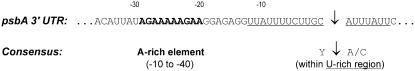Abstract
Eukaryotic cells arose through the uptake of free-living bacteria by endosymbiosis and their gradual conversion into organelles (plastids and mitochondria). Capture of the endosymbionts was followed by massive translocation of their genes to the genome of the host cell. How genes were transferred from the (prokaryotic) organellar genome to the (eukaryotic) nuclear genome and how the genes became functional in their new eukaryotic genetic environment is largely unknown. Here, we report the successful experimental reconstruction of functional gene transfer between an organelle and the nucleus, a process that normally occurs only on large evolutionary timescales. In consecutive genetic screens, we first transferred a chloroplast genome segment to the nucleus and then selected for gene activation in the nuclear genome. We show that DNA-mediated gene transfer can give rise to functional nuclear genes if followed by suitable rearrangements in the nuclear genome. Acquisition of gene function involves (1) transcriptional activation by capture of the promoter of an upstream nuclear gene and (2) utilization of AT-rich noncoding sequences downstream of the plastid gene as RNA cleavage and polyadenylation sites. Our results reveal the molecular mechanisms of how organellar DNA transferred to the nucleus gives rise to functional genes and reproduce in the laboratory a key process in the evolution of eukaryotic cells.
INTRODUCTION
Eukaryotes are believed to have arisen more than 1.5 billion years ago (Martin and Müller, 1998; Heckman et al., 2001; Javaux et al., 2001) by endosymbiosis. Uptake of an α-proteobacterium marked the origin of mitochondria, and the additional uptake of a cyanobacterium gave rise to the plastids present in all eukaryotic algae and higher plants as well as in some protozoa. The engulfment of the two eubacterial endosymbionts was followed by massive restructuring of the genomes of both the host and the symbionts. This involved the loss of dispensable and redundant genetic information and, most importantly, the large-scale translocation of genes from the endosymbiont's genome to the host cell's nuclear genome (Martin et al., 1998; Blanchard and Lynch, 2000). As a consequence, contemporary organellar genomes are greatly reduced and contain only a small proportion of the genes that their free-living ancestors had possessed. This gene transfer took place over a timescale of hundreds of millions of years, and phylogenetic evidence suggests that it is still an ongoing process (Grohmann et al., 1992; Millen et al., 2001; Adams et al., 2002). The infA gene encoding the plastid translation initiation factor 1 provides a particularly striking example of gene transfer events that occurred relatively recently in evolution. It had long been known that infA, while being a functional gene in the plastid genomes of the liverwort Marchantia polymorpha and rice (Oryza sativa) (Ohyama et al., 1986; Hiratsuka et al., 1989), exists only as a pseudogene in the tobacco (Nicotiana tabacum) plastid DNA (Shinozaki et al., 1986; Shimada and Sugiura, 1991). A systematic phylogenetic study of infA structure in plastid genomes of angiosperm species revealed that the gene has repeatedly become nonfunctional in ∼24 separate lineages of angiosperm evolution. Search for nuclear infA copies in four of these lineages resulted in identification of expressed nuclear infA genes whose gene products are targeted to plastids. Molecular analysis of the nuclear loci (exon-intron structure and transit peptide sequence) provided strong evidence for four independent gene transfer events (Millen et al., 2001).
Recent transgenic experiments have demonstrated that mitochondrial and chloroplast DNA escapes to the nucleus at relatively high frequency (Thorsness and Fox, 1990; Huang et al., 2003a; Stegemann et al., 2003), but if and how this DNA can give rise to functional nuclear genes has remained enigmatic. In fact, pieces of chloroplast and mitochondrial DNA are regularly found in nuclear genomes (Farrelly and Butow, 1983; Yuan et al., 2002; Timmis et al., 2004; Bock, 2005; Matsuo et al., 2005). These sequences lack any apparent function and are commonly referred to as promiscuous DNA. They have been hypothesized to provide the raw material for converting organellar genes into functional nuclear genes (Timmis et al., 2004), but evidence in support of this assumption is largely lacking. Thus, while there is ample evidence for organellar genes having been successfully transferred to the nucleus during evolution (Adams et al., 2000, 2002; Millen et al., 2001), two key questions about the mechanisms involved are still unanswered: (1) What were the relative contributions of DNA-mediated versus RNA/cDNA-mediated gene transfer events? (2) Can transferred (promiscuous) organellar DNA fragments be turned into functional nuclear genes?
As genome organization and gene expression mechanisms in organelles are clearly of prokaryotic type, conversion of an organellar gene into a functional (that is, eukaryotic type) nuclear gene must be considered difficult. Here, we have attempted to reproduce this key process in evolution and obtained a functional nuclear gene by first selecting for its transfer from the plastid to the nuclear genome and subsequently selecting for its functional activation in the nucleus.
