Abstract
Centromere identity is determined by the formation of a specialized chromatin structure containing the centromere-specific histone H3 variant CENP-A. The precise molecular mechanism(s) accounting for the specific deposition of CENP-A at centromeres are still poorly understood. Centromeric deposition of CENP-A, which is independent of DNA replication, might involve specific chromatin assembly complexes and/or specific interactions with kinetochore components. However, transiently expressed CENP-A incorporates throughout chromatin indicating that CENP-A nucleosomes can also be promiscuously deposited during DNA replication. Therefore, additional mechanisms must exist to prevent deposition of CENP-A nucleosomes during replication and/or to remove them afterwards. Here, using transient expression experiments performed in Drosophila Kc cells, we show that proteasome-mediated degradation restricts localization of Drosophila CENP-A (CID) to centromeres by eliminating mislocalized CID as well as by regulating available CID levels. Regulating available CID levels appears essential to ensure centromeric deposition of transiently expressed CID as, when expression is increased in the presence of proteasome inhibitors, newly synthesized CID mislocalizes. Mislocalization of CID affects cell cycle progression as a high percentage of cells showing mislocalized CID are reactive against αPSer10H3 antibodies, enter mitosis at a very low frequency and show strong segregation defects. However, cells showing reduced amounts of mislocalized CID show normal cell cycle progression.
INTRODUCTION
Eukaryotic centromeres are characterized by the presence of a specific histone H3 variant (CENP-A) [reviewed in (1)], which replaces canonical H3.1 in nucleosomes both in vivo and in vitro (2–6). CENP-A appears to dictate centromere identity as it is exclusively found at centromeres, recruits kinetochore components and is required for centromere function (7–12). The precise molecular mechanisms accounting for the specific deposition of CENP-A at centromeres are not well understood. It is known that targeting to centromeres is mediated by the LI/α2 region of the histone-fold domain (HFD) (3,6,13) and, contrary to canonical nucleosomes, deposition of CENP-A containing nucleosomes at centromeres is not linked to replication (14–17). However, CENP-A containing nucleosomes can also be deposited during DNA replication as expression during S phase, or over-expression, leads to its mislocalization throughout chromatin (3,11–13,18). These observations suggest that expression of CENP-A must be tightly regulated during cell cycle progression to prevent replication-dependent deposition at non-centromeric sites during S phase and, in fact, mammalian CENP-A is expressed during G2 phase (3,16). However, expression of the Drosophila homolog of CENP-A (CID) appears to take place early during S phase (18). Therefore, additional mechanisms must exist to either avoid deposition of CENP-A containing nucleosomes at non-centromeric sites during DNA replication and/or to remove them afterwards. In this paper, a CID–YFP fusion was transiently expressed from the cid promoter in Drosophila Kc cells. Our results show that proteolytic degradation restricts localization of transiently expressed CID-YFP to centromeres by, on one hand, eliminating mislocalized CID-YFP and, second, regulating available CID-YFP levels. These results are consistent with previous findings showing that, in Saccharomyces cerevisiae, a proteolysis-resistant CENP-A (Cse4–351) mutant mislocalizes throughout chromatin (19).
MATERIALS AND METHODS
Plasmid construction
cid Promoter (nucleotide position +1 to −412) (18) and cid cDNA were obtained from Drosophila melanogaster genomic DNA by PCR-amplification using appropriate primers and cloned into pEYFP-N1 (Clontech) to generate plasmid pYFP-CID, which expresses CID-YFP under the control of the own cid promoter. HFDCID (amino acid position 127 to 221) and NCID (amino acid position 1 to 124 of CID) were obtained by PCR-amplification with appropriate primers and then, cloned into pEYFP-N1 (Clontech) to generate plasmids expressing HFDCID-YFP and NCID-YFP under the control of the cid promoter. HFDH3 (amino acid position 41 to 136 of H3.1) and NH3 (amino acid position 1 to 40 of H3.1) were obtained from D.melanogaster genomic DNA by PCR-amplification using appropriate primers and cloned into the corresponding pYFP plasmids to generate NCIDHFDH3-YFP and NH3HFDCID-YFP fused proteins. All constructs were confirmed by DNA sequencing. For a description of the plasmids used in these experiments see Supplementary Figure S1.
