Abstract
NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) is conserved from yeast to human and was proposed to act as a histone chaperone. While budding yeast contains a single NAP1 gene, multicellular organisms, including plants and animals, contain several NAP1 and NAP1-RELATED PROTEIN (NRP) genes. However, the biological role of these genes has been largely unexamined. Here, we show that, in Arabidopsis thaliana, simultaneous knockout of the two NRP genes, NRP1 and NRP2, impaired postembryonic root growth. In the nrp1-1 nrp2-1 double mutant, arrest of cell cycle progression at G2/M and disordered cellular organization occurred in root tips. The mutant seedlings exhibit perturbed expression of ∼100 genes, including some genes involved in root proliferation and patterning. The mutant plants are highly sensitive to genotoxic stress and show increased levels of DNA damage and the release of transcriptional gene silencing. NRP1 and NRP2 are localized in the nucleus and can form homomeric and heteromeric protein complexes. Both proteins specifically bind histones H2A and H2B and associate with chromatin in vivo. We propose that NRP1 and NRP2 act as H2A/H2B chaperones in the maintenance of dynamic chromatin in epigenetic inheritance.
INTRODUCTION
The nucleosome is the basic repeating unit of chromatin and consists of 146 bp of DNA wrapped in roughly two superhelical turns around a histone octamer containing two molecules each of histones H2A, H2B, H3, and H4 (Luger et al., 1997). During DNA replication in S phase of the cell cycle, the passage of the fork displaces parental histones, which are then redistributed on the daughter strands, and new histones are deposited de novo for replicated DNA to be fully assembled (Krude and Keller, 2001). In addition, nucleosome disassembly-reassembly processes, including histone replacement (by newly synthesized histones and/or histone variants) or recycling, also occur during transcription, DNA repair, and recombination (Jin et al., 2005). Together with chromatin remodeling and covalent modification of histones and DNA (Hsieh and Fischer, 2005; Martin and Zhang, 2005), nucleosome assembly is likely to contribute significantly to epigenetic regulation and inheritance. Histone chaperones play a crucial role in nucleosome assembly and are thought to be necessary for prevention of nonproductive aggregation between highly positive charged histones and highly negative charged DNA in a dense protein environment. The current list of histone chaperones includes nucleoplasmin, N1/N2, chromatin assembly factor-1 (CAF-1), antisilencing factor 1 (Asf1), histone regulatory homolog A (HIRA), Spt6, NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1), and nucleolin (Haushalter and Kadonaga, 2003; Angelov et al., 2006; Polo and Almouzni, 2006).
NAP1 represents the primary chaperone of H2A and H2B and is highly conserved from yeast to human (Ishimi et al., 1984; Dong et al., 2003; Ohkuni et al., 2003). In Saccharomyces cerevisiae, NAP1 is encoded by a single gene, and its deletion altered the expression of ∼10% of all nuclear genes, but the mutant cells are still viable (Ohkuni et al., 2003). In Drosophila melanogaster, knocking out NAP1 dramatically reduced viability (Lankenau et al., 2003). In mammals, NAP1 belongs to a multigene family, and the knockout of the mouse neuron-specific NAP1-homolog-2 gene is embryo lethal (Rogner et al., 2000). In plants, NAP1 also belongs to a multigene family (e.g., four genes encoding close NAP1 homologues are present in the genome of Arabidopsis thaliana) (Arabidopsis Genome Initiative, 2000; Dong et al., 2003). The different members of the tobacco (Nicotiana tabacum) and rice (Oryza sativa) NAP1 group proteins have distinct subcellular localizations and appear to have specific functions (Dong et al., 2005).
In addition, plants contain genes encoding more distantly NAP1-related proteins (hereafter called NRPs), which form a distinct phylogenic group more closely related to the animal SET/TAF-I/I2PP2A proteins (Dong et al., 2003). SET was identified first as the product of a translocated gene in acute undifferentiated leukemia (von Lindern et al., 1992), but its role in oncogenesis is not established. In biochemical assays, the SET homologue TAF-I and I2PP2A proteins stimulate replication of the adenovirus genome (Nagata et al., 1995) and inhibit protein phosphatase 2A (PP2A) (Li et al., 1996). The SET/TAF-I/I2PP2A group proteins were also identified in protein complexes with histone acetylation and methylation enzymes (Adler et al., 1997; Shikama et al., 2000), with B-type cyclins (Kellogg et al., 1995), with the granzyme A–activated DNase NM23-H1 (Fan et al., 2003), and with transcription factors (Telese et al., 2005). Some of these interacting proteins seem to be common with NAP1-interacting proteins, whereas others appear to be more specific for SET/TAF-I/I2PP2A. However, specific function of SET/TAF-I/I2PP2A compared with NAP1 is not clear, and the role of SET/TAF-I/I2PP2A proteins in animal growth and development are not yet known.
By reverse genetics, we directly investigated the role of NRPs in Arabidopsis. We show that the two Arabidopsis genes NRP1 (At1g74560) and NRP2 (At1g18800) are required for maintaining cell proliferation and cellular organization in root tips. While the single mutants nrp1-1 and nrp2-1 have a wild-type phenotype, the nrp1-1 nrp2-1 double mutant has a short-root phenotype. The embryonic root of the double mutant appears normal but, shortly after seed germination, G2/M-arrested cells cumulate in root tips. In the nrp1-1 nrp2-1 double mutant seedlings, expression of ∼100 genes is affected. Among the affected genes, GLABRA2 (GL2), encoding a homeodomain transcription factor, is required for repression of root hair formation (Ohashi et al., 2003), and PLETHORA2 (PLT2), encoding an AP2-type transcription factor, is required for root quiescent center specification and stem cell activity (Aida et al., 2004). The nrp1-1 nrp2-1 double mutant plants are also hypersensitive to genotoxic stress and show release of silencing of TRANSCRIPTIONALLY SILENT INFORMATION (TSI) and of transposon Ta3. NRP1 and NRP2 proteins bind histones H2A and H2B and are localized in the nucleus, supporting the idea that they are bona fide H2A/H2B chaperones. Chromatin immunoprecipitation (ChIP) shows that NRP1 and NRP2 bind to genes located in actively transcribed euchromatin and to the pericentromeric TSI. Taken together, our results provide evidence for specific functions of NRPs, which cannot be replaced by NAP1 group proteins, and demonstrate that NRP-mediated H2A/H2B dynamics play a critical role in epigenetic inheritance.
RESULTS
Phenotype of nrp1-1, nrp2-1, and nrp1-1 nrp2-1 Mutants
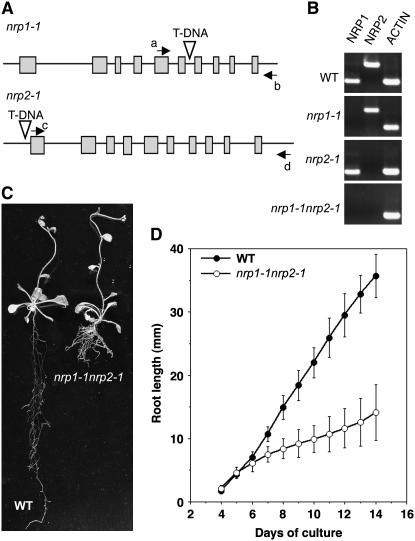
Both NRP1 and NRP2 are located on chromosome 1 in Arabidopsis. NRP1 and NRP2 amino acid sequences show 88% identity and 93% similarity to each other but show <30% identity and <48% similarity with four members of the Arabidopsis NAP1 group proteins, NAP1;1 (At4g26110), NAP1;2 (At2g19480), NAP1;3 (At5g56950), and NAP1;4 (At3g13782). RT-PCR analysis revealed that NRP1 and NRP2 as well as NAP1;1, NAP1;2, and NAP1;3 are ubiquitously expressed in all material examined, including seedlings, roots, stems, and flower buds (see Supplemental Figure 1 online). To investigate the function of NRPs in Arabidopsis, we searched for lines with T-DNA insertions within NRP1 and NRP2. From the SALK collection (Alonso et al., 2003), two independent lines were identified and named nrp1-1 and nrp2-1. PCR analysis confirmed that nrp1-1 contains a T-DNA insertion in the sixth intron of NRP1, and nrp2-1 contains a T-DNA insertion at −162 bp (from the ATG codon) in the promoter of NRP2 (Figure 1A; data not shown). In both mutants, the T-DNA segregated as a single locus. Homozygous (hereafter called mutant) plants were obtained for both T-DNA insertion lines by self-pollination. RT-PCR analysis showed that the transcripts of the corresponding genes were absent in the mutant plants (Figure 1B). These mutant plants with loss of function of either NRP1 or NRP2 did not show any phenotype under our in vitro culture and greenhouse growth conditions.