RESULTS
Selection for Functional Gene Transfer from the Plastid to the Nuclear Genome
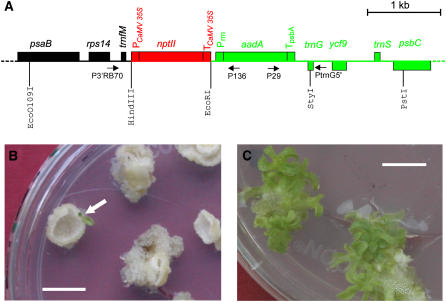
We have previously described a genetic screen facilitating the selection of DNA exchange events that transfer genes from the plastid to the nuclear genome (Stegemann et al., 2003). In this screen, a kanamycin resistance gene (nptII) was fused to nuclear expression signals (cauliflower mosaic virus 35S promoter and terminator; Figure 1A) and integrated into the plastid genome by chloroplast transformation. When subjected to selection on kanamycin-containing plant regeneration medium, kanamycin-resistant cell lines appear only when the nptII gene is transferred into the nuclear genome, which we found to happen at a surprisingly high frequency of approximately one in five million cells (Stegemann et al., 2003). The selected plant lines carry large pieces of silent chloroplast sequence (Huang et al., 2003a, 2004; Stegemann et al., 2003) and, in this way, reproduce the appearance of promiscuous organellar DNA in the nuclear genome. Likewise, earlier experiments in yeast had shown that mitochondrial DNA can escape to the nucleus at high frequency (Thorsness and Fox, 1990). These findings are consistent with the presence of large tracts of plastid and mitochondrial sequence in the nuclear genomes of higher plants (reviewed in Timmis et al., 2004; Bock, 2005; Leister, 2005). Whether these promiscuous organellar sequences can give rise to functional nuclear genes or rather represent junk DNA condemned to evolutionary deterioration is unknown and has been hotly debated. We therefore sought to design an experimental strategy that would allow us to directly test for functionalization of transferred organellar genes in the nucleus. To this end, we used plants from a previously conducted genetic screen for DNA transfer from the plastid to the nucleus (Stegemann et al., 2003) and, in a new screen, selected several additional lines with chloroplast DNA integrated in the nuclear genome. We determined the structure of the transferred antibiotic resistance locus (Figure 1A) in the tobacco nuclear genome, and for further analysis, we chose three lines (subsequently referred to as Nt-GT16, Nt-GT21, and Nt-GT31; Table 1) that (1) contained the complete nptII cassette, including the entire 35S promoter (verified by PCR with primer pair P3′RB70/P136; Figure 1A), and (2) had the complete aadA cassette, including downstream chloroplast sequences cotransferred with nptII (determined by PCR with primer pair P29/PtrnG5′; Figure 1A). The aadA gene encodes an enzyme inactivating the aminoglycoside antibiotics spectinomycin and streptomycin and was originally used to generate the chloroplast transformants subjected to the screen for transfer of the nptII to the nucleus (Stegemann et al., 2003). The transferred segment mimics the landing of a transferred plastid gene and its surrounding plastid sequence (green in Figure 1A) downstream of a nuclear gene (red in Figure 1A) in that the aadA gene is driven by plastid promoter and terminator sequences, whereas the upstream nptII carries eukaryotic promoter and terminator sequences.
Figure 1.
A Genetic Screen for Functional Gene Transfer from the Plastid Genome to the Nucleus.
(A) Physical map of the region in the plastid genome transferred to the nuclear genome by selection for kanamycin resistance (Stegemann et al., 2003). The chloroplast-type spectinomycin/streptomycin resistance gene aadA and flanking chloroplast sequences are shown in green. The eukaryotic-type nptII gene mimics an upstream nuclear gene (red) in that it simulates the landing of the chloroplast aadA gene downstream of a resident nuclear gene. Genes above the line are transcribed from the left to the right, and genes below the line are transcribed in the opposite orientation. Relevant restriction sites and primers are also shown, and the orientation of PCR primers is indicated by arrows. PCaMV 35S, cauliflower mosaic virus 35S promoter; TCaMV 35S, cauliflower mosaic virus 35S terminator; Prrn, chloroplast rRNA operon promoter; TpsbA, terminator from the chloroplast psbA gene.
(B) Selection for functional activation of the transferred aadA gene. Primary lines with an activated aadA gene (arrow) were identified by large-scale selection experiments on plant regeneration medium containing both spectinomycin and streptomycin. Resistance to these antibiotics is dependent on the aadA gene product, an enzyme inactivating aminoglycoside antibiotics.
(C) Shoot regeneration from tissue pieces of the primary line in the presence of 500 μg/mL spectinomycin indicates high-level antibiotic resistance. Bars = 1 cm.
Table 1.
Selection for Activation of the Chloroplast-Type aadA Gene in the Nucleus following Transfer from the Plastid to the Nuclear Genome
| Line | Zygosity | Number of Selection Plates | Selected Spec/Strp- Resistant Lines | Confirmed Lines with aadA Activation |
|---|---|---|---|---|
| Nt-GT16 | Heterozygous | 130 | 2 | 0 |
| Nt-GT21 | Homozygous | 137 | 8 | 4 |
| Nt-GT31 | Heterozygous | 161 | 6 | 4 |
| Total | 428 (= 5564 explants) | 16 | 8 |
Note that lines Nt-GT16 and Nt-GT31 are hetereozygous for the nptII-aadA cassettes in the nucleus (Figure 1A), whereas line Nt-GT21 is homozygous.
From all three selected lines, the transgenic chloroplasts were crossed out and replaced by wild-type chloroplasts (Stegemann et al., 2003). When tested on media containing spectinomycin and/or streptomycin, the three lines proved sensitive to both aminoglycosides, indicating that the plastid, prokaryotic-type aadA gene is not expressed in the nucleus (which is in line with the Prrn-driven aadA cassette acting as a chloroplast-specific selectable marker; Figure 1; Svab and Maliga, 1993). To directly determine whether transferred plastid genes can acquire functionality in the nucleus, we subjected all three lines to a second genetic screen by conducting large-scale selection experiments on plant regeneration media containing spectinomycin (spec) and streptomycin (strp). To largely suppress the appearance of spontaneous resistance mutants that can arise from specific point mutations in the plastid 16S rRNA or the plastid gene for ribosomal protein S12 (rps12; Svab and Maliga, 1991; Hsu et al., 1993), media with both drugs were used for selection. From >5500 leaf explants (each ∼5 × 5 mm in size), altogether 16 candidate lines were selected (Figure 1B, Table 1). To distinguish between spontaneous mutants that show elevated levels of tolerance to spectinomycin and/or streptomycin, all lines were subsequently tested for their resistance to 500 mg/L spec, 500 mg/L strp, and 500 mg/L spec + 500 mg/L strp (Figure 1C; data not shown). Lines showing identical levels of resistance to both antibiotics were considered candidates for aadA activation in the nucleus, whereas lines showing high-level resistance to one of the two aminoglycosides but low-level tolerance to the other (and the combination of both) were suspected to represent spontaneous chloroplast mutants. This test tentatively eliminated eight out of the selected 16 lines (Table 1). To ultimately confirm that the eight eliminated lines originated from spontaneous mutations in the plastid genome and to obtain additional genetic evidence for the remaining eight candidate lines representing events in which the transferred aadA gene was functionally activated in the nucleus, we regenerated plants from all lines that were then selfed and also reciprocally crossed to wild-type plants. Segregation assays of the progeny from these crosses demonstrated that the eight suspected spontaneous chloroplast mutants inherited the antibiotic resistance uniparentally maternally, as expected for a plastid-encoded trait, and, moreover, confirmed that these lines displayed high-level resistance only to one of the two antibiotics, indicating that they do not express the aadA gene (Table 1; data not shown). Interestingly, the remaining eight candidate lines all displayed Mendelian inheritance of the spec/strp resistance, strongly suggesting that they express a nuclear-encoded aminoglycoside resistance gene (Tables 1 and 2, Figure 2; data not shown). These lines are subsequently referred to as Nt-GT-A lines (e.g., Nt-GT21-A3 for N. tabacum gene transfer line 21/activated line 3).