Cell culture techniques
D.melanogaster Kc167 cells were grown in Schneider's medium (Sigma) supplemented with 10% FBS (Gibco), 100 μg/ml Streptomycin and 100 μg/ml Penicillin at 25°C. For transfection, 2 × 106 cells in 5 ml of medium were plated onto 6 cm diameter tissue culture dishes 24 h before transfection and then, transfected using the calcium phosphate method as described (20) using 10 μg of plasmid DNA. Cells were recovered at different times after transfection and analyzed by fluorescence microscopy (see below).
For treatment with Triton X-100, 24 h after transfection cells were grown in cover slips treated with Concanavalin-A (Sigma) and, after 24 h, were treated with 0.05% Triton X-100 during 10 min and visualized by fluorescence microscopy (see below).
When cells were treated with the proteasome inhibitor MG132 (Sigma), 5 μg/ml of inhibitor was added to the culture 39, 42 and 45 h after transfection and cells were recovered at 48 h post-transfection, i.e. after 3, 6 and 9 h of treatment with the inhibitor.
For in vivo time-lapse analysis, 1.2 × 106 cells were plated 48 h post-transfection on 35 mm glass-bottomed culture dishes (Mattek Inc.), treated with Concanavalin-A (Sigma), and after 4 h at 25°C, the media was replaced by fresh media supplemented with 0.3% soft-agar (Pronadisa). Cells were left for 1 h at 25°C and visualized as described below.
FACS sorting
For FACS sorting, cells were recovered 48 h after transfection by centrifugation at 1300 r.p.m. for 2 min, counted in a Neubauer chamber and resuspended in fresh medium at a concentration of 107 cells/ml. Then, cells were sorted in a MoFlo cytometer with a 488 Argon laser. After sorting, cells were collected by centrifugation at 1300 r.p.m. for 2 min, resuspended in fresh media and plated at a concentration of 1 × 106 cell/ml on cover slips treated with Concanavalin-A (Sigma) in 24-multiwell plates. Cells were grown at 25°C for an additional 24 h and visualized by fluorescence microscopy (see below).
RT–PCR analysis
For RT–PCR analysis, total RNA was prepared using ULTRASPEC™ RNA Isolation System (Biotecx), resuspended in 50 μl of RNase free water, treated with 0.1 U/ml of DNase I (Boehringer), precipitated and redissolved in 10 μl of RNase free water.
RT–PCRs were carried out with the QIAGEN® One Step RT–PCR kit using 0.5 and 5 ng of RNA obtained as described above. Two sets of primers were added for the simultaneous detection of CID-YFP and actin5C mRNAs. For detection of CID-YFP mRNA, primers that specifically amplify the YFP region were used: CidCt5 (5′-GCCAATCATATGAGCAGAGCC-3′) and EGFP-N (5′-CGTCGCCGTCCAGCTCGACCAG-3′). Actin5C mRNA was used as a control and it was amplified with primers Actforward (5′-TCTGGCACCACACCTTCTACAATG-3′) and Actreverse (5′-GCTCTGGCGGGGCAATGAT-3′). As negative controls, similar reactions were run but primers were added after completion of the RT-step and before PCR-amplification.
Fluorescence microscopy analysis
For fluorescence microscopy visualization of transfected cells, 1 ml of hypotonic MAC buffer (50 mM glycerol, 5 mM KCl, 10 mM NaCl, 0.8 mM CaCl2 and 10 mM sucrose) were added to 300 μl of transfected cells and, after incubation at room temperature for 5 min, 200 μl of this mix were spun down for 10 min at 500 r.p.m. on low acceleration in a TermoShandon Cytospin 4 using a single-chamber Cytospin funnel. Cells were then fixed in 4% paraformaldehyde for 10 min, washed with phosphate-buffered saline (PBS) for 15 min, mounted in Mowiol (Calbiochem-Novabiochem) containing 0.2 ng/ml DAPI (Sigma) and visualized.
For immunolocalization with specific antibodies, cells were washed with PBS and blocked in PBS, 0.2% Tween-20, 0.1% BSA for 20 min. Primary antibodies were diluted in blocking solution and the incubation was carried out for 1 h at room temperature and overnight at 4°C. The antibodies used were rabbit αNCID, at a 1:250 dilution, and αPSer10H3 (Upstate), at a 1:500 dilution. After incubation, slides were washed twice with blocking solution for 10 min and incubated for 45 min at room temperature with αrabbit Cy3 secondary antibody (Jackson ImmunoResearch) diluted 1:400 in blocking solution. Finally, slides were washed twice for 10 min in blocking solution, twice in PBS, mounted in Mowiol (Calbiochem-Novabiochem) containing 0.2 ng/ml DAPI (Sigma) and visualized.