Figure 1.
Simultaneous Knockout of NRP1 and NRP2 Results in a Short-Root Phenotype.
(A) Genomic structure of NRP1 and NRP2. Boxes indicate the coding sequences. Triangles, T-DNA insertion sites in nrp mutants; arrows, PCR primer location sites.
(B) RT-PCR analysis of NRP1 and NRP2 expression in wild-type, single mutants nrp1-1 and nrp2-1, and double mutant nrp1-1 nrp2-1 plants 14 DAG. ACTIN serves as an internal control.
(C) Wild-type and double mutant nrp1-1 nrp2-1 plants 32 DAG in the in vitro culture medium.
(D) Root growth curves for the wild-type and the double mutant nrp1-1 nrp2-1 plants. The mean value from 20 plants is shown. Error bars represent standard deviations.
We then crossed nrp1-1 with nrp2-1 and obtained the double mutant, named nrp1-1 nrp2-1. RT-PCR analysis confirmed that both NRP1 and NRP2 transcripts were absent in the double mutant plants (Figure 1B). In contrast with single mutant plants, the double mutant plants showed a short-root phenotype (Figure 1C). A detailed study revealed that the double mutant roots grew like the wild-type roots until 6 d after germination (DAG). However, after 7 DAG, the elongation of the double mutant roots was dramatically reduced compared with the wild-type roots (Figure 1D). The aerial organs (leaves, rosettes, inflorescences, flowers, fruits, and embryos) developed normally in the double mutant plants.
Rescue of the nrp1-1 nrp2-1 Mutant Phenotype
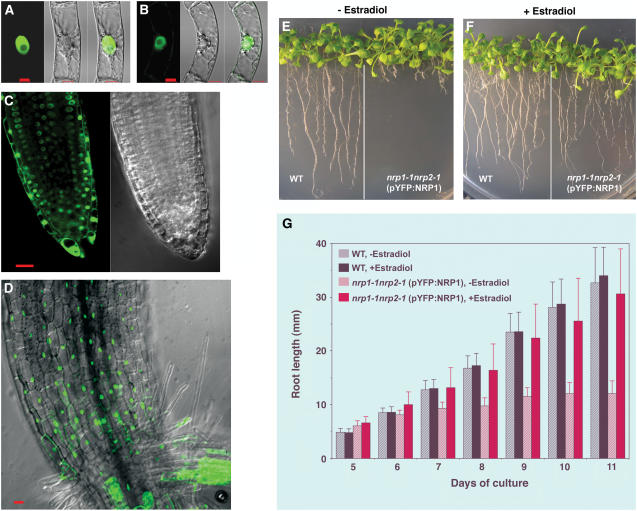
To obtain direct evidence that the short-root phenotype is caused by loss of function of NRPs, the NRP1 and NRP2 cDNAs were cloned and fused in frame to the 3′-end of the cDNA encoding the yellow fluorescent protein (YFP), resulting in YFP:NRP1 and YFP:NRP2. The fusion constructs under the control of an estradiol-inducible promoter (Zuo et al., 2000) were introduced into both tobacco BY-2 cells and Arabidopsis plants. The YFP:NRP1 and YFP:NRP2 fusion proteins were localized primarily in the nuclei in both transgenic tobacco BY-2 cells (Figures 2A and 2B) and Arabidopsis plants (Figures 2C and 2D). A significant amount of YFP:NRP1 and YFP:NRP2 was also observed in the cytoplasm in some cells of transgenic Arabidopsis plants, suggesting that intracellular localization of these proteins is affected by the physiological state of the cells. In the absence of the estradiol inducer, the YFP:NRP1-transgenic double mutant plants maintained the short-root mutant phenotype (Figure 2E). In the presence of estradiol, however, they showed root growth similar to the wild-type plants (Figure 2F). Root length measurements showed that induced expression of YFP:NRP1 rescued root elongation of the mutant plants (Figure 2G). Rescue by YFP:NRP2 expression was also observed, while no rescue was obtained by expressing YFP alone (data not shown). These results show that the YFP fusion proteins are functional and that deficiency in both NRP1 and NRP2 caused the mutant short-root phenotype, arguing for a significant degree of functional redundancy of the two proteins.
Figure 2.
Subcellular Localization of YFP:NRP1 and YFP:NRP2 Proteins and Rescue of the Mutant Phenotype.
(A) and (B) Transgenic tobacco BY-2 cells expressing YFP:NRP1 and YFP:NRP2, respectively, were visualized by fluorescence confocal microscopy. YFP fluorescence image (left panels), bright-field differential interference contrast image (middle panels), and their merged image (right panels) are shown. Note that green fluorescence is concentrated in the spherical nucleus but absent from the nucleolus inside the nucleus. Bars = 10 μm.
(C) and (D) Root tip and stem-root junction region, respectively, from a transgenic Arabidopsis plant expressing YFP:NRP1. YFP fluorescence (in green) and differential interference contrast images are shown. Bar = 20 μm.
(E) and (F) Wild-type and transgenic double mutant nrp1-1 nrp2-1 (pYFP:NRP1) plants were grown in the in vitro culture medium in the absence or presence of the transgene expression inducer estradiol. Images were taken at 14 DAG.
(G) Comparison of root elongation between wild-type and the rescue-transgenic mutant nrp1-1 nrp2-1 (pYFP:NRP1) plants. The mean value from 20 plants is shown. Error bars represent standard deviations. Both YFP:NRP1 and YFP:NRP2 constructs were under the control of the induced estradiol-inducible promoter.
Cellular Defects in the nrp1-1 nrp2-1 Mutant Roots
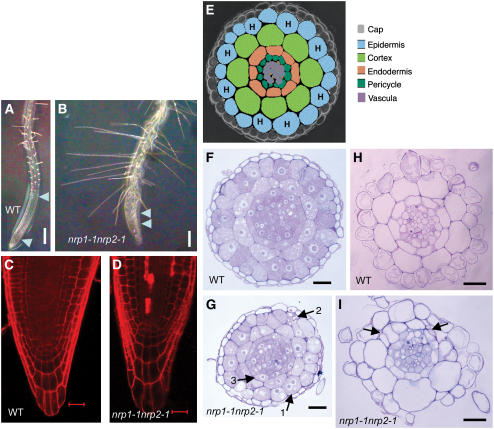
The nrp1-1 nrp2-1 mutant roots contained irregularly positioned long hairs, and the elongation zone was greatly reduced (Figure 3B) compared with that of wild-type roots (Figure 3A). Longitudinal sections revealed that the nrp1-1 nrp2-1 mutant root tips contain larger cells (Figure 3D) compared with the wild-type root tips (Figure 3C), ruling out the possibility that the short-root phenotype is caused by a reduced cell size in the mutant. Furthermore, dead cells, which accumulate dye, are found in the mutant root tips (Figure 3D). Transverse sections showed that the radial organization of the mature tissues in wild-type roots consists of the epidermis, cortex, endodermis, and pericycle, which surround the vascular cylinder (Figures 3E and 3F) (Dolan et al., 1993). Root hair cells arise from the epidermal cells that contact two underlying cortical cells (the so-called H position), whereas the epidermal cells overlying a single cortical cell develop into nonhair cells (Figures 3E and 3H) (Galway et al., 1994). In nrp1-1 nrp2-1 mutant roots, cell division was abnormal and cell organization in the epidermis, cortex, and endodermal layers was irregular (Figure 3G). Irregular cell size and number in these layers were much more pronounced in sections of the differentiation zone (Figure 3I), whereas the regular radial organization was maintained in wild-type roots (Figure 3H). Taken together, these results suggest that NRP1 and NRP2 are necessary for the maintenance of cell proliferation and differentiation in postembryonic root growth.