Table 2.
Segregation of the Spectinomycin/Streptomycin Resistance in the Progeny from Crosses of Gene Transfer Lines That Were Selected for Functional Activation of the Transferred aadA Gene in the Nucleus
| Line | Cross | Expected Segregation | Observed Segregation | |
|---|---|---|---|---|
| Nt-GT21-A3 | GT × wild type | 1:1 | 92:85 | (1.08:1) |
| GT × GT | 3:1 | 734:299 | (2.45:1) | |
| Wild type × GT | 1:1 | 136:120 | (1.13:1) | |
| Nt-GT21-A5 | GT × wild type | 1:1 | 320:304 | (1.05:1) |
| GT × GT | 3:1 | 317:110 | (2.88:1) | |
| Wild type × GT | 1:1 | 350:389 | (0.90:1) | |
| Nt-GT21-A6 | GT × wild type | 1:1 | 152:139 | (1.09:1) |
| GT × GT | 3:1 | 266:90 | (2.96:1) | |
| Wild type × GT | 1:1 | 314:313 | (1:1) | |
| Nt-GT21-A7 | GT × wild type | 1:1 | 137:117 | (1.17:1) |
| Nt-GT31-A1 | GT × wild type | 1:1 | 295:264 | (1.11:1) |
| GT × GT | 3:1 | 230:74 | (3.11:1) | |
| Wild type × GT | 1:1 | 162:176 | (0.92:1) | |
| Nt-GT31-A2 | GT × wild type | 1:1 | 280:270 | (1.04:1) |
| GT × GT | 3:1 | 607:212 | (2.86:1) | |
| Wild type × GT | 1:1 | 309:279 | (1.11:1) | |
| Nt-GT31-A3 | GT × wild type | 1:1 | 1060:824 | (1.29:1) |
| GT × GT | 3:1 | 780:253 | (3.08:1) | |
| Wild type × GT | 1:1 | 663:653 | (1.02:1) | |
| Nt-GT31-A4 | GT × wild type | 1:1 | 664:665 | (1:1) |
| GT × GT | 3:1 | 287:88 | (3.26:1) | |
| Wild type × GT | 1:1 | 215:205 | (1.05:1) | |
Line Nt-GT21-A7 was male sterile and thus could not be used as pollen donor. GT, gene transfer line.
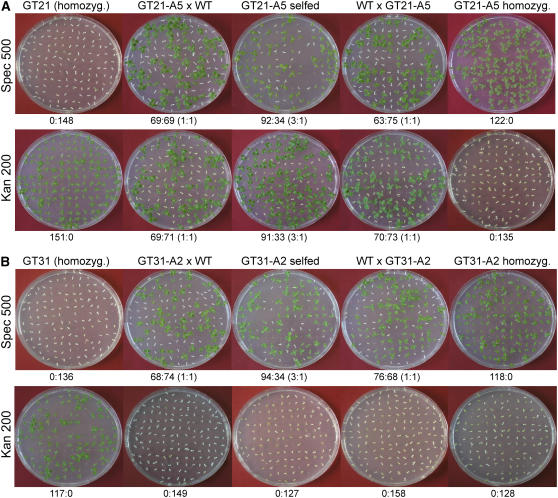
Figure 2.
Analysis of the Inheritance of the Resistance to Spectinomycin and Kanamycin in Lines Selected for Functional Activation of the aadA Gene Transferred from the Chloroplast Genome to the Nucleus.
Crosses are denoted at the top, the antibiotic is indicated at the left (concentration in mg/L), and the segregation data are given below each row.
(A) Segregation data for line Nt-GT21-A5 selected from the homozygous gene transfer plant Nt-GT21 (Table 1).
(B) Segregation data for line Nt-GT31-A2 selected from the heterozygous gene transfer plant Nt-GT31 (Table 1). Note that this line, as it was selected from a heterozygous plant, produced progeny that was uniformly sensitive to kanamycin in all crosses.
Transcriptional Activation of the Transferred Gene in the Nucleus
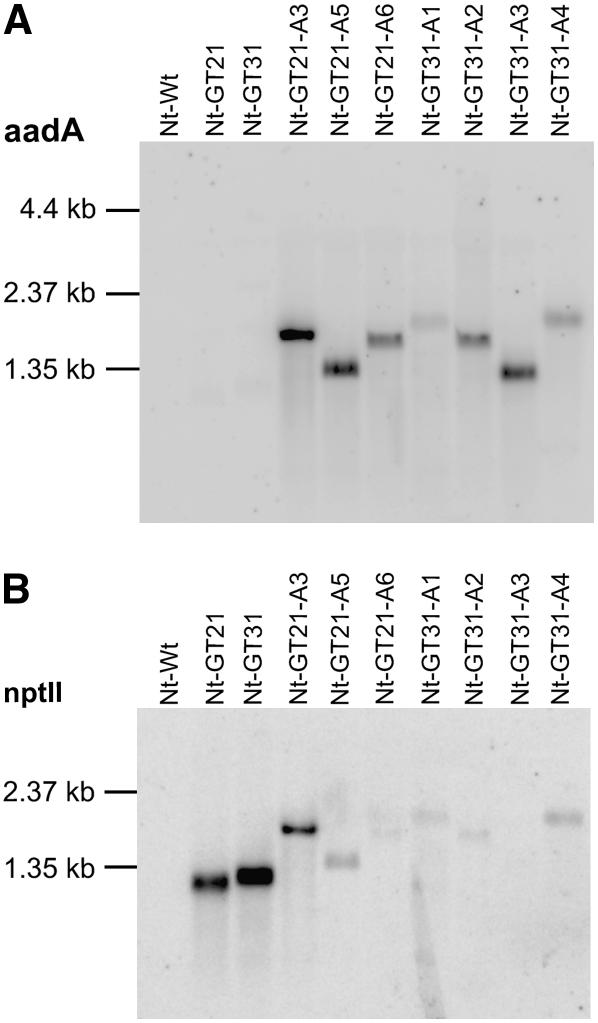
To test if functional activation of the transferred aadA gene had indeed occurred in the eight candidate lines that showed Mendelian segregation, we analyzed them for the accumulation of aadA mRNAs. Using a specific radiolabeled probe, aadA transcripts were readily detected in all lines (Figure 3A), indicating that the transferred aadA gene was transcriptionally activated in the nucleus. Remarkably, aadA transcripts of widely different sizes accumulated in different lines (Figure 3A), suggesting that different rearrangements had occurred in the nuclear genomes of the different lines.