All images were collected with a Colorview 12 camera mounted on a Nikon Eclipse E-800 inverted microscope with a 40× objective lens. Acquisition parameters were controlled by analysis software and images were analyzed with Adobe Photoshop software.
For in vivo time-lapse analysis, cells were prepared as described before and live cell images were taken on a confocal microscope (Leica TCS SP2-AOBS). Images were acquired as 9 z-stacks of 1.5 μm increments for each channel (YFP and transmission) at 1 frame/5 min by using a 40× oil-immersion objective, processed with Adobe Photoshop and ImageJ software, and displayed as quick projections.
RESULTS
Transiently expressed CID mislocalizes throughout chromatin but, during cell culture, its localization is progressively constrained to centromeres
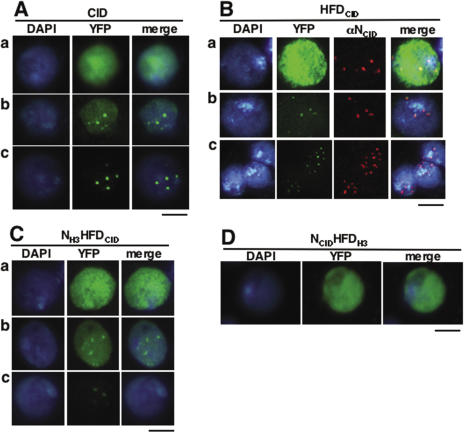
As shown in Figure 1, transient expression of CID results in two patterns of nuclear distribution that coexist. In these experiments, a CID-YFP fusion was transiently expressed in Kc cells under the control of the own cid promoter. In some cells, CID-YFP shows a diffuse nuclear distribution [Figure 1A (a)] while, in others, a distinct punctuated pattern is observed [Figure 1A (c)]. Cells showing CID-YFP foci on top of a diffuse background are also detected [Figure 1A (b)], indicating that both patterns of distribution can coexist. The punctuated pattern of distribution corresponds to CID-YFP localized at centromeres as it is also observed with truncated forms containing the histone-fold domain of CID, which is known to mediate targeting to centromeres, either by itself (HFDCID-YFP) (Figure 1B) or in combination to the N-terminal domain of canonical histone H3 (NH3HFDCID-YFP) (Figure 1C). Moreover, HFDCID-YFP foci fully co-localize with the signals obtained with a αNCID antibody that specifically recognizes the N-terminal domain of CID, but not the HFDCID, and, therefore, marks the positions corresponding to endogenous CID bound to centromeres (Figure 1B). On the other hand, the diffuse pattern of nuclear distribution arises from binding of CID-YFP throughout chromatin as it is resistant to treatment with Triton X-100 (see Supplementary Figure S2) that, otherwise, solubilizes unbound nuclear proteins, such as a truncated NCID-YFP form, containing exclusively the N-terminal domain of CID, which does not bind chromatin and gives rise only to the diffuse pattern of nuclear distribution (Supplementary Figure S2). Moreover, in some cases, binding of CID-YFP through chromatin is also observed in condensed metaphase chromosomes [Figure 3B (b)]. These observations indicate that, upon transient expression, CID shows two coexisting patterns of localization, at centromeres and throughout chromatin. Others have previously reported similar results (12,13,18).
Figure 1.
Transiently expressed CID-YFP shows two coexisting patterns of localization, at centromeres and throughout chromatin. CID-YFP (A), HFDCID-YFP (B) and NH3HFDCID-YFP (C) can localize throughout chromatin (a), only at centromeres (c) and both, at centromeres and throughout chromatin (b). NCIDHFDH3-YFP (D) localizes only throughout chromatin. YFP is shown in green. DAPI-staining is shown in blue. The immunolocalization pattern obtained with αNCID, which specifically recognizes the N-terminal domain of CID, is shown in red only in (B). Bars correspond to 5 μm, except in row c of (B) where it corresponds to 10 μm.