Figure 3.
nrp1-1 nrp2-1 Roots Show Defects in Cell Division, Viability, and Cellular Organization.
(A) and (B) Primary roots of 12-d-old seedlings of the wild type and the double mutant nrp1-1 nrp2-1, respectively. The region between two arrowheads indicates the elongation zone. Bars = 0.5 mm. (C) and (D) Longitudinal confocal sections of the wild-type and the double mutant nrp1-1 nrp2-1 roots of 7-d-old seedlings, respectively. Propidium iodide (red fluorescence) only stains the cell wall in living cells but stains the entire cell in dead cells. Dead cells were observed in 8/10 mutant but 0/10 wild-type primary root tips. Bars = 20 μm.
(E) Schema of transverse section of root apex with different colors indicating different cell types. H, root hair cell.
(F) and (G) Transverse sections within the root elongation zone of wild-type and double mutant nrp1-1 nrp2-1 10-d-old-seedlings, respectively. Arrows indicate different types of abnormal cells having either two nuclei (1) possibly due to a defect in cytokinesis, granular and shrunken form (2) because of death, or irregular size and misplacement in cell layers (3). Bars = 50 μm.
(H) and (I) Transverse sections within the differentiation zone of wild-type and the double mutant nrp1-1 nrp2-1 roots from 10-d-old-seedlings, respectively. Arrows indicate extra cell divisions. Note that the wild-type root contains the invariant eight cortical and endodermal cell files. Bars = 50 μm.
Impaired Expression of the Cell Division Marker CYCB1:GUS in the nrp1-1 nrp2-1 Mutant
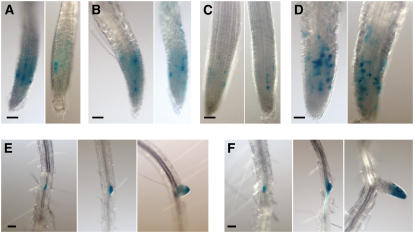
To analyze the role of NRP1 and NRP2 in cell cycle progression in proximal meristem cells and in lateral root initiation, we crossed the nrp1-1 nrp2-1 mutant with a transgenic plant expressing CYCB1:GUS (Colon-Carmona et al., 1999) and obtained CYCB1:GUS-containing wild-type and nrp1-1 nrp2-1 mutant lines. The CYCB1:GUS gene contains the β-glucuronidase (GUS) reporter fused to the mitotic destruction sequence (D-box) and the promoter of the cyclin CYCB1;1. This fusion gene is expressed upon entry into G2 (via the CYCB1:1 promoter), and its protein product is degraded upon exit from metaphase (via D-box) (Criqui et al., 2001), and subsequently the GUS activity marks cells in G2 and early M phase (Colon-Carmona et al., 1999). As shown in Figure 4, GUS-positive cells were detected in the root tips and at the initiation sites of lateral root formation. At early stages after germination, the wild-type (Figure 4A) and the mutant (Figure 4B) siblings showed similar GUS activity. However, at later stages, a significantly higher number of GUS-positive cells were observed in the mutant root tips (Figure 4D) compared with the wild-type root tips (Figure 4C), suggesting that G2/M arrest occurred in the mutant. Examination of 6- to 12-d-old seedlings revealed that lateral roots initiated similarly in the wild type (Figure 4E) and the mutant (Figure 4F). The number of lateral roots and primordia per seedling was similar in the mutant and the wild type, 19 ± 6 (15) compared with 20 ± 5 (15). However, the distance separating two lateral roots/primordia was significantly shorter in the mutant than in the wild-type, 1.85 ± 0.34 (15) compared with 2.58 ± 0.48 (15) mm. Thus, it appears that NRP1 and NRP2 could play a repressive role in spatial lateral root initiation.
Figure 4.
Cell Cycle Arrest and Lateral Root Initiation in the Double nrp1-1 nrp2-1 Mutant.
The wild-type and the nrp1-1 nrp2-1 mutant plants containing the G2/M marker CYCB1:GUS were analyzed by histochemical staining for GUS activity. Bars = 50 μm.
(A) and (B) Primary root tips of 6-d-old seedlings of the wild type and the mutant, respectively.
(C) and (D) Primary root tips of 8-d-old seedlings of the wild type and the mutant, respectively. Two representative examples are shown for each.
(E) and (F) Lateral root formation in the wild type and the mutant, respectively. Representatives of early to late stages of lateral root formation are shown in panels from left to right. Observations were performed on primary roots of 6- to 12-d-old seedlings.
Alterations in Gene Expression in the nrp1-1 nrp2-1 Mutant
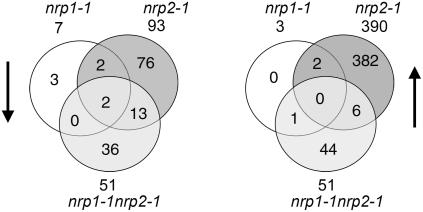
To uncover molecular events in the mutant plants, we first analyzed gene expression profiles in 6-d-old seedlings before any mutant phenotype was visible. We also reasoned that secondary transcriptional changes caused by NRP1- and NRP2-dependent differentially expressed genes would be minimal at this early developmental stage. The complete Arabidopsis transcriptome microarray (CATMA) containing 24,576 genes of the Arabidopsis genome (Crowe et al., 2003) was used for hybridization. Based on a stringent statistical test (see Methods), 10 genes in nrp1-1, 483 genes in nrp2-1, and 102 genes in nrp1-1 nrp2-1 were found to be differentially expressed (see Supplemental Tables 1 to 3 online). These differentially expressed genes belong to both downregulated and upregulated categories, and some were common to the different mutants (Figure 5). A considerably higher number of differentially expressed genes were found in the nrp2-1 mutant, suggesting that NRP2 has a broader activity. The fact that many differentially expressed genes in the single mutants were unchanged in the double mutant suggests complexity of molecular interactions between NRP1 and NRP2, which is beyond simple redundancy as viewed by plant phenotype. Since only the nrp1-1 nrp2-1 double mutant has a morphological phenotype, we focused on genes that are differentially expressed specifically in this mutant but not in the single mutants. Among this group of genes, it is interesting to note that while several genes encoding cell wall or extracellular matrix proteins are downregulated, the expression of several genes encoding transcription factors, including a zinc-finger protein, a bZIP protein, and three ethylene-responsive proteins, are upregulated to more than twofold in the mutant (Table 1). The cell wall or extracellular matrix is important not only for structure but also for providing positional information that determines root cell fate (Kwak et al., 2005). The bZIP transcription factor PosF21 is likely to play an important role in vascular development (Jakoby et al., 2002). The ethylene-responsive transcription factors are involved in phytohormone ethylene signaling (Broekaert et al., 2006), and ethylene plays a crucial role in root growth and root hair development (Stepanova et al., 2005). It is reasonable to speculate that the difference in expression of these genes contributes to the nrp1-1 nrp2-1 mutant phenotype.
Figure 5.
Transcriptome Analysis of nrp1-1, nrp2-1, and nrp1-1 nrp2-1 Mutants.
Venn diagram shows the number of downregulated (left) and upregulated (right) genes in the nrp1-1, nrp2-1, and nrp1-1 nrp2-1 mutants compared with wild-type seedlings.
Table 1.
