea, we analyzed recruitment of TBP and RNA polymerase II to the core promoters of two other SAGA-dependent genes, namely ADH1 and PHO84 (6). Figure 3A shows that recruitment of TBP and RNA polymerase II to the ADH1 and PHO84 core promoters was significantly reduced in Δsgf73. Consistently, transcription of these genes was also altered in the SGF73 deletion mutant strain (Figure 3B). As a negative control, we demonstrate that the formation of the PIC assembly (and hence transcription) at a SAGA-independent gene, RPS5, was not altered in Δsgf73 (Figure 3C). Thus, SAGA-associated Sgf73p regulates gene expression by stimulating formation of the PIC assembly in vivo. However, such a function of Sgf73p in PIC formation seems to be mediated by the regulatory role of Sgf73p in recruitment of SAGA or its integrity. In addition, Sgf73p may also contact with the component(s) of the PIC assembly to promote PIC formation.
Figure 3.
Sgf73p is required for PIC formation (and hence transcriptional activation) at the ADH1 and PHO84 promoters. (A) The analysis of the PIC assembly at the ADH1 and PHO84 core promoters in Δsgf73. The yeast strains were grown in YPD at 30°C up to an OD600 of 1.0 prior to crosslinking. IPs were performed as described in Figure 2B. Primer-pairs located at the ADH1 and PHO84 core promoters were used for PCR analysis of the immunoprecipitated DNA samples. (B) Transcription. Total cellular RNA was prepared from the wild-type or Δsgf73, and mRNA levels from the ADH1 and PHO84 genes were quantitated by primer-extension. (C) Sgf73p is not required for PIC formation (and hence transcription) at the RPS5 promoter. Left panel: Analysis of recruitment of RNA polymerase II at the RPS5 core promoter in the wild-type and SGF73 deletion mutant strains. Both the wild-type and deletion mutant strains were grown, crosslinked and immunoprecipitated as in panel A. Primer-pair located at the RPS5 core promoter was used for PCR analysis of the IP DNA samples. Right panel: Transcription. Total cellular RNA was prepared from the wild-type or Δsgf73, and mRNA level from the RPS5 gene was quantitated by primer-extension.
Formation of the PIC assembly is differentially regulated by HAT or histone H3 acetylation in vivo
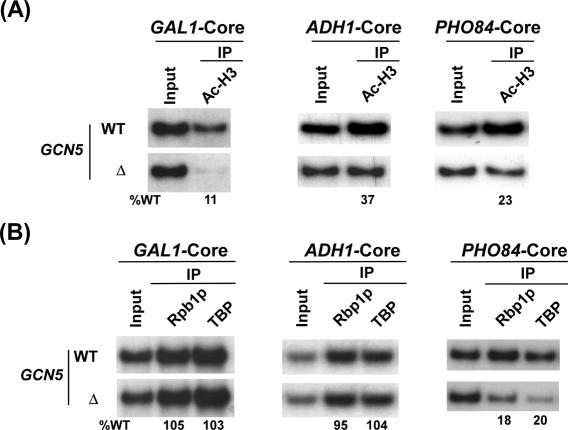
SAGA-associated Gcn5p acetylates primarily histone H3 at K9 (lysine 9) and K14. Histone H3 acetylation or the HAT activity of SAGA has been previously implicated in regulation of transcriptional initiation (3,36–41). However, how histone H3 acetylation or HAT regulates PIC formation and hence transcriptional initiation is not clearly known in vivo. Thus, we asked whether formation of Sgf73p-mediated PIC assembly at the core promoters of the aforementioned SAGA-dependent genes is dependent on histone H3 acetylation or HAT. To address this question, we analyzed recruitment of TBP and RNA polymerase II to the core promoters of the GAL1, PHO84 and ADH1 genes in the GCN5 deletion mutant and its isogenic wild-type equivalent. Figure 4A shows that the GAL1, PHO84 and ADH1 core promoters were acetylated at histone H3. Histone H3 acetylation at these core promoters were significantly reduced in Δgcn5. Interestingly, such a reduction in the level of histone H3 acetylation did not alter recruitment of RNA polymerase II and TBP to the GAL1 and ADH1 core promoters (Figure 4B). Thus, at these two promoters, Sgf73p facilitates formation of the PIC assembly in a HAT-independent-manner. In contrast, recruitment of RNA polymerase II and TBP to the PHO84 core promoter was dependent on histone H3 acetylation or Gcn5p-HAT (Figure 4A and B), demonstrating the requirement of histone H3 acetylation or HAT for formation of the PIC assembly. Thus, our data demonstrate that histone H3 acetylation or the HAT activity of SAGA is differentially required for formation of the PIC assembly in vivo, revealing a complex regulation of the PIC formation.