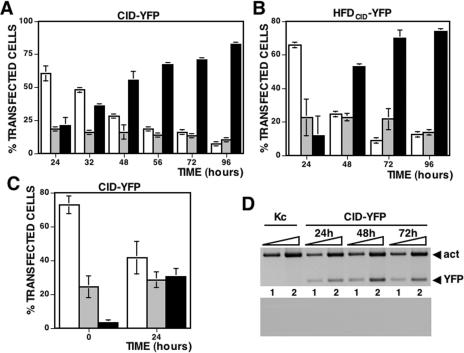
During cell culture, the percentage of cells showing CID exclusively localized at centromeres increases (Figure 2). In these experiments, the percentage of cells showing mislocalized CID-YFP [Figure 1A (a)] or where CID-YFP is exclusively found at centromeres [Figure 1A (c)] were determined at increasing times after transfection of the plasmid expressing CID-YFP. The percentage of cells showing a mixed localization pattern, at centromeres as well as throughout chromatin [Figure 1A (b)], was also determined. As shown in Figure 2A, early after transfection (24 h), most cells show mislocalized CID-YFP but, as the culture progresses, the percentage of cells where CID-YFP is exclusively found at centromeres increases so that, at 48–96 h, they are majority. Similar results were obtained when HFDCID-YFP (Figure 2B) or NH3HFDCID-YFP (data not shown) were expressed. These observations suggest that, during cell culture, cells showing mislocalized CID-YFP progress to cells were CID-YFP is exclusively found at centromeres. To confirm this hypothesis, cells showing mislocalized CID-YFP were sorted by FACS 48 h post-transfection taking advantage of their much higher intensity of fluorescence and, then, cultured for an additional 24 h. As shown in Figure 2C, the percentage of cells showing mislocalized CID-YFP that, immediately after sorting, account for close to 75% of the total transfected cells decrease to around 40% after 24 h of additional culture while, in parallel, cells showing localized CID-YFP increase from 3 to 30%. These results indicate that transiently expressed CID-YFP, which at early times post-transfection mislocalizes throughout chromatin, is progressively constrained to centromeres during cell culture.
Figure 2.
Upon cell culture, CID-YFP localization is constrained to centromeres. (A) The percentage of total transfected cells showing CID-YFP mislocalized throughout chromatin (white columns), localized exclusively at centromeres (black columns) or both, at centromeres and throughout chromatin, (grey columns) are presented as a function of increasing time after transfection. Results are the average of three independent experiments. (B) As in (A) but for HFDCID-YFP. In (C), cells showing mislocalized CID-YFP were sorted by FACS 48 h after transfection and cultured for an additional 24 h. The percentage of cells showing mislocalized CID-YFP (white columns), CID-YFP localized only at centromeres (black columns) or a mixed localization pattern (grey columns) is presented as a function of increasing time after sorting. Results are the average of three independent experiments. In (D), the levels of CID-YFP mRNA are analyzed by RT–PCR at different times after transfection using primers specific for YFP (bands labelled YFP) and, as a control, primers specific for actin5C (bands labelled act). Kc corresponds to untransfected cells. Reactions were performed with 0.5 ng (lane 1) and 5 ng (lane 2) of total RNA. As a negative control, the results of similar reactions where the two sets of primers were added after the RT-step but before PCR-amplification are shown at the bottom.
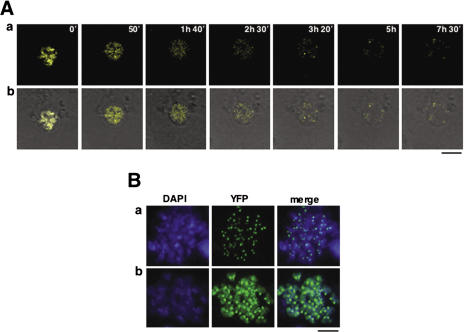
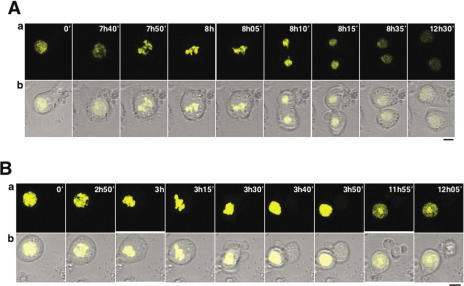
The process of clearance of mislocalized CID-YFP was followed in vivo by time-lapse analysis. In these experiments, living cells were immobilized in soft-agar 48 h post-transfection and, then, monitored by confocal microscopy for an additional 24 h. In most cases, clearance of mislocalized CID-YFP precedes chromosome condensation (Figure 3A and Supplementary Movie 1). Several observations, however, indicate that clearance can also occurs after mitosis: (i) though most metaphase chromosomes show CID-YFP exclusively localized at centromeres [Figure 3B (a)], in some cases, CID-YFP is also found bound throughout the entire condensed chromosome [Figure 3B (b)] and (ii) cells showing a mixed pattern of CID-YFP localization progress normally through mitosis (Figure 6A and Supplementary Movie 3).