Genes Differentially Expressed in the Double Mutant nrp1-1 nrp2-1 but Not in the Single Mutant nrp1-1 or nrp2-1
| Probe Set | Gene ID | Annotated Function |
|---|---|---|
| Down (less than twofold) | ||
| CATMA2A31980 | At2g33790 | Pollen Ole e 1 allergen and extensin family protein |
| CATMA5A49170 | At5g53250 | Arabinogalactan protein (AGP22) |
| CATMA5A50230 | At5g54370 | Late embryogenesis abundant protein-related protein |
| CATMA1A43140 | At1g52060 | Jacalin lectin family protein |
| CATMA3A00185 | At3g01190 | Peroxidase 27 (PER27; P27; PRXR7) |
| CATMA5A36740 | At5g41080 | Glycerophosphoryl diester phosphodiesterase family protein |
| CATMA3A47440 | At3g54500 | Expressed protein |
| CATMA3A39300 | At3g46280 | Protein kinase-related |
| CATMA5A35160 | At5g39580 | Peroxidase, putative |
| CATMA1A22610 | At1g23720 | Pro-rich extensin-like family protein |
| Up (more than twofold) | ||
| CATMA3A48950 | At3g55980 | Zinc-finger (CCCH-type) family protein, putative transcription factor |
| CATMA4A19530 | At4g18440 | Adenylosuccinate lyase/adenylosuccinase, putative |
| CATMA4A02710 | At4g02410 | Lectin protein kinase family protein |
| CATMA5A41290 | At5g45340 | Cytochrome P450 family protein |
| CATMA2A29600 | At2g31370 | bZIP transcription factor (PosF21) |
| CATMA1A33350 | At1g35210 | Expressed protein |
| CATMA1A04730 | At1g05730 | Expressed protein |
| CATMA2A43300 | At2g44840 | Ethylene-responsive element binding protein, putative transcription factor |
| CATMA1A09360 | At1g10522 | Expressed protein |
| CATMA5A43215 | At5g47230 | Ethylene-responsive element binding factor 5 (ERF5), transcription factor |
| CATMA3A29010 | At3g29000 | Calcium binding EF hand family protein |
| CATMA3A55720 | At3g62550 | Universal stress protein (USP) family protein |
| CATMA2A36760 | At2g38480 | Integral membrane protein, putative |
| CATMA3A43500 | At3g50440 | Hydrolase, α/β fold family protein |
| CATMA4A08330 | At4g08540 | Expressed protein |
| CATMA4A38005 | At4g36430 | Peroxidase, putative |
| CATMA5A57200 | At5g61600 | Ethylene-responsive element binding family protein, putative transcription factor |
| CATMA3A14260 | At3g14900 | Expressed protein |
| CATMA4A30165 | At4g28510 | Prohibitin, putative |
| CATMA3A01430 | At3g02480 | Abscisic acid–responsive protein-related |
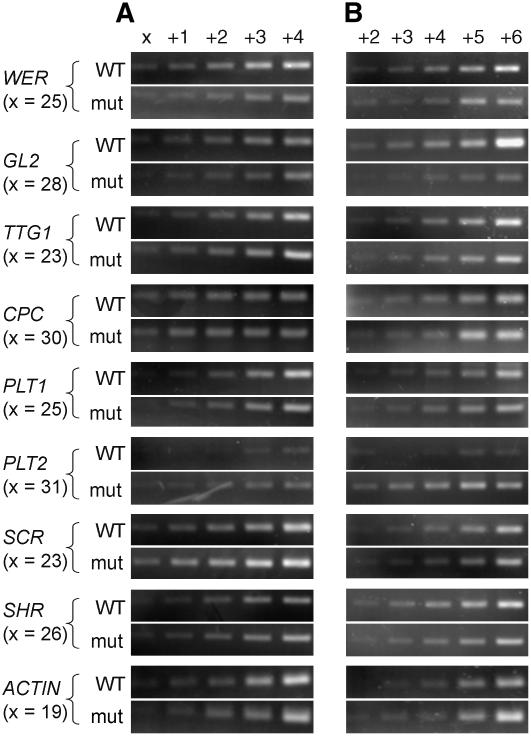
We also investigated the expression level of several patterning genes in the wild-type and nrp1-1 nrp2-1 mutant roots by semiquantitative RT-PCR. In Arabidopsis roots, GL2, WEREWOLF (WER), TRANSPARENT-TESTA-GLABRA1, and CAPRICE (CPC) are involved in epidermal patterning, PLT2 and PLT1 are involved in proximo-distal patterning, and SCARECROW and SHORT-ROOT are involved in radial patterning (for a review, see Ueda et al., 2005). No significant differences were observed between the wild-type and the mutant roots in 6-d-old seedlings (Figure 6A). This is consistent with previous microarray data where these genes were not among the differentially expressed genes. In 12-d-old seedlings, however, the level of GL2 was significantly lower, whereas that of PLT2 was higher in the mutant roots compared with the wild-type roots, while the other patterning genes showed unchanged expression levels (Figure 6B). These RT-PCR results were reproducible in two independent experiments. GL2 represses root hair formation (Ohashi et al., 2003), so a decrease in expression level correlates with the increased proliferation of root hairs in the nrp1-1 nrp2-1 mutant. Since the ectopic overexpression of PLT2 enhances root stem cell formation (Aida et al., 2004), the increased level of PLT2 expression is consistent with the capacity of root primordia formation in the nrp1-1 nrp2-1 mutant.
Figure 6.
Semiquantitative RT-PCR Analysis of Expression of Root Patterning Genes in the Wild-Type and the Double Mutant nrp1-1 nrp2-1 Roots.
RNA was isolated from roots collected from 6-d-old (A) and 12-d-old (B) seedlings. The increasing number of PCR cycles is given at the top of lanes, and the basal number of cycles (x) is given for each gene at the left of the panels. mut, mutant.
Taken together, our data indicate that NRP1 and NRP2 are required for the maintenance of correct expression of several genes involved in root proliferation and patterning.
Increased Sensitivity to DNA Damage and Release of Transcriptional Gene Silencing in the nrp1-1 nrp2-1 Mutant
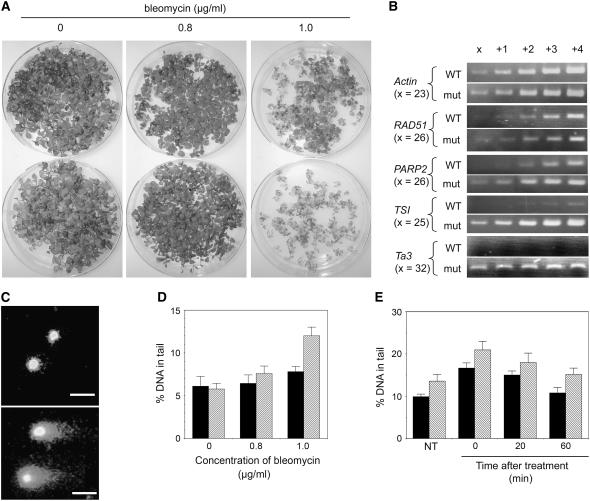
The G2/M arrest and the perturbed expression of some stress-responsive genes prompted us to investigate DNA damage response in the nrp1-1 nrp2-1 mutant. We first examined sensitivity of plant growth to the genotoxic agent bleomycin, an inducer of DNA strand breaks (Menke et al., 2001). The nrp1-1 nrp2-1 mutant plants were significantly more sensitive to bleomycin treatment than the wild-type plants (Figure 7A). The genes encoding poly(ADP-ribose) polymerase 2 (PARP2) and RAD51, which is involved in meiotic recombination and in homologous recombination repair (Schuermann et al., 2005), are transcriptionally induced by increased levels of DNA breaks (Chen et al., 2003). Consistent with their hypersensitivity to bleomycin, the nrp1-1 nrp2-1 mutant plants showed higher levels of PARP2 expression than the wild-type plants (Figure 7B). On the other hand, expression levels of RAD51 were quite similar in the mutant and wild-type plants.
Figure 7.
Response to Bleomycin, DNA Damage, and Release of Gene Silencing in the Double nrp1-1 nrp2-1 Mutant.
(A) Plant growth in the culture medium containing different concentrations of bleomycin. The top plates contain wild-type plants, and the bottom plates contain the mutant plants. Images were taken at 21 DAG.
(B) Semiquantitative RT-PCR analysis of gene expression in the wild-type and the mutant (mut) plants grown with 1.0 μg/mL bleomycin. The increasing number of PCR cycles is given on top of lanes, and the basal number of cycles (x) is given for each gene at the left of the panels.
(C) Representative examples of nuclei seen in the comet assay from the wild-type (top panel) and the mutant (bottom panel) plants grown with 1.0 μg/mL bleomycin.
