Abstract
We previously showed that Arabidopsis thaliana histone acetyltransferase TAF1/HAF2 is required for the light regulation of growth and gene expression, and we show here that histone acetyltransferase GCN5 and histone deacetylase HD1/HDA19 are also involved in such regulation. Mutation of GCN5 resulted in a long-hypocotyl phenotype and reduced light-inducible gene expression, whereas mutation of HD1 induced opposite effects. The double mutant gcn5 hd1 restored a normal photomorphogenic phenotype. By contrast, the double mutant gcn5 taf1 resulted in further loss of light-regulated gene expression. gcn5 reduced acetylation of histones H3 and H4, mostly on the core promoter regions, whereas hd1 increased acetylation on both core and more upstream promoter regions. GCN5 and TAF1 were both required for H3K9, H3K27, and H4K12 acetylation on the target promoters, but H3K14 acetylation was dependent only on GCN5. Interestingly, gcn5 taf1 had a cumulative effect mainly on H3K9 acetylation. On the other hand, hd1 induced increased acetylation on H3K9, H3K27, H4K5, and H4K8. GCN5 was also shown to be directly associated with the light-responsive promoters. These results suggest that acetylation of specific histone Lys residues, regulated by GCN5, TAF1, and HD1, is required for light-regulated gene expression.
INTRODUCTION
Many physiological modifications induced by light signaling are caused by changes in gene expression. Although recent studies have led to the identification of a large number of regulators involved in light-signaling pathways, the molecular mechanisms of light activation of gene transcription remain obscure. Promoter analyses have defined a number of light-responsive elements. For instance, the ACGT core–containing G-box identified in the light-responsive genes CAB2 (or LHCB1.1, hereafter referred to as CAB2) and RBCS-1A is bound by the basic domain/Leu zipper transcription factor HY5 in vitro (Chattopadhyay et al., 1998), which acts downstream of both phytochromes and cryptochromes (Oyama et al., 1997). Further studies have shown that a combination of several light-responsive elements is required for light activation, suggesting that interaction between the cognate DNA binding transcription factors is necessary for light-induced gene activation (Martinez-Hernandez et al., 2002; Yadav et al., 2002, 2005; Maxwell et al., 2003). However, it is not known how DNA binding proteins function to activate light-responsive gene transcription.
Chromatin structure plays a critical role in gene transcription. The basic structural unit of chromatin is the nucleosome. The positioning of nucleosomes on promoters is inhibitory to transcription in that they prevent the transcriptional initiation complex or transcription factors from binding to a promoter (Horn and Peterson, 2002). Covalent modifications of the N-terminal tails of the core histones affect nucleosome positioning and compaction; thus, they play important roles in gene regulation (Lusser, 2002; Loidl, 2004). Histone acetylation involves the transfer of acetyl groups from acetyl-CoA to the N-terminal Lys residues of histones H3 and H4, as well as H2A and H2B. Hyperacetylation of histones relaxes chromatin structure and is associated with transcriptional activation, whereas hypoacetylation of histones induces chromatin compaction and gene repression (Sterner and Berger, 2000; Marmorstein and Roth, 2001; Carrozza et al., 2003).
Histone acetylation is catalyzed by histone acetyltransferases (HATs), whereas histone deacetylation is catalyzed by histone deacetylases (HDACs). Recent work has shown that several HATs are associated with transcription coactivators and that HDACs are associated with corepressor complexes in yeast and animal cells, indicating that histone acetylation is an integral part of transcriptional regulatory systems (Marmorstein and Roth, 2001; Berger, 2002; Carrozza et al., 2003). The HATs found in Arabidopsis thaliana are grouped into four types based on primary homology with yeast and mammalian HATs: GNAT, MYST, CBP, and TAF1 (Sterner and Berger, 2000; Pandey et al., 2002). The mutation of Arabidopsis HAT genes induces defects in many aspects of plant development and growth (Bertrand et al., 2003, 2005; Vlachonasios et al., 2003).
Plant HDACs can be grouped into four subclasses. Three of them have primary homology with three yeast HDACs: RDP3, HDA1, and SIR2 (Pandey et al., 2002). In addition, a specific class of HDAC (known as the HD2 class) is found only in plants (Lusser et al., 1997; Pandey et al., 2002). The RDP3-like protein is the major HDAC in yeast and mammals. Several RDP3-type HDACs have been found in Arabidopsis and maize (Zea mays) (Pandey et al., 2002; Varotto et al., 2003). Arabidopsis RDP3-type HDACs include HDA6, HDA7, HDA9, and HDA19 (also known as HD1; hereafter, we use HD1). Inactivation of HD1 in transgenic plants either expressing an antisense At HD1 construct or containing a T-DNA insertion mutation (athd1-t1) induces pleiotropic developmental abnormalities (Wu et al., 2000; Tian and Chen, 2001; Tian et al., 2003).
We have previously shown that an Arabidopsis gene encoding the HAT-containing TAF1 was required for leaf greening and light-activated gene transcription, and its absence led to additional developmental abnormalities (Bertrand et al., 2005). In this work, we show that GCN5 and HD1 are also required for light-regulated gene expression and growth. Genetic and molecular analyses suggest that GCN5 and HD1 function antagonistically, whereas GCN5 and TAF1 act agonistically to control the histone acetylation balance required to switch on light-regulated gene transcription.