Figure 3.
Clearance of mislocalized CID-YFP does not take place at any given time during cell cycle progression. (A) Time-lapse analysis of the process of clearance in interphase nuclei is presented as a function of increasing time of visualization. YFP is shown in yellow (a) and merged to transmission (b). (B) Condensed metaphase chromosomes showing CID localized exclusively at centromeres (a) and both, at centromeres and throughout chromatin (b). YFP is shown in green. DAPI-staining is shown in blue. Bars correspond to 5 μm.
Centromeric localization of transiently expressed CID is impaired in the presence of the proteasome inhibitor MG132
As determined from the intensity of fluorescence, CID-YFP protein levels are several orders of magnitude higher incells showing mislocalized CID-YFP than in cells where the protein is exclusively localized at centromeres that, as shown above, arise from the former. This observation suggeststhat, during cell culture, expression of CID-YFP is regulated. It might be argued, however, that decreased protein levels of cells showing centromeric localization of CID-YFP is simply the consequence of dilution of the corresponding expression vector during cell division. Several observations argue against this possibility. First, centromeric localization of CID-YFP is an early event after transfection, which occurs before any significant growth of the culture is detected, and does not correlate with cell division. For instance, from 24 to 32 h after transfection, the percentage of cells showing centromeric localization of CID-YFP increased by ∼75% in the absence of any significant growth of the culture (Supplementary Figure S3). Even at 48 h after transfection, when significant growth was observed (50%), centromeric localization of CID-YFP exceeded growth of the culture by more than 3-fold (Supplementary Figure S3). Similarly, when cells expressing high levels of CID-YFP were sorted by FACS, centromeric localization of CID-YFP was observed 24 h after sorting (Figure 2C) though, during this period, no significant cell growth was observed. These results are in good agreement with the time-lapse analysis described above (Figure 3A and Supplementary Movie 1), where strong reduction of CID-YFP protein levels was observed in the absence of cell division.
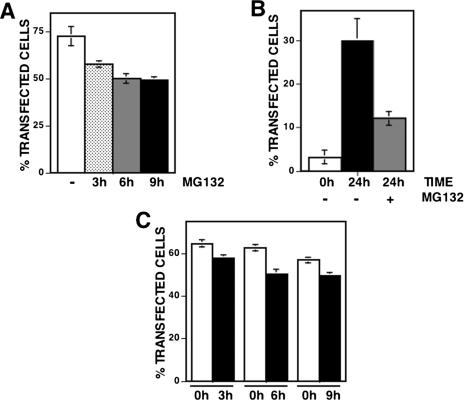
Altogether, these results indicate that expression of CID-YFP is actually regulated during cell culture. This regulation, however, does not take place at the transcriptional level since, as determined by RT–PCR using primers specific for YFP, the levels of CID-YFP mRNA do not change significantly from 24 to 72 h post-transfection (Figure 2D). CID-YFP expression must, therefore, be regulated at the post-transcriptional level. It is possible that, as in yeast (19), CID-YFP expression is regulated by proteolytic degradation of excess protein. Consistent with this hypothesis, centromeric localization of CID-YFP is impaired when cells are cultured in the presence of the proteasome inhibitor MG132 (Figure 4), which is known to be effective in insect cells (21,22). As shown in Figure 4A, the percentage of cells that, at 48 h post-transfection, show CID-YFP exclusively localized at centromeres decreases significantly when they are cultured in the presence of the inhibitor for 3–9 h before harvesting of the cells. Similar results were obtained when the effects of MG132 on the localization of CID-YFP were determined on FACS-sorted cells showing mislocalized CID-YFP (Figure 4B). As mentioned above, after sorting, the percentage of cells showing CID-YFP exclusively localized at centromeres is ∼3%, which increases to 30% after 24 h of additional culture in the absence of any, added inhibitor. However, the percentage of cells that, 24 h after sorting, show centromeric localization of CID-YFP is strongly reduced, by >60%, when cells are cultured in the presence of MG132 (Figure 4B). Altogether, these observations indicate that proteasome-mediated degradation contributes to restrict localization of transiently expressed CID-YFP to centromeres. Proteolytic degradation of CID-YFP does not depend on the YFP-tag as no significant change in proteins levels were detected upon culturing of cells expressing NCID-YFP or NCIDHFDH3-YFP fusion proteins, that do not localize to centromeres (Figure 1D and Supplementary Figure S2), or YFP itself (data not shown).