(D) Levels of DNA damage as measured by the percentage of DNA in the tail of comet in the comet assay for the wild-type (black bars) and the mutant (gray bars) plants grown at different concentrations of bleomycin.
(E) Time course of DNA repair after a 1-h bleomycin treatment in the wild-type (black bars) and the mutant (gray bars) plants. NT, not treated with bleomycin. Each column in (D) and (E) represents the mean value together with the standard deviation bar from three independent experiments in which 200 comets on four gels were evaluated.
We compared the level of DNA damage in the nrp1-1 nrp2-1 mutant with the wild-type plants using the comet assay (Menke et al., 2001). When plants were grown in the absence or at low concentration of bleomycin, the mutant showed similar levels of DNA damage compared with the wild type. When plants were grown at higher concentration of bleomycin, however, the mutant showed a significant increase in DNA damage compared with the wild type (Figures 7C and 7D). To evaluate repair capability, we compared DNA damage levels within a 60-min recovery period after a 1-h bleomycin treatment using seedlings germinated on filter papers (see Methods). In the absence of bleomycin treatment, the nrp1-1 nrp2-1 mutant seedlings showed an increase in DNA damage compared with wild-type seedlings (Figure 7E), indicating that the nrp1-1 nrp2-1 mutant is hypersensitive to growth stress. Bleomycin treatment elevated levels of DNA damage in both the mutant and the wild-type seedlings. Interestingly, DNA damage was repaired in the mutant as rapidly as in the wild type (Figure 7E), indicating that the mutant is proficient in repair. This is consistent with the observation that the repair gene, RAD51, was well expressed in the mutant (Figure 7B). It is likely that the mutant genome is more sensitive to DNA damage and that additional genotoxic treatment leads to an accumulation of DNA damage that simply exceeds the capacity of the repair pathways, resulting in plant growth inhibition.
Release of transcription gene silencing (TGS) at TSI and/or transposons was previously reported in several Arabidopsis mutants defective in DNA/chromatin replication and assembly, including bru1, fas1, fas2, and rnr2 (Takeda et al., 2004; Ono et al., 2006; Schonrock et al., 2006; Wang and Liu, 2006). In the nrp1-1 nrp2-1 mutant, we also found that the silencing of the pericentromeric repeat TSI and the transposon Ta3 was released (Figure 7B). However, when plants were grown in the absence of bleomycin, release of the expression of TSI and Ta3 was not observed (data not shown). By contrast, the bru1, fas1, fas2, and rnr2 mutants released TGS under standard growth conditions. Thus, the defects in maintaining the silencing of heterochromatic chromatin are less severe in the nrp1-1 nrp2-1 mutant.
Histone and Chromatin Binding Activity of the NRP1 and NRP2 Proteins
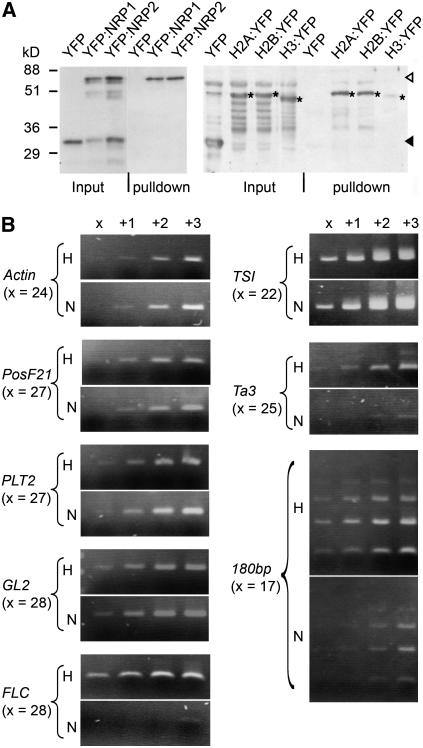
The histone binding activity of NRP1 and NRP2 proteins was investigated using pull-down assays. We found that NRP1 and NRP2 form homo- and heteromeric protein complexes (Figure 8A, left panel) and that NRP1 (Figure 8A, right panel) and NRP2 (data not shown) bind histones H2A and H2B but very little histone H3. This is in agreement with the proposal that NRP1 and NRP2 are histone H2A/H2B chaperones.
Figure 8.
NRP1 Protein Binds Histones H2A and H2B and Chromatin.
(A) Protein–protein interactions examined by pull-down assays. Fractions pulled down by NRP1-coated beads from total protein extracts of transgenic Arabidopsis plants expressing YFP, YFP:NRP1, YFP:NRP2, H2A:YFP, H2B:YFP, or H3:YFP were analyzed by protein gel blotting with a polyclonal anti-GFP antibody (which cross-reacts with YFP). The input fraction represents 5% of the total protein used in pull down. The closed and open arrowheads show the position of YFP and YFP:NRP1/YFP:NRP2, respectively. Positions of the histone fusion proteins H2A:YFP, H2B:YFP, and H3:YFP are indicated by asterisks.
(B) ChIP analysis of NRP1-associated chromatin at different genes. Transgenic Arabidopsis plants expressing H2A:YFP (H) or YFP:NRP1 (N) were analyzed with the anti-GFP antibody. The increasing number of PCR cycles is given at the top of the lanes, and the basal number of cycles (x) is given for each gene at the left of the panels.
Since YFP:NRP1 and YFP:NRP2 proteins are primarily localized in the nucleus (Figures 2A to 2D), we tested whether they could bind to chromatin in vivo. Transgenic Arabidopsis plants expressing YFP:NRP1 or H2A:YFP were first selected in ChIP analyses. We assumed that the histone H2A:YFP fusion protein is incorporated in nucleosomes and is distributed over the genome. The immunoprecipitates of the H2A:YFP-derived fraction (H) and the YFP:NRP1-derived fraction (N) were analyzed by PCR using gene-specific primers (Figure 8B). Actin was positively detected in both H and N fractions and subsequently used to normalize quantity of the two fractions. In the normalized H and N fractions, the levels of PosF21, PLT2, and GL2 were similar, indicating that YFP:NRP1 binds well to these genes. This is consistent with a previous observation showing that the expression of these genes was affected in the nrp1-1 nrp2-1 mutant (Table 1, Figure 6). The expression of FLOWERING LOCUS C (FLC), a gene that is regulated by chromatin environment (Zhao et al., 2005; Baurle and Dean, 2006), was unchanged in the nrp1-1 nrp2-1 mutant (data not shown). Interestingly, binding of YFP:NRP1 with the FLC region was negligible (Figure 8B). Since YFP:NRP1 also binds to genes whose expression was unchanged in the nrp1-1 or nrp1-1 nrp2-1 mutant (data not shown), binding is not restricted to NRP1-regulated genes. In pericentromeric heterochromatin, YFP:NRP1 bound to TSI and to a lesser extent, to Ta3 and the centromeric 180-bp repeats (Figure 8B). These ChIP results were obtained reproducibly in three independent experiments. In addition, YFP:NRP2 gave similar results as did YFP:NRP1, and H2B:YFP gave similar results as did H2A:YFP, whereas the negative control (YFP or in the absence of antibody) did not give signals (see Supplemental Figure 2 online). Our data thus clearly establish that YFP:NRP1 and YFP:NRP2 bind chromatin and are not uniformly distributed over the genome.
DISCUSSION
Our genetic, phenotypic, cellular, and molecular characterization of mutants demonstrates that NRPs are critical in epigenetic regulation in plant development. To our knowledge, this is the first report on cellular function of NRPs. We have shown that in Arabidopsis NRP1 and NRP2 encode histone H2A/H2B chaperones, are required for the maintenance of correct genome function, and play crucial roles in cell proliferation and differentiation in roots.