Figure 4.
Histone H3 acetylation differentially regulates formation of the PIC assembly at the core promoters of the SAGA-dependent genes. (A) The levels of histone H3 acetylation at the GAL1, ADH1 and PHO84 promoters are significantly reduced in the GCN5 deletion mutant. The wild-type and GCN5 deletion mutant strains were grown and crosslinked as described in Materials and Methods section. IP was performed using a rabbit polyclonal antibody against Lys-9/14 di-acetylated H3 (06–599, Upstate Biotechnology, Inc.). (B) Gcn5p is required for formation of the PIC assembly at the PHO84 core promoter, but not GAL1 and ADH1 core promoters. Yeast strains were grown and crosslinked as in (A). IPs were performed as in Figure 2B.
Role of Sgf73p in regulation of histone H3 acetylation or histone H3 eviction at the SAGA-dependent promoters
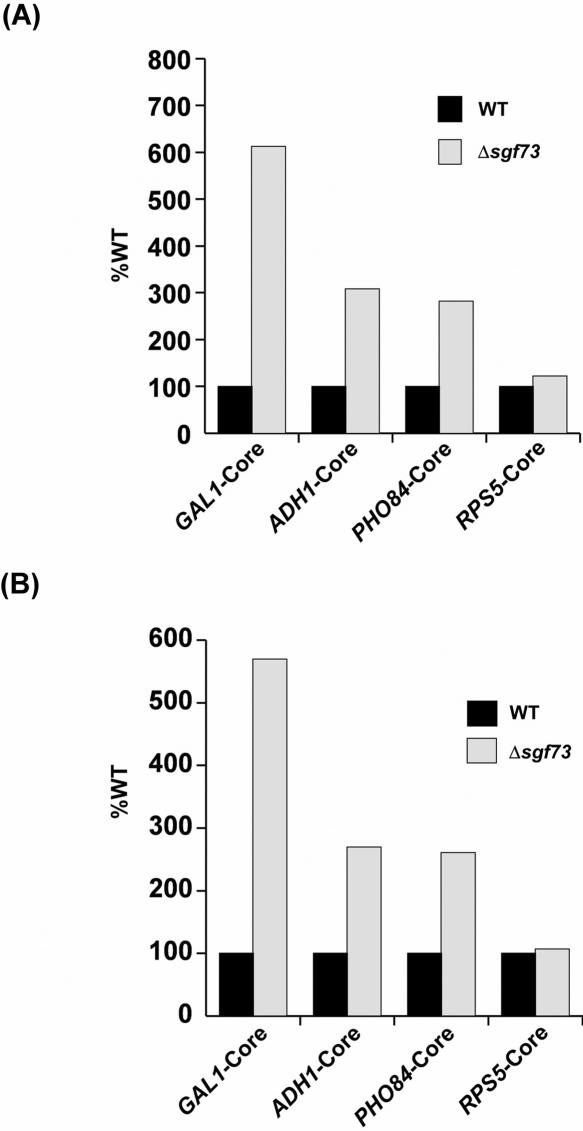
We next asked whether Gcn5p-HAT activity of SAGA is regulated by Sgf73p in vivo. Previous biochemical studies (36) have demonstrated that Gcn5p-HAT activity on nucleosomal histone H3 is dependent on the Ada2p and Ada3p components of SAGA. Similarly, deletion of SGF73 has been shown to biochemically impair the HAT activity of SAGA (10). However, it is not known whether Sgf73p regulates histone H3 acetylation at the promoters of the SAGA-dependent genes in vivo. To address this question, we analyzed the levels of histone H3 acetylation at the GAL1 core promoter in the SGF73 wild-type and deletion mutant strains. Since Sgf73p is required for recruitment of Gcn5p as well as SAGA, histone H3 acetylation at the GAL1 core promoter is expected to be significantly impaired in Δsgf73. Interestingly, our ChIP data demonstrate the elevated level of histone H3 acetylation at the GAL1 core promoter in Δsgf73 when compared with wild-type equivalent (Figure 5A). Likewise, the elevated levels of histone H3 acetylation were also observed at the ADH1 and PHO84 core promoters in Δsgf73 (Figure 5A). In contrast, histone H3 acetylation level at the core promoter of a SAGA-independent RPS5 gene was not altered in the absence of Sgf73p (Figure 5A). Since H3-K9/14 acetylation antibody also recognizes unacetylated histone H3 (Upstate biotechnology, Inc.; Figure 4A), the increase in the levels of histone H3 acetylation at the core promoters of GAL1, ADH1 and PHO84 in Δsgf73 may be caused by the elevated levels of histone H3 at the core promoters of these genes, possibly because of reduction of histone H3 eviction in the absence of the PIC formation or active transcription (37). To