Figure 4.
Centromeric localization of transiently expressed CID-YFP is perturbed in the presence of the proteasome inhibitor MG132. (A) The percentage of total transfected cells showing CID-YFP localized exclusively at centromeres 48 h after transfection are presented as a function of increasing time of treatment with 5 μg/ml of MG132: no treatment (white columns), 3 h (dotted columns), 6 h (grey column) and 9 h (black columns). Results are the average of three independent experiments. In (B), cells showing mislocalized CID-YFP were sorted by FACS 48 h after transfection and cultured for an additional 24 h either in the presence of 5 μg/ml of MG132 or in the absence of any added inhibitor. The percentage of cells showing CID-YFP localized exclusively at centromeres is presented immediately after sorting (white column) and after 24 h of additional culture in the presence of MG132 (grey column) or in its absence (black column). Results are the average of three independent experiments. (C) The percentage of total transfected cells that, 48 h post-transfection, show CID-YFP exclusively localized at centromeres after treatment with 5 μg/ml of MG132 for 3 h, 6 h and 9 h (black columns) is compared with the percentage of cells showing CID-YFP localized exclusively at centromeres at the time when the inhibitor was added (white columns). Results are the average of three independent experiments.
After treatment with MG132 for 3–9 h, the percentage of cells showing CID-YFP localized at centromeres is lower than at the start of the treatment when the inhibitor was added (Figure 4C) indicating that, when proteolysis is inhibited in the presence of MG132, high CID-YFP expression levels are restored in cells showing low CID-YFP levels and that, under these conditions, newly synthesized CID-YFP mislocalizes throughout chromatin. These results strongly support the hypothesis that proteolytic degradation regulates available CID-YFP levels to ensure its preferential deposition at centromeres.
Cell cycle progression is affected in cells showing mislocalized CID
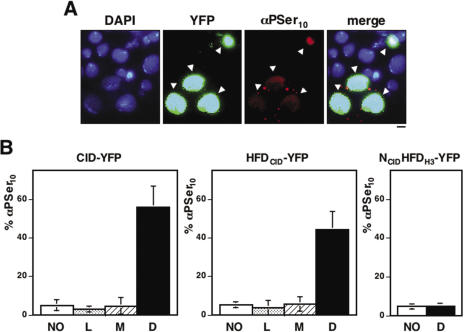
CID is known to play an essential role in centromere organization and kinetochore assembly. Therefore, CID mislocalization is likely to affect cell cycle progression. Consistent with this hypothesis, a high percentage of cells showing mislocalized CID are reactive against antibodies that specifically recognize phosphorylation of S10 of the histone H3 tail (αPSer10H3), an event that occurs in G2 preceding chromosome condensation and is detected all through mitosis (23). As shown in Figure 5, up to 60% of cells showing mislocalized CID-YFP are reactive to αPSer10H3, which is in contrast with the low percentage of αPSer10H3 positive cells (5%) observed among untransfected cells or those showing CID-YFP exclusively localized at centromeres (Figure 5B, panel CID-YFP). A similar behaviour is observed when HFDCID-YFP was expressed. Also in this case, a high proportion of cells where HFDCID-YFP is mislocalized are positive for αPSer10H3 (Figure 5B, panel HFDCID-YFP). This effect is specific, as it is not observed when an NCIDHFDH3-YFP fusion protein is expressed. NCIDHFDH3-YFP shows only the diffuse pattern of localization (Figure 1D) as it binds throughout chromatin. Nevertheless, in this case, the percentage of cells positive for αPSer10H3 is not different from that observed with untransfected cells (Figure 5B, panel NCIDHFDH3-YFP). These results indicate that mislocalization of CID-YFP delays cell cycle progression. No mitotic cells were detected among all αPSer10H3 positive cells analyzed showing mislocalized CID-YFP (N = 260) strongly suggesting that they enter mitosis at a very low frequency. In vivo time-lapse analysis is consistent with this hypothesis as cells showing mislocalized CID-YFP do not normally enter mitosis and, when they do so, strong aberrations on chromosome segregation are observed that, in extreme cases, result in daughter cells receiving no chromosomes at all (Figure 6B and Supplementary Movie 2).