NRPs as H2A/H2B Chaperones
NRP1 and NRP2 can form homo- and heteromeric protein complexes and preferentially bind histones H2A and H2B rather than histone H3. This is consistent with the high degree of sequence homology of NRP1 and NRP2 to the NAP1 group proteins in the dimerization and the histone binding regions (Park and Luger, 2006). The current models propose that NAP1 binds to newly synthesized H2A and H2B in the cytoplasm, transports the histones to the nucleus, and deposits H2A/H2B on preformed H3/H4-DNA complex during nucleosome formation (Krude and Keller, 2001; Haushalter and Kadonaga, 2003; Polo and Almouzni, 2006). In vitro, the animal NRP group protein TAF-I can substitute for NAP1 in chromatin-based activation of replication and transcription as well as in chromatin assembly (Nagata et al., 1995; Kawase et al., 1996; Gamble et al., 2005). Dimerization of TAF-I was shown to be important for its activity in activating replication of the adenovirus genome in vitro (Miyaji-Yamaguchi et al., 1999). Our in vivo data demonstrate that NRPs associate with chromatin and regulate global expression of the Arabidopsis genome. It indicates that NRP group proteins act as H2A/H2B chaperones in chromatin assembly and regulate transcription on a chromatin template.
YFP:NRP1 and YFP:NRP2 are primarily localized in the nucleus, in contrast with our previous observation that showed that YFP fusions of several tobacco and rice NAP1 group proteins were predominantly localized in the cytoplasm (Dong et al., 2003, 2005). Compartmentalization could be involved in functional specification of the NRP and NAP1 groups' proteins. In some cells of transgenic Arabidopsis plants, YFP:NRP1 and YFP:NRP2 also accumulate in the cytoplasm. It is not yet known how the physiological state of the cells influences intracellular localization of these proteins. In addition to histones, a variety of other protein complexes contain the SET/TAF-I/I2PP2A proteins. Among these, B-type cyclins bind to SET and to NAP1, likely playing functional roles (Kellogg et al., 1995; Canela et al., 2003). While the tobacco B-type cyclin, Nicta;CYCB1;1, binds to NAP1 group proteins (Dong et al., 2005), it did not bind to NRP1 or to NRP2 in our pull-down assays (data not shown). The mammalian SET/TAF-I/I2PP2A proteins have PP2A inhibitor activity (Li et al., 1996). However, in our physiological tests, the PP2A-specific inhibitor cantharidin (Zhou et al., 2004) did not rescue the nrp1-1 nrp2-1 mutant short-root phenotype (data not shown). Whether or not NRP1 and NRP2 have functions in addition to that of histone chaperones is thus unclear at this stage.
Chaperoning H2A/H2B in Replication, Repair, and Transcription
The defects observed in the nrp1-1 nrp2-1 mutant underline crucial functions of dynamic H2A/H2B in chromatin-based genome function. Nucleosome assembly primarily occurs during DNA replication in the S phase of the cell cycle. The best-characterized histone chaperone is CAF-1, a heterotrimeric complex of CAC1/p150/FAS1, CAC2/p60/FAS2, and CAC3/p48/MSI1 in yeast, mammals, and plants, respectively. CAF-1 binds newly synthesized histones H3/H4 and deposits them onto replicating DNA through an interaction with proliferating cell nuclear antigen, the DNA polymerase sliding clump (for reviews, see Krude and Keller, 2001; Haushalter and Kadonaga, 2003; Polo and Almouzni, 2006). Our observation that cells arrest at G2/M in the nrp1-1 nrp2-1 mutant implies that H2A/H2B deposition also is actively involved in chromatin duplication. In support of this view, similar to that observed in the nrp1-1 nrp2-1 mutant, CYCB1:GUS expression accumulates in G2/M-arrested cells in mutants fas1 and fas2 (Schonrock et al., 2006) as well as in tonsoku (Suzuki et al., 2005), which is allelic to bru1 and thought to be involved in histone chaperoning together with CAF-1 (Takeda et al., 2004). The G2/M checkpoint ensures correct and complete DNA synthesis and chromatin duplication before the entry into mitosis and cytokinesis. B-type cyclins are involved in G2/M checkpoint activation (Stark and Taylor, 2006). The accumulation of CYCB1:GUS expression could be interpreted as a result of inhibition of chromatin duplication in the fas1, fas2, bru1, and nrp1-1 nrp2-1 mutants. In support of this view, CYCB1:GUS expression accumulates when S phase is inhibited by aphidicolin. Such an effect was not detected with hydroxyurea, which inhibits the G1/S and G2/M transitions, nor in mutants of RETINOBLASTOMA-RELATED, which regulates G1/S transition (Culligan et al., 2004; Wildwater et al., 2005).
Although to a lesser extend than the bru1 mutant (Takeda et al., 2004), the nrp1-1 nrp2-1 mutant also shows significant hypersensitivity to genotoxic stress and increased level of DNA damage. It appears that timely completion of chromatin assembly is critical to maintain the genome integrity. In mammals, SET displays inhibitor activity to NM23-H1, a nucleoside diphosphate kinase (NDPK) implicated in DNA single-strand nicks and in the suppression of tumor metastasis (Fan et al., 2003). NDPK homologues exist in plants and are likely to play a role in UV response (Zimmermann et al., 1999). It will be interesting to investigate whether NRP1, NRP2, and NDPK proteins are involved in the same pathway. Phosphorylation of H2A and its variant H2A.X rapidly occurs after DNA damage in yeast, mammals, and plants, and histone exchange and chromatin remodeling appear to play roles in DNA repair (Friesner et al., 2005; Jin et al., 2005; Polo and Almouzni, 2006). NRP1 and NRP2 are, however, unlikely to be critical in these processes since the nrp1-1 nrp2-1 mutant is proficient in repair. Specific chaperones, such as SWR1 in yeast and TIP60 in animals, together with the INO80 nucleosome-remodeling complex are important for DNA repair (Jin et al., 2005; Schuermann et al., 2005; Polo and Almouzni, 2006). In contrast with fas1 and fas2 (Schonrock et al., 2006), the nrp1-1 nrp2-1 mutant did not show significant perturbation of expression of DNA repair genes.
The number of differentially expressed genes in the nrp1-1 nrp2-1 mutant is significantly lower than that reported in the fas1 and fas2 mutants (Schonrock et al., 2006). In addition, there is very little overlap of the genes identified in these two categories of mutants. It is likely that the NRP1 H2A/H2B chaperone and the CAF1 H3/H4 chaperone act independently. Both transcriptional repression (Shikama et al., 2000) and activation (Telese et al., 2005) have been previously reported for animal SET/TAF-I/I2PP2A proteins. Our results provide additional data on NRP1- and NRP2-regulated gene expression of the complete Arabidopsis genome. The genes identified in this study will provide a framework for understanding the events that occur at these gene-specific chromatin locations. Our observation of release of TGS in the nrp1-1 nrp2-1 mutant and binding of NRP1 and NRP2 to pericentromeric TSI and Ta3 suggests that NRP1 and NRP2 are also involved in heterochromatin formation.
Chromatin Remodeling in Root Development
The nrp1-1 nrp2-1 mutant has a specific short root phenotype. This is in contrast with the majority of the mutants affecting chromatin remodeling factors that exhibit pleiotropic phenotypes, particularly in aerial organs of the plants. The fas1, fas2, and bru1 mutants all show stem fasciation and abnormal phyllotaxy (Kaya et al., 2001; Takeda et al., 2004). The knockout of HIRA, a H3/H4 chaperone involved in replication-independent nucleosome assembly, is embryo lethal. However, a decrease in the level of HIRA resulted in dramatic modifications in leaves in transgenic Arabidopsis (Phelps-Durr et al., 2005). A short-root phenotype was also previously observed in the fas1, fas2, and bru1 mutants, but the underlying molecular mechanism is not clear. We demonstrated that expression of several transcription factors, including GL2 and PLT2, which are important in root cell fate determination (Ohashi et al., 2003; Aida et al., 2004), are regulated by NRP1 and NRP2. The chromatin at GL2 was recently shown to be dynamic and reorganized upon cell division in response to local positional information in roots (Costa and Shaw, 2006). Histone chaperones likely play important roles in this chromatin remodeling process. Histone acetylation also is involved in expression of the root patterning genes CPC, GL2, and WER (Xu et al., 2005). Taken together, these results indicate that chromatin assembly and remodeling contribute to the maintenance of a correct pattern of gene transcription during root growth.