Figure 3.
Transcription of aadA and nptII in Gene Transfer Lines before and after Activation of the Spectionmycin/Streptomycin Resistance Gene aadA.
(A) Detection of aadA transcripts by hybridization with a probe spanning the aadA coding region. While no aadA transcripts are detectable in the two gene transfer lines Nt-GT21 and Nt-GT31, all lines selected for spectinomycin and streptomycin resistance show transcriptional activation of the nuclear aadA gene. Different transcript sizes suggest that different rearrangements have occurred in different lines.
(B) Detection of nptII transcripts by hybridization with a probe spanning the nptII coding region. Most lines selected for activation of the aadA gene lack the prominent 1.2-kb nptII transcript present in the two original gene transfer lines Nt-GT21 and Nt-GT31. Detection of weakly hybridizing bands of different sizes in most spectionmycin/streptomycin-resistant lines suggests the presence of rearranged nptII sequences that either are only weakly transcribed or have only limited homology to the probe.
We next wanted to know whether the rearrangements leading to aadA activation had changed the structure of the upstream eukaryotic-type gene, the nptII locus (Figure 1A). We therefore analyzed the seed progeny from selfed Nt-GT-A plants and from Nt-GT-A plants reciprocally crossed to the wild type for phenotypic expression of the kanamycin resistance. Interestingly, the kanamycin resistance was found to be lost in all lines (Figure 2; data not shown), indicating that the acquisition of aadA expression is accompanied by loss of nptII expression. This may indicate that the rearrangements resulting in transcriptional activation of the transferred aadA gene involve mutations inactivating the upstream nptII. To provide additional molecular evidence for nptII inactivation, we assayed the Nt-GT-A lines for accumulation of nptII transcripts (Figure 3B). While nptII transcripts were completely absent from one line (Nt-GT31-A3), the other lines showed at least some weakly hybridizing RNA species, indicating that either nptII transcription has fallen to very low levels or large parts of the nptII coding region have been deleted that would reduce complementarity to the hybridization probe and thus result in poor detection of the nptII remnant sequences.
Rearrangements Leading to Functional Activation of the Transferred Gene
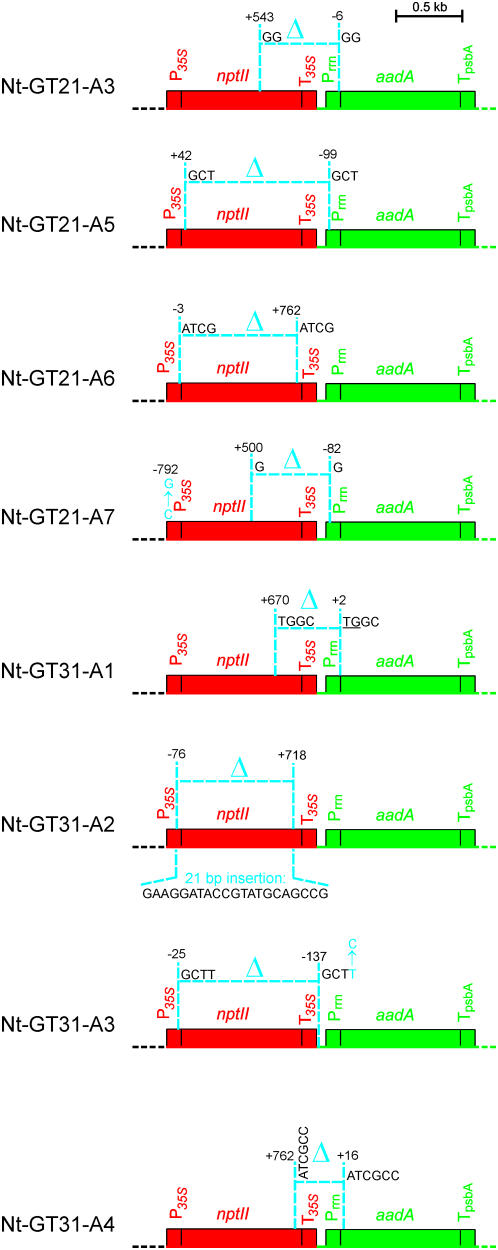
To identify the molecular rearrangements that led to aadA activation in the nucleus, we employed thermal asymmetric interlaced PCR (TAIL-PCR), a strategy suitable to determine unknown flanking DNA sequences (Liu et al., 1995). As our genetic and molecular analyses had suggested major rearrangements upstream of aadA, we first wanted to map the 5′ flanking region. In two to three rounds of TAIL-PCR followed by complete sequencing of bands appearing in reactions with Nt-GT-A DNA but not in control reactions with Nt-GT DNA or wild-type DNA as template, the structure of the region upstream of the aadA could be elucidated in all lines (Figure 4).
Figure 4.
Overview of the Molecular Rearrangements Leading to Functional Activation of the Transferred aadA Gene in the Nucleus.
Nucleotide positions at break points of deletions and/or insertions and point mutations refer to the start of the nptII or aadA coding regions, with negative numbers denoting positions upstream and positive numbers indicating positions downstream of the first nucleotide of the respective coding region (EMBL accession number AM235741). As in Figure 1, prokaryotic-type (chloroplast) sequences are shown in green, whereas eukaryotic-type sequences are in red. Δ indicates a deletion, point mutations are shown by the respective nucleotide substitution (in blue), and the nucleotide sequence of the insertion in Nt-GT31-A2 is shown underneath the map. Promoters and terminators are abbreviated as in Figure 1. Short directly repeated sequences at deletion break points are also indicated. The line Nt-GT31-A1 has the second break point within the translation initiation codon, the two remaining nucleotides of which are underlined. See text for details.