Figure 5.
Cell cycle progression is altered in cells showing mislocalized CID-YFP. (A) Cells were stained with αPSer10 48 h after transfection of CID-YFP. YFP is shown in green. αPSer10 is shown in red. DAPI-staining is shown in blue. Bar corresponds to 5 μm. (B) The percentage of cells that 48 h after transfection of CID-YFP, HFDCID-YFP and NCIDHFDH3-YFP are positive to αPSer10 are presented as a function of the pattern of localization: untransfected cells (white columns); cells showing CID-YFP localized exclusively at centromeres (dotted columns); cells showing mislocalized CID-YFP (black columns) and cells showing CID-YFP at centromeres and throughout chromatin (dashed columns). Results are the average of three independent experiments.
Figure 6.
Cells showing a mixed pattern of CID-YFP localization, at centromeres and throughout chromatin, progress normally through mitosis. Time-lapse analysis of cells showing mislocalized CID-YFP (B) or a mixed pattern of CID-YFP localization (A) are presented as a function of increasing time of visualization. YFP is shown in yellow (a) and merged to transmission (b). Bars correspond to 5 μm.
Interestingly, cells showing a mixed localization pattern, at centromeres and throughout chromatin, appear to progress normally through cell cycle as, in this case, the percentage of αPSer10H3 positive cells is not significantly different from that observed with untransfected cells or with cells showing CID-YFP exclusively localized at centromeres (Figure 5B, panels CID-YFP and HFDCID-YFP). In good agreement with this interpretation, cells showing a mixed pattern of CID-YFP localization were observed to progress normally through mitosis (Figure 6A and Supplementary Movie 3) and metaphase chromosomes are detected where, in addition to a strong centromeric localization, CID-YFP is also found bound throughout the entire condensed chromosome [Figure 3B (b)].
DISCUSSION
Strong evidence indicates that centromere identity is dictated by the deposition at centromeric chromatin of the specific histone H3 variant CENP-A. The molecular mechanism(s) underlying specific deposition of CENP-A at centromeres are, however, poorly understood. Components of the inner-kinetochore complex CENP-I/H are known to be required for centromeric deposition of newly synthesized CENP-A both in chicken and Schizosaccharomyces pombe (24,25). This is, however, an intriguing observation as recruitment of CENP-I to centromeres is reciprocally dependent on CENP-A (25,26). On the other hand, the identification of assembly factors responsible for the specific deposition of CENP-A nucleosomes at centromeres remains elusive. Drosophila CENP-A (CID) was shown to interact with RbAp48/p55 (27), a chaperone protein that participates in known chromatin assembly complexes, CAF-1 and HIRA, as well as in many chromatin remodeling and modifying complexes. The contribution of this interaction to the specific deposition of CENP-A at centromeres appears uncertain as neither CAF-1 nor HIRA appear to be associated to CENP-A chromatin (28) and, in addition, these complexes are not specific for CENP-A as they also load nucleosomes containing other H3 variants (H3.1 for CAF-1 and H3.3 for HIRA). Furthermore, as shown here and elsewhere (3,11–13,18), transiently expressed CENP-A incorporates throughout chromatin indicating that deposition of CENP-A nucleosomes is a promiscuous process. Altogether, these observations suggest that additional mechanisms must exist to guarantee that stable incorporation of CENP-A nucleosomes occurs only at centromeres.
In this paper, we show that proteasome-mediated degradation contributes to restrict localization of Drosophila CENP-A (CID) to centromeres. In these experiments, a CID-YFP fusion was transiently expressed from the cid promoter in Kc cells. Our results show that, though at early times post-transfection CID-YFP mislocalizes throughout chromatin, its localization is progressively constrained to centromeres upon culturing of the cells as: (i) the percentage of cells showing CID-YFP localized exclusively at centromeres increases at late post-transfection times; (ii) FACS-sorted cells showing mislocalized CID-YFP progress to cells where CID-YFP is exclusively localized at centromeres and (iii) in vivo imaging shows clearance of mislocalized CID-YFP in individual living cells. Centromeric localization of transiently expressed CID-YFP is prevented in the presence of inhibitors of the proteasome indicating that clearance of mislocalized CID-YFP requires proteasome-mediated proteolysis. Most likely, mislocalized CID-YFP nucleosomes are exchanged from chromatin prior to degradation as, in general, histones are resistant to proteolysis when incorporated to chromatin (29). As a consequence of the interaction with kinetochore components, centromeric CID-YFP nucleosomes might be exchanged at a much slower rate or, alternatively, be more resistant to proteolytic degradation.