Understanding the epigenetic regulation of root growth is of particular interest in view of the astonishing capacity of plants to cope with environmental stress (Bengough et al., 2006). The nrp1-1, nrp2-1, and nrp1-1 nrp2-1 mutants described in this study will be useful to analyze gene expression maps to gain a better understanding of the transcriptional circuits in the root tissue (Birnbaum and Benfey, 2004). The H2A gene HTA1, but not the other 12 H2A genes, was reported to be involved in T-DNA integration during Agrobacterium tumefaciens–mediated transformation of roots but not of flowers in Arabidopsis (Yi et al., 2006). Additional roles of H2A/H2B in the dynamic chromatin involved in plant growth and development likely await discovery. Our unpublished observation reveals that single mutants of the NAP1 group genes in Arabidopsis have no obvious phenotype. Further analysis of double, triple, and quadruple mutants of this group of genes will help to clarify their biological function.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana alleles were derived from the Columbia ecotype. nrp1-1 and nrp2-1 alleles correspond respectively to Salk_117793 and Salk_030348, T-DNA insertion strains from The Arabidopsis Information Resource (http://arabidopsis.org). The double mutant nrp1-1 nrp2-1 was obtained in our laboratory by crossing the two single mutants. In vitro plant culture was performed on agar-solidified Murashige and Skoog (MS) medium M0255 (Duschefa) supplemented with 0.9% sucrose at 21°C under 16 h light/8 h dark.
Plant Vector Construction and Plant Transformation
The NRP1 and NRP2 cDNAs were obtained by RT-PCR using primer pairs N5P1/N5P2 and N6P1/N6P2 (see Supplemental Table 4 online), respectively. The resulting PCR products were cloned in pEYFP-EYFP vector (Yu et al., 2004) and sequenced to confirm the absence of sequence errors and the in-frame fusion with YFP. The YFP:NRP1 and YFP:NRP2 fragments were subcloned in pER8 vector (Zuo et al., 2000) using SalII-XhoI and SpeI restriction sites, resulting in pYFP:NRP1 and pYFP:NRP2, respectively. The pYFP:NRP1 and pYFP:NRP2 plasmids were introduced into Agrobacterium tumefaciens, and the resulting strains were used to transform Arabidopsis and tobacco (Nicotiana tabacum) BY-2 cells as described previously (Yu et al., 2003). Induction of transgene expression from pER8-based vectors was performed according to Zuo et al. (2000), using 4 μM estradiol.
Microscopy and Histology
Root tips were incubated in 10 μg/mL propidium iodide for 5 to 10 min and then imaged using a Zeiss model LSM510 confocal microscope (Carl Zeiss). Tobacco BY-2 cells were imaged as previously described (Yu et al., 2004). For histological analysis, roots were fixed in 25 mM phosphate buffer, pH 7.2, containing 1% glutaraldehyde, embedded in LR White resin (EMS), and transversely sectioned at 1 μm.
RT-PCR
Total RNA was prepared using the TRIzol kit according to the manufacturer's instructions (Invitrogen). Semiquantitative RT-PCR was performed according to standard procedures using Improm-II reverse transcriptase (Promega). Gene-specific primers used in PCR analysis are given in Supplemental Table 4 online.
GUS Activity Assay
Histochemical GUS activity assay was performed as described (Yu et al., 2003). Essential results were reproducibly obtained from >10 plants for each condition.
Microarray Analyses
Wild-type and mutants seeds were germinated under the same growth conditions. Three independently derived sets of 6-d-old seedlings, 50 to 60 plants per set, were pooled for each genotype. Total RNA was isolated from each sample, and microarray analysis was performed as described (Lurin et al., 2004). Briefly, cRNA was synthesized and fluorescent cDNAs were synthesized from cRNA using cy3-dUTP and cy5-dUTP, respectively, for each sample. The cy3-labeled cDNA derived from mutant seedlings was combined with cy5-labeled cDNA derived from wild-type seedlings and used for hybridization of the CATMA slides (Crowe et al., 2003). Repeated hybridization was performed using combined cy5-labeled mutant cDNA with cy3-labeled wild-type cDNA. Hybridization, scanning of microarrays, and statistical analysis were performed as decribed (Lurin et al., 2004). To further enrich for biologically relevant changes linked with mutant genotype, the whole procedure was repeated in a second experiment with new sets of seeds. Genes were considered as significantly perturbed in the mutant if the change was at least 1.5-fold and the P values inferior to 0.05 from the two independent experiments.
Bleomycin Treatment and Comet Assay
Genotoxic effect of bleomycin was evaluated by growth of plants in the presence of the drug in the agar-solidified MS medium. For DNA repair test, seeds were germinated on filter paper soaked in liquid MS medium. Twelve days after germination, seedlings together with filter paper were transferred to MS medium containing bleomycin and incubated for 1 h. Then, they were rinsed three times in MS medium and placed on bleomycin-free MS medium for recovery. Approximately 150 seedlings were harvested at each assay point from three replicates of treatment and used in RT-PCR and comet assays. Comet assay was performed using the N/N protocol as described (Menke et al., 2001). Images of comets were captured under a Nikon 800 epifluorescence microscope equipped with a DXM1200 digital camera. The comet analysis was performed using CometScore software (http://autocomet.com).
Pull-Down Assay and ChIP Analysis
The coding regions of NRP1 and NRP2 were amplified using primer pairs N5P3/N5P4 and N6P3/N6P4 (see Supplemental Table 4 online), respectively. The resulting PCR products were cloned into pET-14b vector (Novagen) using the NdeI and BamHI restriction sites. Recombinant protein production and pull-down assays were performed as described (Dong et al., 2003, 2005). The anti-GFP rabbit polyclonal antibody (Molecular Probes) was used at a dilution of 1:5000 for protein gel blotting. ChIP analysis was performed as described (Zhao et al., 2005). Gene-specific primers used in ChIP analysis are given in Supplemental Table 4 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_001036205 (NRP1) and NM_101738 (NRP2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Analysis of Expression of the Arabidopsis NAP1 and NRP Group Genes.
Supplemental Figure 2. ChIP Analysis of Chromatin Association of Proteins at Different Genes.
Supplemental Table 1. Genes Differentially Expressed in nrp1-1.
Supplemental Table 2. Genes Differentially Expressed in nrp1-2.
Supplemental Table 3. Genes Differentially Expressed in nrp1-1 nrp2-1.
Supplemental Table 4. List of Primers.