RESULTS
Mutations in GCN5 and HD1 Genes Induce Opposite Hypocotyl Phenotypes
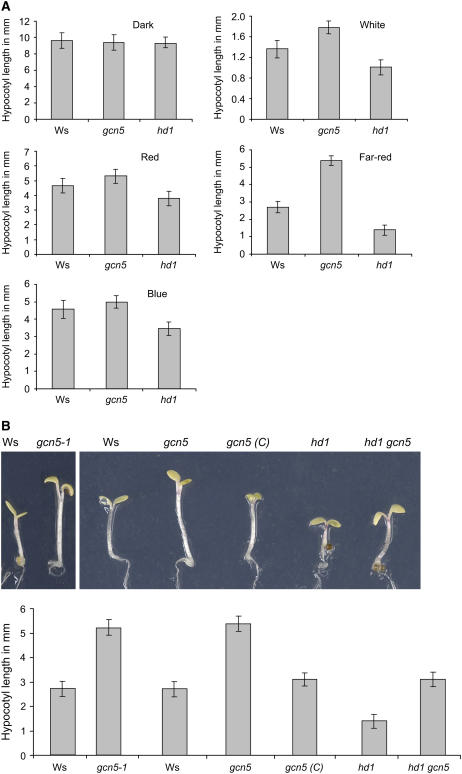
To examine whether HAT- and HDAC-coding genes are involved in light-regulated plant developmental processes, we used previously characterized mutant alleles of GCN5 (dlx8; hereafter called gcn5) and HD1 (athd1-t1; hereafter called hd1) (all in a Wassilewskija [Ws] background) (Bertrand et al., 2003; Tian et al., 2003). Seeds of the wild type (Ws) and the mutants were germinated under different light conditions including complete darkness, white light (16 h/d at 120 μmol·m−2·s−1) and continuous blue (18 μmol·m−2·s−1), far-red (5 μmol·m−2·s−1), and red (10 μmol·m−2·s−1) light. At day 7 after germination, hypocotyl lengths (a characteristic photomorphogenic trait) were measured and statistically analyzed (see Methods) to determine significant differences. In the dark, hypocotyl lengths of the different genotypes were similar. Under different light conditions, gcn5 seedlings showed a light-hyposensitive phenotype with longer hypocotyls compared with those of wild-type plants. By contrast, hypocotyls of hd1 seedlings were shorter than wild-type hypocotyls, showing an enhanced photomorphogenic phenotype. The phenotypes for both gcn5 and hd1 were most pronounced when grown under continuous far-red light (Figure 1A).
Figure 1.
gcn5 and hd1 Mutations Affect the Light Repression of Hypocotyl Elongation.
(A) Seven-day-old seedling hypocotyl lengths of wild-type Ws, gcn5, and hd1plantlets grown in darkness or under white light (16 h/d at 120 μmol·m−2·s−1) or continuous red (10 μmol·m−2·s−1) far-red (5 μmol·m−2·s−1), or blue (18 μmol·m−2·s−1) light.
(B) Seven-day-old far-red light–grown seedling phenotypes. Left, comparison of the gcn5-1 allele with the wild type (Ws); right, comparison of Ws, gcn5, gcn5 complemented with GCN5 cDNA [gcn5 (C)], hd1, and the double mutant gcn5 hd1-1. Means of 20 plantlets are given. Error bars represent sd values. The measures were analyzed by Student's t test at α = 0.05.
To confirm these observations, we analyzed another GCN5 T-DNA insertion allele, called gcn5-1 (in a Ws background) (Vlachonasios et al., 2003). The T-DNA insertion is located in the last exon of the gene. The gcn5-1 mutant also showed a long-hypocotyl phenotype (Figure 1B). In addition, complementation of the gcn5 mutation with the GCN5 cDNA restored a normal-hypocotyl phenotype (Bertrand et al., 2003) (Figure 1B). Furthermore, hd1 gcn5 double mutants were made using gcn5 pollen to fertilize hd1. The hypocotyl length of the double mutant was similar to that of the wild type, suggesting that both mutations could mutually compensate for the respective phenotypes.
gcn5 and hd1 Have Opposite Effects on Light-Inducible Gene Expression
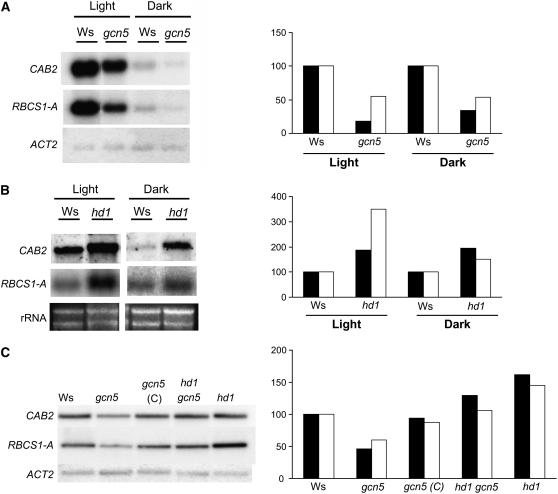
Light-inducible gene expression in gcn5 and hd1 mutants was analyzed by RNA gel hybridization. Total RNA isolated from either dark- or light-grown seedlings was hybridized with probes corresponding to the CAB2 and RBCS-1A genes. As shown in Figure 2A, the transcripts of CAB2 and RBCS-1A in light-grown gcn5 seedlings were seriously reduced compared with those of the wild type. Decreases were also observed in dark-grown plants (Figure 2A). By contrast, the hd1 mutation induced a clear increase of CAB2 expression in both light- and dark-grown plants (Figure 2B). Thus, GCN5 and HD1 also have opposite functions in light-inducible gene expression. The decrease of CAB2 and RBCS-1A expression in gcn5 mutants could be restored by complementation with the GCN5 cDNA or by the hd1 gcn5 double mutation (Figure 2C). Together, these data indicated that the gcn5 mutation induced the downregulation of light-activated gene expression and that the gcn5 and hd1 mutations could compensate mutually with respect to light-induced gene expression.