test this possibility, we analyzed the levels of histone H3 at the GAL1, ADH1 and PHO84 core promoters in the SGF73 deletion mutant strain and its isogenic wild-type equivalent. Figure 5B shows that the levels of histone H3 at the core promoters of GAL1, ADH1 and PHO84, but not RPS5, were elevated in Δsgf73. Interestingly, the increase in the level of histone H3 acetylation is remarkably similar to that of histone H3 at each of the SAGA-dependent promoters in Δsgf73. Thus, Sgf73p does not seem to elevate the relative level of histone H3 acetylation or the HAT activity of Gcn5p in vivo. Nonetheless, our data reveal that the loss of Sgf73p lowers histone H3 eviction from the SAGA-dependent promoters in vivo.
Figure 5.
Elevation of the levels of histone H3 as well as histone H3 acetylation at the core promoters of GAL1, ADH1 and PHO84 in Δsgf73. (A) Analysis of the levels of histone H3 acetylation at the promoters of GAL1, ADH1, PHO84 and RPS5 in the SGF73 deletion mutant strain. The wild-type and SGF73 deletion mutant strains were grown, crosslinked and immunoprecipitated as in Figure 4A. The percentages of DNA immunoprecipitated in Δsgf73 relative to wild-type (%WT) at the core promoters of GAL1, ADH1, PHO84 and RPS5 are plotted in the form of a histogram. (B) Analysis of the levels of histone H3 at the promoters of GAL1, ADH1, PHO84 and RPS5 in the SGF73 deletion mutant strain. The wild-type and SGF73 deletion mutant strains were grown and crosslinked as in panel A. IP was performed using an anti-histone H3 antibody (Ab1791; Abcam) and a modified ChIP protocol as described in Materials and Methods section.
DISCUSSION
Although Sgf73p has been shown to be a new component of SAGA on the basis of the biochemical studies (9,10), it is not known whether Sgf73p is a SAGA component in vivo. If so, it would be recruited along with other SAGA components to the GAL1 UAS but not to its core promoter, since SAGA is specifically recruited to the GAL1 UAS by the activator Gal4p (5,7). Here, we show that, like SAGA, Sgf73p is recruited to the GAL1 UAS, but not core promoter, or even to the minimal Gal4p-binding sites in a plasmid, implicating Sgf73p as a SAGA component in vivo. Furthermore, we show that the deletion of SGF73 significantly impairs recruitment of the SAGA complex, consistent with biochemical requirement of Sgf73p for SAGA integrity (10). Thus, our results support the fact in vivo that Sgf73p is a SAGA component and is required for the structural integrity of SAGA, consistent with recent biochemical data (9,10).