Sequencing of the rearranged locus revealed that, in all eight lines, aadA was transcriptionally activated by capturing the promoter of the upstream eukaryotic-type gene (i.e., the 35S promoter of the nptII gene; Figure 4). However, the mutation types and their functional consequences were widely different. We detected altogether four types of mutations: (1) deletions on short directly repeated sequences, (2) deletions lacking homology at the break points, (3) point mutations, and (4) insertions that apparently patch double-strand breaks (Figure 4). The presence of short direct repeats (of 2 to 6 bp) at the deletion break points in six out of the eight selected lines suggests strongly that these microhomologies are mechanistically involved in the generation of the deletions (Figure 4). The sequence found inserted into a deletion that was presumably caused by double-strand breaks (as it lacks microhomology at the break points; line Nt-GT31-A2) is of unknown origin. It is not derived from the (fully sequenced) tobacco chloroplast or mitochondrial genomes and thus most probably comes from an unsequenced region of the tobacco nuclear genome.
The functional consequences of the mutations and the way they lead to activation of aadA in the nucleus also can be grouped into different categories (Figure 4): In a first group, represented by lines Nt-GT31-A1 and Nt-GT31-A4, the activation occurs by translational fusion. A deletion with one break point in the nptII and the other at the beginning of the aadA coding region (in line Nt-GT31-A1 directly within the translational start codon) generates a chimeric gene that gives rise to an NptII-AadA fusion protein that is active as an aminoglycoside 3″-adenylyltransferase, but not as a neomycin phosphotransferase, due to deletion of the C-terminal region of the NptII protein. In a second group, represented by lines Nt-GT21-A3, Nt-GT21-A5, and Nt-GT21-A7 (Figure 4), the transferred aadA gene becomes expressed by translational read-through. In these lines, the first deletion break point resides within the nptII coding region, whereas the second is in the promoter region of aadA and thus is upstream of the aadA start codon. In all three lines, the deletions result in an in-frame fusion of the aadA reading frame with the residual nptII sequence and translation reads over the remaining aadA promoter sequence. Generation of a contiguous reading frame becomes possible by the second break point being downstream of the first in-frame stop codon upstream of aadA. In the third group, represented by lines Nt-GT21-A6, Nt-GT31-A2, and Nt-GT31-A3, expression of the transferred aadA is most likely facilitated by internal translation initiation. Lines in this group carry the 5′ break point in the cauliflower mosaic virus 35S promoter, thus resulting in deletion of the nptII translational start codon (Figure 4). However, in all three lines, the break point is downstream of the TATA box; thus, the transcriptional activity from the promoter is likely to remain unaffected. In this group, translation most probably initiates from the aadA start codon, as no other in-frame ATG codon is present upstream. As multiple out-of-frame ATGs are located upstream of the aadA start codon, an internal translation initiation mechanism is likely to be used in these lines.
Interestingly, lines Nt-GT21-A6 and Nt-GT31-A2 still contain the complete 35S terminator (Figure 4). The presence of abundant aadA-containing transcripts (Figure 3) that correspond in size to transcription initiation from the 35S promoter suggests that transcription termination by the 35S terminator is sufficiently incomplete to produce enough aadA mRNA for expression of the antibiotic resistance. In general, we observed a good correlation between the sizes of the deletions identified in the eight lines (Figure 4) and the sizes of the aadA transcripts determined by RNA gel blot analysis (Figure 3).
The two point mutations detected in lines Nt-GT21-A7 and Nt-GT31-A3 (Figure 4) are unlikely to be causally involved in aadA activation: The 35S promoter functions well in other lines without the mutation found in line Nt-GT21-A7, and the point mutation downstream of the deletion break point in line Nt-GT31-A3 is unlikely to have functional consequences. Assuming that the point mutations were not selected for, the detection of two such mutations in just a few kilobase pairs of DNA sequence is surprising and by far exceeds the normal mutation rate in the plant nuclear genome (estimated to be ∼5 × 10−9; Wolfe et al., 1987; Clegg et al., 1997). We therefore propose that the accumulation of these point mutations reflects the beginning evolutionary deterioration of organellar DNA sequences transferred to the nucleus (Huang et al., 2005). The high transfer rate of chloroplast sequences to the nucleus (Huang et al., 2003a; Stegemann et al., 2003) poses the intriguing question of how the nucleus prevents the inflation of its genome by the permanent influx of promiscuous DNA. Two of the molecular mechanisms discovered in this work, deletions on short direct repeats and mutational decay by accumulation of point mutations, potentially could contribute to the rapid degeneration and eventual elimination of those transferred sequences that were not converted into functional nuclear genes.
3′ Maturation of aadA mRNAs in the Nucleus
Gene expression in the nucleus does not only require a promoter to initiate transcription but also critically depends on faithful mRNA 3′ end formation. The 3′ maturation involves endonucleolytic cleavage of the primary transcript followed by polyadenylation. Addition of the poly(A) tail is crucial to mRNA stability in that nonpolyadenylated transcripts are highly unstable and condemned to rapid degradation (Sachs, 1993; Sachs and Wahle, 1993; Wu et al., 1995). By contrast, maturation of chloroplast transcripts does not involve polyadenylation, and the 3′ ends of chloroplast mRNAs are formed by prokaryotic-type mechanisms (Herrin and Nickelsen, 2004). We therefore wanted to know how stable mRNAs can be produced from the transferred aadA gene (Figure 3A). To test if rearrangements had occurred also downstream of the aadA coding region (e.g., by capturing cleavage/polyadenylation signals from resident nuclear genes), the region downstream of the coding region was PCR amplified and fully sequenced (using primers P29 and PtrnG5′; Figure 1A). Surprisingly, the sequence of the entire region turned out to be identical with the original chloroplast sequence in all eight lines, suggesting that 3′ maturation of the transcripts from the activated aadA gene does not require additional DNA rearrangements. In the absence of any detectable mutation downstream of the aadA coding region, we wanted to identify the 3′ end of the aadA mRNAs. In view of the apparent stability of the aadA transcripts (Figure 3A), we reasoned that they must be polyadenylated. We therefore designed a reverse transcription–based PCR strategy employing an oligo(dT) primer with a short 5′ anchor sequence to prime cDNA synthesis followed by two rounds of nested PCR amplification. By directly sequencing the amplification products, we mapped the mRNA 3′ cleavage and polyadenylation site to a specific sequence within the psbA 3′ untranslated region (UTR) (Figure 5).