Figure 2.
The gcn5 and hd1 Mutations Have Opposite Effects on Light-Inducible Gene Expression.
(A) RNA gel analysis of CAB2 and RBCS-1A gene expression in both light- and dark-grown gcn5 and Ws seedlings. Actin mRNA levels were analyzed to normalize gel loading.
(B) RNA gel analysis of CAB2 and RBCS-1A gene expression in both light- and dark-grown hd1 and Ws seedlings. rRNA transferred to the membranes was used to normalize gel loading.
(C) RNA gel analysis of CAB2 and RBCS-1A gene expression in light-grown Ws, gcn5, gcn5 complemented (C), the gcn5-2 hd1 double mutant, and hd1 seedlings.
Signal quantifications relative to the wild type (arbitrarily given as 100) are presented at right of the gels. Black bars, RBCS-1A; white bars, CAB2.
Effects of the Double Mutation of gcn5 and taf1
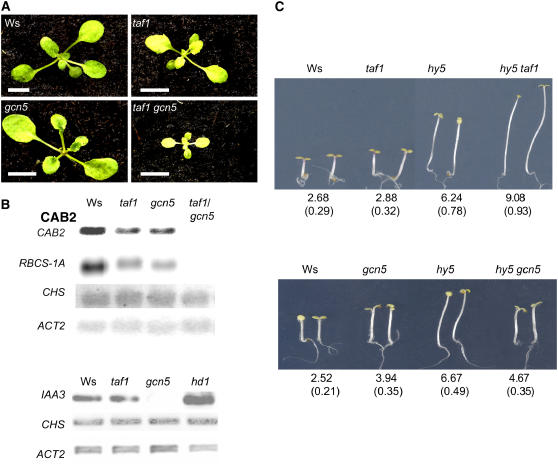
We have shown previously that the mutation of TAF1 (haf2; hereafter called taf1) also impairs CAB2 and RBCS-1A gene expression (Bertrand et al., 2005). To assess any functional relationship between TAF1 and GCN5, we made taf1 gcn5 double mutants by genetic crosses. The double mutant exhibited a severe growth phenotype but managed to yield mature plants (Figure 3A). RNA gel blot analyses showed that the transcripts of CAB2 and RBCS-1A were further decreased to background levels in the double mutant (Figure 3B), suggesting that gcn5 and taf1 mutations had cumulative effects on both plant growth and light-responsive gene expression. However, the expression of the chalcone synthase gene (CHS), another light-responsive gene, was not affected significantly by either mutation, except for a slight decrease in the double mutant (Figure 3B), suggesting that the two HAT genes are involved only in the expression of a subset of light-inducible genes. By contrast, the expression of a highly light-activated gene, IAA3 (Tian et al., 2002), was repressed dramatically in gcn5 and gcn5-1 (Figure 3B) (Vlachonasios et al., 2003) but not in taf1 (Figure 3B), indicating that the two HAT genes have distinct light-inducible target genes. This finding is in agreement with the different photomorphogenic phenotypes observed in the respective mutants (Figure 3C) (Bertrand et al., 2005). The hd1 mutation also induced the expression of IAA3 but did not affect that of CHS (Figure 3B).
Figure 3.
Genetic Relationship between gcn5 and taf1 Mutations.
(A) Phenotypes of taf1 gcn5 double mutants with rosette leaves (3 weeks) grown in the greenhouse. Bars = 0.5 cm.
(B) Top, expression of the light-inducible genes CAB2, RBCS-1A, and CHS in the double mutant compared with that in Ws and the single mutants. Bottom, comparison of expression levels of IAA3 and CHS in Ws, gcn5, taf1, and hd1.
(C) Comparison of 7-d-old seedling hypocotyl lengths of taf1 hy5 (top) and gcn5 hy5 (bottom) double mutants grown under continuous far-red light (5 μmol·m−2·s−1). Average lengths (in millimeters) are indicated at bottom with sd values from at least 20 plantlets (in parentheses).