Since Sgf73p is essential for recruitment of SAGA, the PIC would thus not be formed at the SAGA-dependent promoters in Δsgf73. Indeed, our study demonstrates that recruitment of the PIC components such as TBP and RNA polymerase II is significantly impaired at the core promoters of the three representative SAGA-dependent genes, GAL1, ADH1 and PHO84, in the absence of Sgf73p. Consistently, transcription of these genes is altered in Δsgf73. However, whether Sgf73p plays a dual role in assembling PIC by interacting with the component(s) of PIC as well as maintaining structural integrity of SAGA to regulate gene expression remains to be elucidated.
Previous studies (3,38–41) have demonstrated the functional role of histone H3 acetylation at the promoters in regulation of transcriptional initiation. Here, we demonstrate that all three SAGA-dependent promoters are acetylated at histone H3. Acetylation of histone H3 at these promoters is significantly lost in Δgcn5. However, formation of the PIC assembly at the GAL1 and ADH1 promoters is not altered in Δgcn5. Thus, our study reveals that histone H3 acetylation status does not regulate PIC formation at GAL1 and ADH1. In contrast, PIC formation is significantly reduced at the PHO84 promoter in Δgcn5, demonstrating the role of histone H3 acetylation in regulation of the PIC formation, consistent with previous studies (3,38–41). Thus, our study reveals that histone H3 acetylation differentially regulates formation of the PIC assembly. However, the molecular basis for such differential roles of histone H3 acetylation in regulation of the PIC formation remains unknown. According to the ‘histone-code’ hypothesis (42), it is likely that histone H3 acetylation in combination with other histone modifications at the GAL1 and ADH1 promoters may contribute to the role of chromatin structure in PIC formation and hence transcriptional activation.
Histones are evicted during active transcription in vivo (37). Since Sgf73p is required for formation of the PIC assembly (and hence transcriptional activation), histone eviction from the SAGA-dependent promoters would thus be significantly reduced in Δsgf73. Indeed, our study demonstrates the elevated levels of histone H3 at the SAGA-dependent promoters in the absence of Sgf73p. Thus, impairment of PIC formation in Δsgf73 decreases histone H3 eviction and hence alters chromatin structure at the SAGA-dependent promoters.
In summary, we provide evidence that Sgf73 is a component of the SAGA complex in vivo, consistent with previous biochemical data (9,10). Sgf73p regulates SAGA recruitment, and is required for the assembly of PIC at the core promoters of the SAGA-dependent genes. However, formation of the PIC assembly at the core promoters is differentially regulated by histone H3 acetylation or Gcn5p-HAT activity of SAGA. Further, impairment of PIC formation in Δsgf73 lowers histone H3 eviction from the SAGA-dependent promoters, and thus alters chromatin structure. These results provide valuable information on the functional role of yeast Sgf73p (and possibly human ataxin-7 or Sca7p) in regulation of gene expression in vivo.