Figure 5.
mRNA 3′ End Formation in Transcripts from the Activated aadA Gene in the Nucleus.
The identified polyadenylation site within the psbA 3′ UTR is indicated by a vertical arrow. The consensus sequence elements known to mediate mRNA 3′ end formation in plants are indicated underneath the psbA sequence. The A-rich element upstream of the cleavage/polyadenylation site in psbA sequence is marked in bold, and the U-rich region surrounding the site is underlined.
When we compared the mapped cleavage and polyadenylation site with the loose consensus sequence for mRNA 3′ end formation in plants (Li and Hunt, 1997), the sequence surrounding the processing site in the psbA 3′ UTR turned out to match the consensus (Figure 5). At first sight, the presence of a functional cleavage and polyadenylation motif within the 3′ UTR of a chloroplast gene may seem a strange coincidence. However, the consensus sequence for mRNA 3′ end formation in plants is highly AU rich (Li and Hunt, 1997), and AT richness is also a hallmark of chloroplast genomes. AT richness is particularly pronounced in non-protein-coding regions, such as UTRs and intergenic spacers, where it regularly reaches values of >80% AT (Ohyama et al., 1988). It is therefore not all too surprising that a sequence element in the psbA 3′ UTR matches the rather loose AU-rich consensus sequence for mRNA 3′ processing and polyadenylation in the nucleus (Figure 5). Moreover, this finding may suggest that the high AT richness of non-protein-coding regions in chloroplast genomes has contributed significantly to the success rate of gene transfer during evolution by limiting the requirements for functional gene transfer to promoter capture and eliminating the need to acquire specific sequences for faithful mRNA 3′ cleavage and polyadenylation.
DISCUSSION
In the course of this work, we have developed an experimental model system to reconstruct evolutionary events that have decisively shaped the eukaryotic cell since its birth more than 1.5 billion years ago. The transfer of organellar genes to the nucleus and the conversion of such prokaryotic-type genes into functional eukaryotic genes occurs only on a large evolutionary timescale and thus has largely escaped rigorous experimental analysis. The design of stringent selection schemes for gene transfer from the chloroplast genome to the nucleus (Huang et al., 2003a; Stegemann et al., 2003) followed by a second genetic screen selecting for functional activation of the transferred gene in the nucleus has now made it possible to reproduce such extremely rare events in the laboratory within a time frame of just a few years. As some antibiotics, including streptomycin, which was used in our screen, are known to have mutagenic effects (Balashov and Humayun, 2002), it is possible that, in our experiments, evolution of a functional nuclear aadA gene was additionally accelerated by screening for antibiotic resistance.
In our experimental system, nuclear activation of the transferred chloroplast gene occurred at a frequency of eight events in 5564 leaf explants subjected to selection (Table 1), which, if a rough estimate of the cell numbers in the total leaf area subjected to selection is taken into account (Stegemann et al., 2003), amounts to a frequency of ∼3 × 10−8. It seems possible that this frequency is somewhat variable depending on the chromosomal location of the transferred organellar DNA sequence, in that different chromosomal regions in the nucleus may differ in their mutation rates. In fact, our finding that no activated line could be selected from Nt-GT16 plants (Table 1) may lend preliminary support to this idea. It is also noteworthy that in all lines, the transferred chloroplast gene captured the promoter of the gene located immediately upstream. The absence of long-distance deletions trapping the promoter of a more distantly located nuclear gene suggests that the probability of a successful functional activation in the nuclear genome strongly increases with decreasing distance of the gene's landing site from an upstream promoter sequence.
It seems conceivable that the probability of functional gene transfer as determined in this work would be lower if acquisition of a transit peptide sequence for rerouting of the gene product into the chloroplast was additionally required. However, a systematic analysis of the evolutionary fate of nuclear genes that stem from the cyanobacterial endosymbiont has revealed that the gene products from the majority of transferred genes are targeted to cell compartments other than the chloroplast (Martin et al., 2002). On the other hand, our results show that the functional activation of a transferred gene can easily be brought about by generation of fused reading frames with upstream nuclear genes (Figure 4). Thus, it seems conceivable that, if required for functionality of the transferred gene, a transit peptide sequence for rerouting into the organelle can easily be captured by a similar mechanism.
In theory, other mechanisms than the ones identified here could also lead to functional activation of a transferred chloroplast gene. One such possibility would be, for example, the accumulation of point mutations that make the prokaryotic-type chloroplast promoter more eukaryotic like and allow for its recognition by RNA polymerase II. The fact that such events were not recovered in our screen suggests that this mechanism, if it occurs at all, is significantly rarer than gene activation by promoter capture. Likewise, no lines were obtained in which a back-transfer of the gene from the nucleus to the chloroplast genome had occurred. This may indicate that genes flow on a one-way street from the chloroplast to the nuclear genome, with no way back to the chloroplast.
The recent discovery that chloroplast DNA sequences can escape to the nuclear genome at high frequency (Huang et al., 2003a; Stegemann et al., 2003) has sparked controversial discussions about the level of transgene containment provided by plants that harbor transgenes in their plastid genome rather than their nuclear genome (Daniell and Parkinson, 2003; Huang et al., 2003b). Maternal inheritance of chloroplasts can prevent uncontrolled transgene spreading via pollen, but if chloroplast transgenes escape to the nucleus and become functional there, containment would be limited. If the probability of nuclear activation of a transferred chloroplast gene as determined in this study is multiplied by the probability of transgene transfer to the nuclear genome determined earlier (Stegemann et al., 2003), the frequency of a chloroplast transgene escaping to the nucleus and becoming functional there would be in the range of 10−14 to 10−15. Although the frequency may be higher in male generative cells (possibly due to chloroplast degradation resulting in increased DNA release from the chloroplast; Huang et al., 2003a; Bock, 2005), the number indicates that the risk of escape of a functional transgene from the chloroplast genome to the nucleus is very low, underscoring that chloroplast transformation provides a valuable tool for increasing transgene containment. Another aspect that should be considered here is that we have determined the frequency of gene activation for somatic cells. At what frequency these mutations would be passed into the germline (in the presence versus absence of selective pressure) remains to be investigated.