The mutation of the light-responsive positive transcription factor HY5 affects the plant perception of light signals and induces constitutive long hypocotyls (Oyama et al., 1997). We have shown previously that the taf1 mutation could enhance the hy5 long-hypocotyl phenotype irrespective of light conditions (Bertrand et al., 2005), suggesting that the TAF1 pathway may interact with that of HY5. In this study, we obtained gcn5 hy5 double mutants and, in contrast with the enhanced hypocotyl elongation observed in hy5 taf1 plants, the hypocotyl lengths of gcn5 hy5 were similar to those of the gcn5 single mutants. Such data suggest that GCN5 and TAF1 might interact differently with HY5.
gcn5 and hd1 Affect Histone Acetylation on Light-Inducible Promoters
We have shown previously that the taf1 mutation reduces the acetylation of histones H3 and H4 on the promoters of CAB2 and RBCS-1A (Bertrand et al., 2005). To study whether the deregulation of light-inducible gene expression observed in gcn5 and hd1 mutants was also linked to alterations in histone acetylation on the genes, we performed chromatin immunoprecipitation (ChIP) assays. Chromatin fragments isolated from 12-d-old light-grown seedlings of the wild type and gcn5 or hd1 mutants were immunoprecipitated with antibodies raised against acetylated histone H3 or acetylated histone H4 (Upstate Biotechnology; see Methods). The precipitated chromatin DNA was analyzed by real-time PCR to test for enrichment relative to nonprecipitated (input) genomic DNA according to Frank et al. (2001). The enrichment of promoter fragments relative to input chromatin DNA in the wild type (arbitrarily given as 100%) was compared with that found for each mutant. Primer sets used corresponded to upstream promoter regions (∼1.5 to 2 kb from the initiation ATG codon), TATA box proximal or core promoters, coding regions, and 3′ untranslated sequences of CAB2, RBCS-1A, and IAA3 (Figure 4). Because the intergenic space between CAB2 and the upstream gene CAB1 constitutes <1.5 kb, no upstream promoter primer set was designed for CAB2. Because the expression of CHS was not affected by the different mutations, a primer set corresponding only to the core promoter region of CHS was used as a control. From the results of these experiments (Figure 4), several observations could be made. (1) The gcn5 mutation induced a clear decrease of histone H3 acetylation on the core promoter region of CAB2, RBCS-1A, and IAA. Histone H4 acetylation was decreased on the core promoter of RBCS-1A. However, the mutation had no or much less effect on the other analyzed regions of the three genes. (2) By contrast, the hd1 mutation increased the acetylation levels of both histones H3 and H4, not only on the core promoter of the three genes but also on the upstream region of IAA3 and, to a lesser extent, RBCS-1A. (3) Acetylation of histones H3 and H4 on the core promoter of CHS was not significantly affected by the mutations. These data suggested that the gcn5 and hd1 mutations had opposite effects on the acetylation of histones H3 and H4 on the promoters of a subset of light-inducible genes.
Figure 4.
Acetylation State of Histones H3 and H4 of CAB2, RBCS-1A, IAA3, and CHS in Ws, gcn5, and hd1 Seedlings.
Nuclei were extracted from cross-linked 12-d-old light-grown seedlings, sonicated, and immunoprecipitated with antibodies specific to total histone H3, acetylated histone H3, and acetylated histone H4. The immunoprecipitates were analyzed by real-time PCR. The primer sets (arrowheads) are numbered for each gene (open boxes represent the translated regions), and the positions of the primers relative to the initiation ATG codon are indicated. The relative amounts of the PCR products compared with input chromatin from wild-type extracts (arbitrarily given as 100) are shown below the genes. Black bars, acetylated histone H3; white bars, acetylated histone H4. Error bars represent sd values from at least three repetitions.
Requirement of GCN5, TAF1, and HD1 for the Acetylation/Deacetylation of Specific Histone Lys Residues
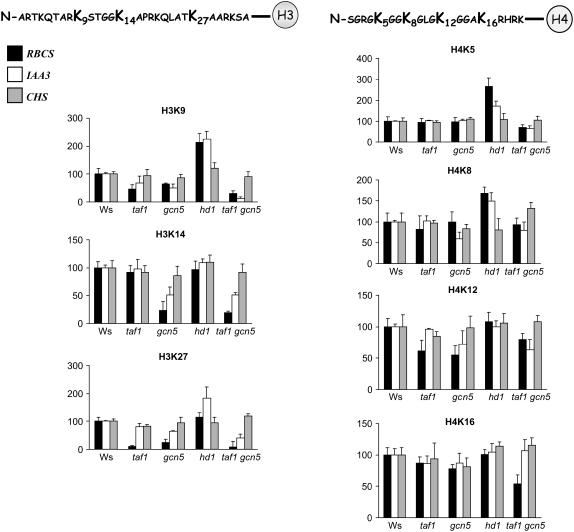
To know which Lys residues in the N-terminal end of histones H3 and H4 were the respective targets of GCN5, TAF1, and HD1, we performed ChIP assays with antibodies raised against specific acetylated Lys residues of histones H3 and H4. The N-terminal tail of histone H3 has five acetylated lysines, K9, K14, K18, K23, and K27, whereas that of H4 has four (K5, K8, K12, and K16). Specific antibodies against H3K9, H3K14, H3K27, H4K5, H4K8, H4K12, and H4K16 (Upstate Biotechnology) were used in this set of ChIP assays. The core promoter regions of three representative genes (RBCS-1A, IAA3, and CHS) were quantified by real-time PCR using the primer sets indicated in Figure 4. The light activation of RBCS-1A was shown to be regulated by both GCN5 and TAF1, whereas that of IAA3 was dependent only on GCN5. The mutation of HD1 increased the expression from both promoters. The expression of CHS was not affected by the three mutations. The enrichment of the promoter fragments relative to input chromatin DNA in the wild type (arbitrarily given as 100%) was compared with that found in each mutant. As shown in Figure 5, the gcn5 mutation significantly decreased the acetylation of H3K9, H3K14, H3K27, and H4K12 on both promoters, with the decreases most pronounced on the H3 Lys residues. The taf1 mutation significantly impaired the acetylation of H3K9, H3K27, and H4K12 on the RBCS-1A, but not on the IAA3, promoter, in agreement with its nonrequirement for the activation of this gene. Thus, acetylation of H3K9, H3K27, and H4K12 depended on both GCN5 and TAF1, whereas acetylation of H3K14 depended only on GCN5. By contrast, the hd1 mutation highly increased the acetylation levels of H3K9, H4K5, and H4K8 on both promoters, whereas a significantly increased acetylation on H3K27 was detected on the IAA3 promoter. Interestingly, compared with the single mutations, the taf1 gcn5 double mutation induced a further loss of acetylation of H3K9, and to a lesser extent of H3K27 and H4K5, of both RBCS-1A and IAA3 core promoters. With the CHS core promoter primer set, no significant changes were detected in the mutants with respect to the analyzed Lys residues, except for the taf1 gcn5 double mutant that showed some increase of H4K8 acetylation (Figure 5).