Our results show that proteolytic degradation not only eliminates mislocalized CID-YFP but, in addition, it also regulates CID-YFP expression levels as, upon culturing of the cells, protein levels are drastically reduced though, at the mRNA level, no significant change in expression is detected. The reduction in protein levels does not correlate with cell division indicating that it is not due to dilution of the expression plasmid during growth. The regulation of available CID-YFP appears essential to ensure centromeric deposition of CID-YFP nucleosomes as newly synthesized CID-YFP mislocalizes throughout chromatin when cells showing CID-YFP localized exclusively at centromeres are cultured in the presence of proteasome inhibitors.
In all experiments described in this paper, a CID–YFP fusion was transiently expressed in Kc cells from the cid promoter. Whether proteolysis also contributes to the centromeric localization of endogenous CID remains to be determined. However, previous work by others showed that, in S.cerevisiae, the levels of CENP-A (Cse4) are actually regulated by the proteasome and that a proteolysis-resistant mutant mislocalizes throughout chromatin (19). Moreover, in some human cancers, CENP-A is over-expressed and, also in this case, it is found mislocalized throughout chromatin (30). Proteasome-mediated degradation of endogenous CENP-A was also reported in human cells infected with herpes simplex virus type 1 (HSV-1) (31). Altogether, these observations strongly suggest that proteasome-mediated degradation might be an evolutionarily conserved mechanism that regulates available CENP-A levels to favour its preferential deposition at centromeres.
As shown here and elsewhere (12), mislocalization of CID-YFP induces cell cycle arrest indicating that clearance of mislocalized CID-YFP nucleosomes is required for normal cell cycle progression. Cell cycle arrest likely reflects functional interference due to recruitment of kinetochore proteins at ectopic sites (12). However, cells showing a mixed pattern of CID-YFP localization, at centromeres and throughout chromatin, show normal cell cycle progression indicating that functional interference of mislocalized CID-YFP occur only above a given threshold, perhaps similar to that found at endogenous centromeres.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
O.M.-M. acknowledges receipt of a doctoral fellowship from CIRIT. This work was supported by grants from the MCyT (BMC2003-243), the CIRIT (2001SGR00344 and 2005SGR00678) and the EU (LSHB-CT-2004-511965). Funding to pay the Open Access publication charges for this article was provided by MCyT grant BMC2003-243.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smith M.M. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell. Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 2.Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisia. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 3.Shelby R.D., Vafa O., Sullivan K.F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell. Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blower M.D., Sullivan B.A., Karpen G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Cleveland D.W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 7.Blower M.D., Karpen G.H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell. Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone H3-like protein in C.elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 9.Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H.A. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 11.Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yonomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell. Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 12.Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., Karpen G.H. Mislocalization of the Drosophila centromer-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermaak D., Hayden H.S., Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad K., Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc. Natl Acad. Sci. USA. 2002;99:16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad K., Henikoff S. Centromeres are specilized replication domains in heterochromatin. J. Cell. Biol. 2001;153:101–109. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelby R.D., Monier K., Sullivan K.F. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell. Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan B., Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell. Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S., Ahmad K., Platero J.S., van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins K.A., Furuyama S., Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Krasnov M.A., Saffman E.E., Kornfeld K., Hogness D.S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Muro I., Hay B.A., Clem R.J. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- 22.Lundgren J., Masson P., Mirzaei Z., Young P. Identification and characterization of a Drosophila proteasome regulatory network. Mol. Cell. Biol. 2005;25:4662–4675. doi: 10.1128/MCB.25.11.4662-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Y., Mizzen C.A., Cook R.G., Gorovsky M.A., Allis C.D. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl Acad. Sci. USA. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K., Chen E.S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 25.Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., 3rd, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell. Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 26.Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell. Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuyama T., Dalal Y., Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl Acad. Sci. USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foltz D.R., Jansen L.E.T., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nature Cell. Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 29.Gunjan A., Paik J., Verreault A. The emergence of regulated histone proteolysis. Curr. Opin. Genet. Dev. 2006;16:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 31.Lomonte P., Sullivan K.F., Everett R.D. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.