Acknowledgments
We thank Michael R. Green for TBP and TAF antibodies; Fred Winston for yeast strains; and Praveena Krishnan and Kathy Yee for technical assistance. This work was supported by a National Scientist Development Grant (0635008N) from American Heart Association, a Research Scholar Grant (06–52) from American Cancer Society, three Central Research Committee Grants from SIU School of Medicine, a Faculty Seed Grant from SIU Office of Research Development and Administration, an RFA Grant from SIU Cancer Institute, and a start-up fund from SIU School of Medicine. Funding to pay the Open Access publication charges for this article was provided by the Department of Biochemistry and Molecular Biology of SIU School of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee T.I., Causton H.C., Holstege F.C., Shen W.C., Hannett N., Jennings E.G., Winston F., Green M.R., Young R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 2.Roberts S.M., Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sterner D.E., Grant P.A., Roberts S.M., Duggan L.J., Belotserkovskaya R., Pacella L.A., Winston F., Workman J.L., Berger S.L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudley A.M., Rougeulle C., Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik S.R., Green M.R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaumik S.R., Green M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaumik S.R., Raha T., Aiello D.P., Green M.R. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larschan E., Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders S.L., Jennings J., Canutescu A., Link A.J., Weil P.A. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahon S.J., Pray-Grant M.G., Schieltz D., Yates J.R., 3rd, Grant P.A. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl Acad. Sci. USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevanin G., Giunti P., Belal G.D., Durr A., Ruberg M., Wood N., Brice A. De novo expansion of intermediate alleles in spinocerebellar ataxia 7. Hum. Mol. Genet. 1998;7:1809–1813. doi: 10.1093/hmg/7.11.1809. [DOI] [PubMed] [Google Scholar]
- 12.David G., Durr A., Stevanin G., Cancel G., Abbas N., Benomar A., Belal S., Lebre A.S., Abada-Bendib M., Grid D., et al. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Hum. Mol. Genet. 1998;7:165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- 13.Del-Favero J., Krols L., Michalik A., Theuns J., Lofgren A., Goossens D., Wehnert A., Van den Bossche D., Van Zand K., Backhovens H., et al. Molecular genetic analysis of autosomal dominant cerebellar ataxia with retinal degeneration (ADCA type II) caused by CAG triplet repeat expansion. Hum. Mol. Genet. 1998;7:177–186. doi: 10.1093/hmg/7.2.177. [DOI] [PubMed] [Google Scholar]
- 14.Kaytor M.D., Duvick L.A., Skinner P.J., Koob M.D., Ranum L.P., Orr H.T. Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum. Mol. Genet. 1999;8:1657–1664. doi: 10.1093/hmg/8.9.1657. [DOI] [PubMed] [Google Scholar]
- 15.Yvert G., Lindenberg K.S., Devys D., Helmlinger D., Landwehrmeyer G.B., Mandel J.L. SCA7 mouse models show selective stabilization of mutant ataxin-7 and similar cellular responses in different neuronal cell types. Hum. Mol. Genet. 2001;10:1679–1692. doi: 10.1093/hmg/10.16.1679. [DOI] [PubMed] [Google Scholar]
- 16.Palhan V.B., Chen S., Peng G.H., Tjernberg A., Gamper A.M., Fan Y., Chait B.T., La Spada A.R., Roeder R.G. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl Acad. Sci. USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmlinger D., Hardy S., Abou-Sleymane G., Eberlin A., Bowman A.B., Gansmuller A., Picaud S., Zoghbi H.Y., Trottier Y., Tora L., et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:432–445. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez E., Kundu T.K., Fu J., Roeder R.G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 19.Martinez E., Palhan V.B., Tjernberg A., Lymar E.S., Gamper A.M., Kundu T.K., Chait B.T., Roeder R.G. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Li X.-Y., Bhaumik S.R., Green M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 24.Bhaumik S.R., Green M.R. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 2003;370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 25.Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S.R. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla A., Stanojevic N., Duan Z., Shadle T., Bhaumik S.R. Functional analysis of H2B-K123 ubiquitination in regulation of H3-K4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J. Biol. Chem. 2006;281:19045–19054. doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 27.Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 28.Grant P.A., Sterner D.E., Duggan L.J., Workman J.L., Berger S.L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 29.Brown C.E., Lechner T., Howe L., Workman J.L. The many HATs of transcription coactivators. Trends Biochem. Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 30.Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniel J.A., Torok M.S., Sun Z.W., Schieltz D., Allis C.D., Yates Y.R., 3rd, Grant P.A. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 32.Powell D.W., Weaver C.M., Jennings J.L., McAfee K.J., He Y., Weil P.A., Link A.J. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K.K., Florens L., Swanson S.K., Washburn M.P., Workman J.L. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingvarsdottir K., Krogan N.J., Emre N.C., Wyce A., Thompson N.J., Emili A., Hughes T.R., Greenblatt J.F., Berger S.L. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., 3rd, Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 36.Balasubramanian R., Pray-Grant M.G., Selleck W., Grant P.A., Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 37.Schwabish M.A., Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 39.Kurdistani S.K., Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell. Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 40.Kurdistani S.K., Tavazoie S., Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]