Figure 5.
Acetylation Profiles of the N-Terminal Tails of Histones H3 and H4 on Light-Responsive Promoters in taf1, gcn5, hd1, and taf1 gcn5 Compared with Wild Type Ws.
As indicated, ChIP with specific antibodies against histones H3K9, H3K14, and H3K27 and H4K5, H4K8, H4K12, and H4K16 were used. The immunoprecipitated promoter fragments of RBCS-1A (black boxes), IAA3 (white boxes), and CHS (gray boxes) were quantified by real-time PCR. The relative amounts of the promoter fragments compared with input chromatin from wild-type extracts were arbitrarily given as 100. Error bars represent sd values from at least three repetitions.
GCN5 Is Recruited to the Light-Inducible Promoters
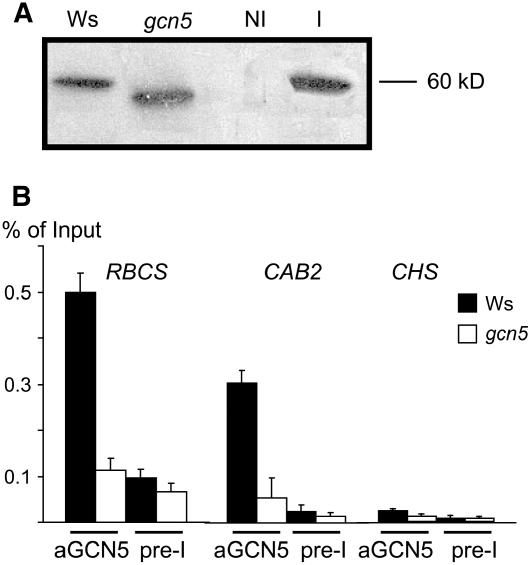
The results described above suggested that GCN5 might act directly on the light-responsive promoters. We tested this hypothesis by ChIP assays using a polyclonal antibody raised against GCN5 (made using two peptide epitopes located between amino acids 85 and 99 and between amino acids 136 to 150, corresponding to the N-terminal region of the protein; see Methods). On protein gel blots, the antibody recognized the recombinant GCN5 protein produced in Escherichia coli (Figure 6A), which showed a molecular mass close to 60 kD (the calculated molecular mass being 62 kD). In wild-type nuclear protein extracts, a protein band was detected that migrated to a position similar to that of the recombinant protein. In the gcn5 mutant, a protein band with a lower molecular mass was detected, suggesting that the T-DNA insertion (located in the 3′ end of the gene) might have produced a truncated or hybrid GCN5 protein (Bertrand et al., 2003). The precipitated chromatin fragments were analyzed by real-time PCR to test for enrichment relative to input chromatin. For CAB2 and RBCS-1A, the GCN5 antibody precipitated ∼0.3 and 0.5% of the input in the wild type, whereas <0.1% was precipitated in gcn5 (Figure 6B). The CHS promoter was not precipitated significantly by the GCN5 antibody in either the wild type or gcn5. These data indicate that GCN5 was recruited specifically to the target light-responsive promoters.
Figure 6.
GCN5 Was Associated with the Target Light-Responsive Promoters.
(A) Protein gel blot analysis of nuclear extracts from wild-type (Ws) and gcn5 rosette leaves and protein extracts of E. coli transformed by a plasmid expressing GCN5 (I, induced by isopropylthio-β-galactoside; NI, noninduced).
(B) Real-time PCR quantification of RBCS-1A, CAB2, and CHS core promoter regions (as in Figure 4) immunoprecipitated by the GCN5 antibodies or by the preimmune serum. The signals are given as percentage of the input chromatin. Error bars represent sd values from at least three repetitions.
DISCUSSION
Arabidopsis has at least 12 HAT and 18 HDAC genes (Pandey et al., 2002), and the members may have either redundant or specific functions. Our data have shown that at least three of them are required for normal photomorphogenesis and light-regulated gene expression. Thus, the control of histone acetylation levels is an important mechanism for light-regulatory processes in plants.