In sum, our work uncovers several principles and mechanisms involved in the functional gene transfer from organellar genomes to the nucleus. First, it demonstrates that direct, DNA-mediated gene transfer can give rise to functional nuclear genes if quickly followed by suitable rearrangements in the nuclear genome. Second, it reveals that capturing the promoter of an upstream nuclear gene represents the predominant mode of how transferred organellar genes become transcriptionally activated. Third, we demonstrate that promoter capture is accomplished by either deletions occurring on short directly repeated sequences or, alternatively, by DNA double-strand breaks into which nonhomologous alien sequences can be pasted. Fourth, we have shown that the inherent AT richness of noncoding regions downstream of plastid genes facilitates their utilization as RNA cleavage and polyadenylation sites in the nucleus, thereby generating stable transcripts and presumably contributing significantly to the success rate of gene transfer events. Finally, we have provided evidence that readily occurring deletions and an unusually high frequency of point mutations in transferred sequences may contribute to the evolutionary decay of those transferred organellar sequences that were not rapidly turned into functional nuclear genes.
METHODS
Plant Material and Selection for DNA Transfer from the Plastid to the Nucleus
Sterile tobacco (Nicotiana tabacum cv Petit Havana) plants were grown on agar-solidified Murashige and Skoog (1962) medium containing 30 g/L sucrose. Primary gene transfer lines Nt-GT16 and Nt-GT31 were isolated from chloroplast-transformed plants in a genetic screen described previously (Stegemann et al., 2003). Line Nt-GT21 was obtained from a similar screen conducted in this work.
Selection for aadA Activation in the Nucleus
A large-scale genetic screen for functional activation of the transferred aadA sequence in the nucleus was conducted by exposing leaf explants from primary gene transfer lines to stringent selection for aadA-mediated resistance to spectinomycin and streptomycin on plant regeneration medium (Svab et al., 1990). To largely suppress the appearance of false positives (i.e., spontaneous antibiotic resistance mutants carrying point mutations in chloroplast genes for components of the ribosome), different combinations of the two aminoglycoside antibiotics ranging from 200 mg/L spec + 100 mg/L strp to 400 mg/L spec + 200 mg/L strp were applied (Bock, 2001). Selected candidate lines were retested on plant regeneration media containing either spec (500 mg/L) or strp (500 mg/L) or a combination of both antibiotics (500 mg/L each). Shoots regenerated from this additional selection round were rooted on phytohormone-free medium, subsequently transferred to soil, and grown under greenhouse conditions.
Crosses and Inheritance Assays
Plants from all lines recovered from the genetic screen for aadA activation in the nucleus were both selfed and reciprocally crossed to wild-type plants. Surface-sterilized seeds from all crosses were sown in Petri dishes with Murashige and Skoog medium containing either spectinomycin (500 mg/L), streptomycin (500 mg/L), or kanamycin (200 mg/L). Seedling phenotypes were scored after approximately 2 weeks.
Isolation of Nucleic Acids and Hybridization Analyses
Total plant nucleic acids were isolated from fresh leaf tissue by a cetyl-trimethyl-ammonium bromide–based method (Doyle and Doyle, 1990). Total cellular RNA was extracted using the peqGOLD TriFast reagent (Peqlab). RNA samples were separated by denaturing gel electrophoresis on 1% formaldehyde-containing agarose gels and transferred onto Hybond nylon membranes (Amersham) by capillary blotting. For the detection of aadA and nptII transcripts, the complete coding regions of the respective genes were excised from plasmid clones, separated by agarose gel electrophoresis, and purified from excised gel slices using the GFX PCR kit (DNA and gel band purification) (Amersham). The fragments were radiolabeled with 32P-dCTP using the MegaPrime kit (Amersham). Hybridizations were performed at 65°C in Rapid-Hyb buffer (Amersham) following the manufacturer's protocol.
PCR and DNA Sequencing
DNA samples were amplified in an Eppendorf thermal cycler using GoTaq Flexi DNA polymerase (Promega) and reaction volumes of 50 μL. Standard amplification reactions were performed according to standard protocols (35 cycles of 45 s at 94°C, 1.5 min at 56°C, and 1.5 min at 72°C, with a 4-min extension of the first cycle at 94°C and a 6-min final extension at 72°C). PCR products were separated by gel electrophoresis in 1.5 to 2% agarose gels, purified from excised gel slices using the GFX PCR kit, and directly sequenced by cycle sequencing reactions. Primer pair P136 (5′-TCGATGACGCCAACTACC-3′)/P3′RB70 (5′-CTTGTTTATCTATTAGTTTTCAGTTC-3′) was used for amplification and control sequencing of the rearrangements upstream of the aadA coding region. To obtain complete sequences, additional primers were derived from internal sequences. Primer pair P29 (5′-CGCTATGGAACTCGCCGCC-3′)/PtrnG5′ (5′-GTGGGTACATATTGTGTATCAA-3′) was used to amplify and determine the sequence downstream of the aadA coding region.
TAIL-PCR
TAIL-PCR was performed according to published protocols (Liu et al., 1995) using the degenerated primers AD1, AD2, AD3, and AD6 (Sessions et al., 2002). These primers were combined with the aadA-specific primers PaadA1 (5′-TCCAAAAGGTCGTTGATCAAAGCTCGCC-3′) for the first round of PCR, PaadA2 (5′-GCGTTGTTTCATCAAGCCTTACGGTCAC-3′) for the second round, and PaadA3 (5′-ACCAGCAAATCAATATCACTGTGTGGCTTCA-3′) for the third round of PCR. Primer PaadA4 (5′-AGGCCGCCATCCACTGCGGA-3′) was used for direct sequencing of TAIL-PCR products. For sequencing of large amplification products, additional internal primers were designed.