Supplementary Material
Acknowledgments
We thank Ludivine Taconnat for assistance in microarray analysis and Jean Canaday and Manfred Heinlein for critical reading of the manuscript. Y.Z. is supported by a foreign student fellowship from the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche. This work was supported in part by the Centre National de la Recherche Scientifique (PICS 2391 to W.-H.S.); by the National Natural Science Foundation of China (Grant NSF30570933 to A.D.), and by the Scientific and Technological Council Foundation of Shanghai (Grant 04JC14017 to A.D.). The InterInstitute confocal microscopy platform was cofinanced by the Centre National de la Recherche Scientifique, the Université Louis Pasteur, the Région Alsace, and the Association pour la Recherche sur le Cancer.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wen-Hui Shen (wen-hui.shen@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
References
- Adler, H.T., Nallaseth, F.S., Walter, G., and Tkachuk, D.C. (1997). HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J. Biol. Chem. 272 28407–28414. [DOI] [PubMed] [Google Scholar]
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Angelov, D., Bondarenko, V.A., Almagro, S., Menoni, H., Mongelard, F., Hans, F., Mietton, F., Studitsky, V.M., Hamiche, A., Dimitrov, S., and Bouvet, P. (2006). Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125 655–664. [DOI] [PubMed] [Google Scholar]
- Bengough, A.G., Bransby, M.F., Hans, J., McKenna, S.J., Roberts, T.J., and Valentine, T.A. (2006). Root responses to soil physical conditions; growth dynamics from field to cell. J. Exp. Bot. 57 437–447. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., and Benfey, P.N. (2004). Network building: Transcriptional circuits in the root. Curr. Opin. Plant Biol. 7 582–588. [DOI] [PubMed] [Google Scholar]
- Broekaert, W.F., Delaure, S.L., De Bolle, M.F., and Cammue, B.P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44 393–416. [DOI] [PubMed] [Google Scholar]
- Canela, N., Rodriguez-Vilarrupla, A., Estanyol, J.M., Diaz, C., Pujol, M.J., Agell, N., and Bachs, O. (2003). The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 278 1158–1164. [DOI] [PubMed] [Google Scholar]
- Chen, I.P., Haehnel, U., Altschmied, L., Schubert, I., and Puchta, H. (2003). The transcriptional response of Arabidopsis to genotoxic stress - A high-density colony array study (HDCA). Plant J. 35 771–786. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Costa, S., and Shaw, P. (2006). Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439 493–496. [DOI] [PubMed] [Google Scholar]
- Criqui, M.C., Weingartner, M., Capron, A., Parmentier, Y., Shen, W.H., Heberle-Bors, E., Bogre, L., and Genschik, P. (2001). Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 28 569–581. [DOI] [PubMed] [Google Scholar]
- Crowe, M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31 156–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Dong, A., Liu, Z., Zhu, Y., Yu, F., Li, Z., Cao, K., and Shen, W.H. (2005). Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 138 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, A., Zhu, Y., Yu, Y., Cao, K., Sun, C., and Shen, W.H. (2003). Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 216 561–570. [DOI] [PubMed] [Google Scholar]
- Fan, Z., Beresford, P.J., Oh, D.Y., Zhang, D., and Lieberman, J. (2003). Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112 659–672. [DOI] [PubMed] [Google Scholar]
- Friesner, J.D., Liu, B., Culligan, K., and Britt, A.B. (2005). Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 16 2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166 740–754. [DOI] [PubMed] [Google Scholar]
- Gamble, M.J., Erdjument-Bromage, H., Tempst, P., Freedman, L.P., and Fisher, R.P. (2005). The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol. Cell. Biol. 25 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haushalter, K.A., and Kadonaga, J.T. (2003). Chromatin assembly by DNA-translocating motors. Nat. Rev. Mol. Cell Biol. 4 613–620. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.F., and Fischer, R.L. (2005). Biology of chromatin dynamics. Annu. Rev. Plant Biol. 56 327–351. [DOI] [PubMed] [Google Scholar]
- Ishimi, Y., Hirosumi, J., Sato, W., Sugasawa, K., Yokota, S., Hanaoka, F., and Yamada, M. (1984). Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 142 431–439. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- Jin, J., Cai, Y., Li, B., Conaway, R.C., Workman, J.L., Conaway, J.W., and Kusch, T. (2005). In and out: Histone variant exchange in chromatin. Trends Biochem. Sci. 30 680–687. [DOI] [PubMed] [Google Scholar]
- Kawase, H., Okuwaki, M., Miyaji, M., Ohba, R., Handa, H., Ishimi, Y., Fujii-Nakata, T., Kikuchi, A., and Nagata, K. (1996). NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1 1045–1056. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131–142. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., Kikuchi, A., Fujii-Nakata, T., Turck, C.W., and Murray, A.W. (1995). Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude, T., and Keller, C. (2001). Chromatin assembly during S phase: Contributions from histone deposition, DNA replication and the cell division cycle. Cell. Mol. Life Sci. 58 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, S.H., Shen, R., and Schiefelbein, J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307 1111–1113. [DOI] [PubMed] [Google Scholar]
- Lankenau, S., Barnickel, T., Marhold, J., Lyko, F., Mechler, B.M., and Lankenau, D.H. (2003). Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Makkinje, A., and Damuni, Z. (1996). The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 271 11059–11062. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251–260. [DOI] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., and Zhang, Y. (2005). The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6 838–849. [DOI] [PubMed] [Google Scholar]
- Menke, M., Chen, I., Angelis, K.J., and Schubert, I. (2001). DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res. 493 87–93. [DOI] [PubMed] [Google Scholar]
- Miyaji-Yamaguchi, M., Okuwaki, M., and Nagata, K. (1999). Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity. J. Mol. Biol. 290 547–557. [DOI] [PubMed] [Google Scholar]
- Nagata, K., Kawase, H., Handa, H., Yano, K., Yamasaki, M., Ishimi, Y., Okuda, A., Kikuchi, A., and Matsumoto, K. (1995). Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92 4279–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Rodrigues-Pousada, R., Possenti, M., Ruberti, I., Morelli, G., and Aoyama, T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300 1427–1430. [DOI] [PubMed] [Google Scholar]
- Ohkuni, K., Shirahige, K., and Kikuchi, A. (2003). Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306 5–9. [DOI] [PubMed] [Google Scholar]
- Ono, T., Kaya, H., Takeda, S., Abe, M., Ogawa, Y., Kato, M., Kakutani, T., Scheid, O.M., Araki, T., and Shibahara, K. (2006). Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11 153–162. [DOI] [PubMed] [Google Scholar]
- Park, Y.J., and Luger, K. (2006). The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 103 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E., and Almouzni, G. (2006). Chromatin assembly: A basic recipe with various flavours. Curr. Opin. Genet. Dev. 16 104–111. [DOI] [PubMed] [Google Scholar]
- Rogner, U.C., Spyropoulos, D.D., Le Novere, N., Changeux, J.P., and Avner, P. (2000). Control of neurulation by the nucleosome assembly protein-1-like 2. Nat. Genet. 25 431–435. [DOI] [PubMed] [Google Scholar]
- Schonrock, N., Exner, V., Probst, A., Gruissem, W., and Hennig, L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281 9560–9568. [DOI] [PubMed] [Google Scholar]
- Schuermann, D., Molinier, J., Fritsch, O., and Hohn, B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21 172–181. [DOI] [PubMed] [Google Scholar]
- Shikama, N., Chan, H.M., Krstic-Demonacos, M., Smith, L., Lee, C.W., Cairns, W., and La Thangue, N.B. (2000). Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20 8933–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, G.R., and Taylor, W.R. (2006). Control of the G2/M transition. Mol. Biotechnol. 32 227–248. [DOI] [PubMed] [Google Scholar]
- Stepanova, A.N., Hoyt, J.M., Hamilton, A.A., and Alonso, J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Nakajima, S., Inagaki, S., Hirano-Nakakita, M., Matsuoka, K., Demura, T., Fukuda, H., Morikami, A., and Nakamura, K. (2005). TONSOKU is expressed in S phase of the cell cycle and its defect delays cell cycle progression in Arabidopsis. Plant Cell Physiol. 46 736–742. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Mittelsten Scheid, O., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telese, F., Bruni, P., Donizetti, A., Gianni, D., D'Ambrosio, C., Scaloni, A., Zambrano, N., Rosenfeld, M.G., and Russo, T. (2005). Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 6 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, M., Koshino-Kimura, Y., and Okada, K. (2005). Stepwise understanding of root development. Curr. Opin. Plant Biol. 8 71–76. [DOI] [PubMed] [Google Scholar]
- von Lindern, M., Fornerod, M., van Baal, S., Jaegle, M., de Wit, T., Buijs, A., and Grosveld, G. (1992). The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., and Liu, Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater, M., Campilho, A., Perez-Perez, J.M., Heidstra, R., Blilou, I., Korthout, H., Chatterjee, J., Mariconti, L., Gruissem, W., and Scheres, B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123 1337–1349. [DOI] [PubMed] [Google Scholar]
- Xu, C.R., Liu, C., Wang, Y.L., Li, L.C., Chen, W.Q., Xu, Z.H., and Bai, S.N. (2005). Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc. Natl. Acad. Sci. USA 102 14469–14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H., Sardesai, N., Fujinuma, T., Chan, C.W., Veena, and Gelvin, S.B. (2006). Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 18 1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Dong, A., and Shen, W.H. (2004). Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 40 699–711. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Steinmetz, A., Meyer, D., Brown, S., and Shen, W.H. (2003). The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15 2763–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Yu, Y., Meyer, D., Wu, C., and Shen, W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7 1156–1160. [DOI] [PubMed] [Google Scholar]
- Zhou, H.W., Nussbaumer, C., Chao, Y., and DeLong, A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, S., Baumann, A., Jaekel, K., Marbach, I., Engelberg, D., and Frohnmeyer, H. (1999). UV-responsive genes of Arabidopsis revealed by similarity to the Gcn4-mediated UV response in yeast. J. Biol. Chem. 274 17017–17024. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.