Function of GCN5 and HD1 in the Light Regulation of Gene Expression
The light-hyposensitive phenotype and the decreases in gene expression and histone acetylation in gcn5 mutants (Figures 1 to 3) indicate that GCN5 is a positive regulator of a subset of light-inducible genes. Two lines of evidence suggest that GCN5 is involved directly in the transcriptional regulation of the light-inducible genes. (1) Decreases of histone acetylation induced by the gcn5 mutation were detected essentially on the core promoter regions (Figure 4). (2) ChIP assays using a GCN5 antibody revealed direct association of the protein with the analyzed photosynthetic gene promoters (Figure 6). The recruitment of GCN5 to the promoters may be mediated by interactions with specific DNA binding proteins, although it could also bind to promoter-associated acetylated histone Lys residues through its C-terminal bromodomain. Association of GCN5 with the promoters would induce either higher levels of overall histone acetylation or specific histone Lys acetylation required for light-activated transcription. These two hypotheses may not be mutually exclusive, because the gcn5 mutation reduced the acetylation of at least four Lys residues in the N-terminal tails of histones H3 and H4, with H3K14 and H3K27 being the most affected (Figure 5). This acetylation profile of histone Lys residues is in agreement with the function of yeast and animal GCN5 proteins that preferentially acetylate histone H3 in vitro (Grant et al., 1997).
Our data show that HD1 negatively regulates light-responsive gene transcription by reducing histone acetylation levels. The hd1mutation induced histone acetylation on both upstream and core promoter regions, suggesting that HD1 might operate on a relatively larger range of the promoters. Tian et al. (2005) have shown that the HD1 mutation induces the acetylation levels of H3K9 and H4K12 on different target promoters. However, our data show that the mutation induced increases of acetylation levels of H3K9, H3K27, H4K5, and H4K8 on the light-responsive promoters studied (Figure 5). Therefore, the regulation of histone acetylation pattern by HD1 might be promoter-specific.
The normal photomorphogenic phenotype and light-responsive gene expression observed in the hd1 gcn5 double mutant (Figures 1B and 2C) suggest that GCN5 and HD1 might form an antagonistic HAT/HDAC couple to control the reversible histone acetylation required for a light-regulated switch of gene expression. However, we have observed that the histone Lys residues, of which the acetylation levels were reduced by the gcn5 mutation, did not correspond completely with those hyperacetylated in the hd1 mutant (Figure 5). For instance, H3K14 acetylation was reduced in gcn5, but it was not altered in hd1. Conversely, H4K5 acetylation was enhanced in hd1, but it was not affected by the gcn5 mutation. However, H3K9 acetylation was increased in hd1 and decreased in gcn5 mutants. It is known that H3K9 acetylation is an essential marker of gene activation (Jenuwein and Allis, 2001). Hypoacetylation of H3K9 induces its methylation, leading to the formation of repressive chromatin. The antagonist function of the GCN5/HD1 couple may operate on the acetylation levels of this Lys residue. Synergistic effects of acetylation exist between neighboring Lys residues (Jenuwein and Allis, 2001). The regulation of light-inducible gene expression by GCN5 and HD1 also could be interpreted by a cumulative mechanism of histone acetylation. We noted that both gcn5 and athd-1 mutations also affected CAB2 and RBCS-1A mRNA levels in the dark (Figure 2). This finding suggests that the balance of histone acetylation and deacetylation maintained by GCN5 and HD1 is also required for the basal transcription of the light-inducible genes. Furthermore, GCN5 has been shown to be involved in the expression of cold-inducible genes (Stockinger et al., 2001; Vlachonasios et al., 2003). Recently, the Arabidopsis FVE gene, which encodes a homolog of a mammalian HDAC complex component, was found to be a negative regulator of the CBF/DREB-dependent cold-responsive pathways (Kim et al., 2004). Thus, distinct HAT/HDAC couples might be formed to carry out reversible histone acetylation in response to different environmental signals.
Functional Relationship between Histone Acetyltransferases in Light Regulation
The regulation of gene transcription involves transcription coactivators and corepressors capable of transmitting signals from enhancer-bound specific transcription factors to the RNA polymerase initiation complex (Sterner and Berger, 2000). We have shown that the disruption of both GCN5 and TAF1 reduced light-responsiveness. In yeast and mammalian cells, GCN5 is a key component of several transcription cofactors, including the SAGA complex (Carrozza et al., 2003). TAF1 within the TFIID complex also functions as a transcriptional coactivator (Sterner and Berger, 2000). Transcriptional cofactors are recruited to promoters by interacting with enhancer-bound transcription factors. A combination of several light-responsive elements is required to construct a light-activating enhancer module, suggesting the possible involvement of transcription coactivators (Martinez-Hernandez et al., 2002; Yadav et al., 2002, 2005; Maxwell et al., 2003). Recent results have shown that the PetE enhancer activates transcription by mediating histone acetylation on the promoter (Chua et al., 2003), suggesting that light-responsive enhancers may interact, via the cognate DNA binding factors, with a HAT-containing transcriptional coactivator. Our results suggest that GCN5 and TAF1 might be the bona fide light-responsive coactivators.