3′-End Mapping of aadA Transcripts
For identification of mRNA 3′ ends, RNA samples were purified by treatment with RNase-free DNase I (Roche). Five micrograms of purified DNA-free RNA was reverse transcribed in a 50-μL reaction at 55°C. cDNA synthesis was primed with primer PC5T17 (5′-CCCCCTTTTTTTTTTTTTTTTT-3′), and elongation reactions were performed with SuperScript III RNase H-free reverse transcriptase (Invitrogen) according to the manufacturer's instructions. First-strand cDNAs were amplified in an Eppendorf thermal cycler using GoTaq Flexi DNA polymerase (Promega) using primers PC5T17 and P29 and the following PCR program: 40 cycles of 45 s at 94°C, 1.5 min at 55.1°C, and 1.5 min at 72°C, with a 4-min extension of the first cycle at 94°C and a 6-min final extension at 72°C. As template for a second amplification, 0.5 μL of the PCR reaction was used with the primer pair P11 (5′-AGCGAAATGTAGTGCTTACG-3′) and P5C19T (5′-CCCCCTTTTTTTTTTTTTTTTTTT-3′). The obtained products were directly sequenced with primer P11.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AM235741.
Acknowledgments
We thank the Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team for plant care and cultivation. This research was supported by the Max Planck Society. This publication is dedicated to Rudolf Hagemann, Halle/Saale, on the occasion of his 75th birthday on October 21, 2006.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ralph Bock (rbock@mpimp-golm.mpg.de).
Open Access articles can be viewed online without a subscription.
References
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfer of a mitochondrial gene to the nucleus in flowering plants. Nature 408 354–357. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Qiu, Y.-L., Stoutemeyer, M., and Palmer, J.D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 99 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov, S., and Humayun, M.Z. (2002). Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J. Mol. Biol. 315 513–527. [DOI] [PubMed] [Google Scholar]
- Blanchard, J.L., and Lynch, M. (2000). Organellar genes: Why do they end up in the nucleus? Trends Genet. 16 315–320. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2001). Transgenic chloroplasts in basic research and plant biotechnology. J. Mol. Biol. 312 425–438. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2005). Extranuclear inheritance: Gene transfer out of plastids. Prog. Bot. 67 75–98. [Google Scholar]
- Clegg, M.T., Cummings, M.P., and Durbin, M.L. (1997). The evolution of plant nuclear genes. Proc. Natl. Acad. Sci. USA 94 7791–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H., and Parkinson, C.L. (2003). Jumping genes and containment. Nat. Biotechnol. 21 374–375. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1990). Isolation of plant DNA from fresh tissue. Focus 12 13–15. [Google Scholar]
- Farrelly, F., and Butow, R.A. (1983). Rearranged mitochondrial genes in the yeast nuclear genome. Nature 301 296–301. [DOI] [PubMed] [Google Scholar]
- Grohmann, L., Brennicke, A., and Schuster, W. (1992). The mitochondrial gene encoding ribosomal protein S12 has been translocated to the nuclear genome in Oenothera. Nucleic Acids Res. 20 5641–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman, D.S., Geiser, D.M., Eidell, B.R., Stauffer, R.L., Kardos, N.L., and Hedges, S.B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293 1129–1133. [DOI] [PubMed] [Google Scholar]
- Herrin, D.L., and Nickelsen, J. (2004). Chloroplast RNA processing and stability. Photosynth. Res. 82 301–314. [DOI] [PubMed] [Google Scholar]
- Hiratsuka, J., et al. (1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of cereals. Mol. Gen. Genet. 217 185–194. [DOI] [PubMed] [Google Scholar]
- Hsu, C.-M., Yang, W.-P., Chen, C.-C., Lai, Y.-K., and Lin, T.-Y. (1993). A point mutation in the chloroplast rps12 gene from Nicotiana plumbaginifolia confers streptomycin resistance. Plant Mol. Biol. 23 179–183. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y., Ayliffe, M.A., and Timmis, J.N. (2003. a). Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422 72–76. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y., Ayliffe, M.A., and Timmis, J.N. (2003. b). Organelle evolution meets biotechnology. Nat. Biotechnol. 21 489–490. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y., Ayliffe, M.A., and Timmis, J.N. (2004). Simple and complex nuclear loci created by newly transferred chloroplast DNA in tobacco. Proc. Natl. Acad. Sci. USA 101 9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.Y., Grünheit, N., Ahmadinejad, N., Timmis, J.N., and Martin, W. (2005). Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 138 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaux, E.J., Knoll, A.H., and Walter, M.R. (2001). Morphological and ecological complexity in early eukaryotic ecosystems. Nature 412 66–69. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2005). Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 21 655–663. [DOI] [PubMed] [Google Scholar]
- Li, Q., and Hunt, A.G. (1997). The polyadenylation of RNA in plants. Plant Physiol. 115 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Martin, W., and Müller, M. (1998). The hydrogen hypothesis for the first eukaryote. Nature 392 37–41. [DOI] [PubMed] [Google Scholar]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393 162–165. [DOI] [PubMed] [Google Scholar]
- Matsuo, M., Ito, Y., Yamauchi, R., and Obokata, J. (2005). The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell 17 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen, R.S., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Ohyama, K., et al. (1988). Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J. Mol. Biol. 203 281–298. [DOI] [PubMed] [Google Scholar]
- Ohyama, K., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322 572–574. [Google Scholar]
- Sachs, A., and Wahle, E. (1993). Ploy(A) tail metabolism and function in eukaryotes. J. Biol. Chem. 268 22955–22958. [PubMed] [Google Scholar]
- Sachs, A.B. (1993). Messenger RNA degradation in eukaryotes. Cell 74 413–421. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, H., and Sugiura, M. (1991). Fine structural features of the chloroplast genome: Comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 19 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann, S., Hartmann, S., Ruf, S., and Bock, R. (2003). High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 100 8828–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab, Z., Hajdukiewicz, P., and Maliga, P. (1990). Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 87 8526–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1991). Mutation proximal to the tRNA binding region of the Nicotiana plastid 16S rRNA confers resistance to spectinomycin. Mol. Gen. Genet. 228 316–319. [DOI] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness, P.E., and Fox, T.D. (1990). Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature 346 376–379. [DOI] [PubMed] [Google Scholar]
- Timmis, J.N., Ayliffe, M.A., Huang, C.Y., and Martin, W. (2004). Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5 123–136. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Li, W.-H., and Sharp, P.M. (1987). Rates of nucleotide substitutions vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., Ueda, T., and Messing, J. (1995). The formation of mRNA 3′-ends in plants. Plant J. 8 323–329. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., Hill, J., Hsiao, J., Moffat, K., Ouyang, S., Cheng, Z., Jiang, J., and Buell, C.R. (2002). Genome sequencing of a 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Genomics 267 713–720. [DOI] [PubMed] [Google Scholar]