In contrast with the gcn5 mutants, the taf1 mutants did not show the typical (light-hyposensitive) long-hypocotyl phenotype under different light conditions. However, this mutation could dramatically enhance the long-hypocotyl phenotype of hy5 mutants, irrespective of light quality (Bertrand et al., 2005) (Figure 3C). Differences between the two mutants also existed at the level of the target genes. For instance, gcn5, but not taf1, severely affected the expression of the light-activated IAA3 gene. This difference suggests that the recruitment of the proteins may be dependent on promoter structure. TAF1 is believed to be recruited by the TATA binding protein TBP, whereas GCN5 proteins can be recruited by interaction with specific DNA binding transcription factors. The enhancement of the hy5 phenotype by the taf1 mutation suggests that TAF1 may function in a transcriptional pathway in parallel with that of the basic domain/Leu zipper protein HY5. The phenotype observed in the gcn5 hy5 double mutant suggests that GCN5 has a different relationship with HY5 compared with TAF1.
The analysis of the taf1 gcn5 double mutant suggests that GCN5 and TAF1 have a cumulative function in light regulation (Figure 3). This hypothesis is supported by the ChIP data showing that the two genes have redundant and distinct requirements for the acetylation of both histones H3 and H4 on the targeted promoters (Figure 5). Therefore, it is likely that both the cumulative and specific mechanisms of acetylation of histone Lys residues proposed by Dion et al. (2005) are involved in light regulation. These observations are in agreement with the redundant and distinct roles of GCN5-containing SAGA and TAF1-containing TFIID complexes in global transcription in yeast (Lee et al., 2000; Huisinga and Pugh, 2004). In addition, both GCN5 and TAF1 proteins contain a bromodomain in their C-terminal ends, which is known to bind to acetylated histone Lys residues. Histone acetylation by one of the HAT proteins would induce the binding of the other to the chromatin, enhancing histone acetylation on the targeted promoters. Interestingly, the ChIP assays presented in Figure 5 showed that the cumulative effect of the taf1 gcn5 double mutation was mostly observed on H3K9 acetylation, which, as mentioned previously, is critical for gene activation (Jenuwein and Allis, 2001).
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana T-DNA insertion mutant plants studied in this work were in the Ws background (for gcn5, see Bertrand et al., 2003; for gcn5-1, see Vlachonasios et al., 2003; for athd-1, see Tian et al., 2003; for taf1, see Bertrand et al., 2005). Three backcrosses were performed on gcn5 and taf1. The hy5-1 mutant (in the Landsberg background) was obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis plants were grown in a greenhouse under long-day conditions (16 h of light) at 19.5°C (day) and 17.5°C (night). For in vitro cultures, seeds were sown on 0.5× Murashige and Skoog medium, incubated at 4°C for 48 h, and then transferred to growth chambers for germination at 20°C under white light (16 h/d at 120 μmol·m−2·s−1) or continuous red (10 μmol·m−2·s−1), far-red (5 μmol·m−2·s−1), or blue (18 μmol·m−2·s−1) light. Hypocotyl lengths were measured 7 d after germination. Student's t tests were performed to determine whether the difference between two sample means was significant at α = 0.05.
Construction of Double Mutants
Double mutants were made from genetic crosses with gcn5 as the male donor and hd1-t, taf1, or hy5-1 as female. Putative double mutants were selected from the F2 generation, genotyped by PCR using a T-DNA–matching primer and gene-specific primers to distinguish the wild type from the mutant DNA sequences for gcn5 or hd1 or by sequencing the genomic region of the hy5-1 locus, and confirmed in the F3 generation based on the mutant phenotypes. The primers used for genotyping were as follows: gcn5, 5′-GGTATCGGGGAGTTGTAAGTTCTAC-3′ and 5′-TTGTGCTAGTCGCTCCATGA-3′; T-DNA in gcn5, 5′-CTACAAATTGCCTTTTATCGAC-3′; hd1, 5′-ATGCTGGAGGATCTGTTGG-3′ and 5′-CCAGACAATGAATCAGCACC-3′.
Genomic DNA and Total RNA Extraction, PCR, and RNA Gel Blots
Arabidopsis leaves were used for genomic DNA extraction. PCR was performed using the Promega Taq polymerase. Total RNA was isolated from 12-d-old light-grown or dark-grown seedlings using TRIzol (Invitrogen). For RNA gel blot analysis, 5 μg of total RNA were separated on 1% denaturing agarose gels, blotted onto a nylon membrane, and hybridized with 32P-labeled gene-specific probes that were prepared from cDNA clones of CAB2, RBCS-1A, CHS, and ACT2. Hybridization signals were scanned with a Molecular Image FX Pro phosphor imager (Bio-Rad) and normalized to actin mRNA or rRNA signals using Bio-Rad Quantity One one-dimensional analysis software.
GCN5 Antibody and Recombinant Protein Production and Protein Gel Blots
Polyclonal antibodies were custom-made in rabbits (Eurogenetic) using two synthetic peptides (H2N-CARGADTDSDPDESED and H2N-SSRNTKLKTESSTVKLC; both peptides are located on the N-terminal region of the protein) and affinity-purified. For recombinant GCN5 production, the cDNA was cloned into pQE30 (Qiagen) and introduced into Escherichia coli strain M15. The conditions used for induction and purification were described by Ayadi et al. (2004). For protein gel blots, nuclear proteins were extracted from the wild type or gcn5 by grinding rosette leaves to a fine powder in liquid nitrogen. The powder was transferred to nuclear extraction buffer (0.4 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 5 mM β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]), and the extracts were filtered twice through Miracloth (Calbiochem) and centrifuged at 4000g for 20 min at 4°C. The nuclei pellets were dissolved in 15% SDS-PAGE gel-loading buffer for electrophoretic analysis along with protein extracts from isopropylthio-β-galactoside–induced or noninduced E. coli cells transformed with the expression plasmid. The blots were incubated with the GCN5 antibody (1:500 dilution) overnight at 4°C and revealed by enhanced chemiluminescence (Amersham).
ChIP Assays
Immunoprecipitations were performed as described previously (Gendrel et al., 2002). Seeds of Ws, gcn5, taf1, hd1, and the double mutants were sterilized, kept for 2 d at 4°C, and grown in vitro under long-day conditions. Twelve-day-old seedlings were harvested and fixed in 1% formaldehyde for 15 min in a vacuum and then neutralized by 0.125 M Gly. After washing with sterilized water, the samples were ground in liquid nitrogen as described above. Nuclei pellets were suspended in a buffer containing 0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors (one minitablet per milliliter; Roche). The suspensions were transferred to microfuge tubes and centrifuged at 12,000g for 10 min. The pellets were suspended in 1.7 M sucrose, 10 mM Tris-HCl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors and centrifuged through a layer of the same buffer in microfuge tubes. The nuclear pellets were lysed in a buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, and protease inhibitors. The lysed nuclei were sonicated four times for 15 s at 4°C followed by centrifugation. The supernatants containing chromatin fragments were diluted 10-fold with 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, and 167 mM NaCl. A 1 mL aliquot of the dilution was used for an immunoprecipitation assay.
The antibodies for histone acetylation tests were purchased from Upstate Biotechnology: anti-histone H3 (05-499), anti-acetylated H3 (06-599), anti-acetylated H4 (06-598), anti-acetyl-histone H3K9 (07-352), anti-acetyl-histone H3K14 (07-353), anti-acetyl-histone H3K27 (07-360), anti-acetyl-histone H4K5 (07-327), anti-acetyl-histone H4K8 (07-328), and anti-acetyl-histone H4K12 (07-323). Immunoprecipitated DNA was analyzed by real-time PCR (Light Cycler; Roche) using the following primers. For CAB2: 1, 5′-CATTCTTGTCACGAG-3′; 2, 5′-CGTTTAGTAGTCTTAC-3′; 3, 5′-GGAACGGAGTCAAGTTTGGA-3′; 4, 5′-AACGGCTCCCATCAAAATAA-3′; 5, 5′-TTTGTGTTTGTGGTGGATGG-3′; 6, 5′-CAATCACGTTGCTCGATTGT-3′. For RBCS-1A: 1, 5′-TGTTTGACTGAGTCTCAAAGTGG-3′; 2, 5′-ACAATCTCGACCACGGAAAA-3′; 3, 5′-CAAGCCGATAAGGGTCTCA-3′; 4, 5′-TGATCGGAGGGTCTAGGATA-3′; 5, 5′-TGATGGACGGTACTGGACAA-3′; 6, 5′-TGGGGTACTCCTTCTTGCAC-3′; 7, 5′-GGATCATCGGATTCGACAAC-3′; 8, 5′-CCGGAAATTAAAACCCAAGA-3′. For IAA3: 1, 5′-CTTACGTTCCACTGACGGATT-3′; 2, 5′-CTTGACAGAACAGGTCATAAGTTTG-3′; 3, 5′-GAAAACAGTTTCTTCTCTCTCTACCA-3′; 4, 5′-TCTTCAAGAATTGCAGGAGAAG-3′; 5, 5′-TGGATGCTCATTGGTGATGT-3′; 6, 5′-GCCTAAACCTTTGGCTTCTG-3′; 7, 5′-GGTCTTAAGCATATGAAACTGGAAC-3′; 8, 5′-GATCAATGAGAACGCAAAACAG-3′. For CHS: 1, 5′-CACAGAAAAGGGGGCTAACA-3′; 2, 5′-AGAGTTTGATGTTGCTGTTGTG-3′.
Quantification of immunoprecipitated chromatin fragments was performed according to Frank et al. (2001).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: GCN5, At3g54610; HD1, At4g38130; TAF1, At3g19040; CAB2, At1g29920; RBCS-1A, At1g67090; CHS, At5g13930; and IAA3, At1g04240.
Acknowledgments
We thank Jeff Z. Chen for providing hd1 mutants, S.J. Triezenberg for providing gcn5-1 mutants, Michael Hodges for critical reading of the manuscript, Jean-Pierre Bouly for plant culture facilities, Roland Boyer for taking photographs, and the Nottingham Arabidopsis Stock Centre for hy5-1 seeds. This work was supported by a grant from the French plant functional genomics program GENOPLANT II (Grant AF2001019).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dao-Xiu Zhou (dao-xiu.zhou@u-psud.fr).
References
- Ayadi, M., Delaporte, V., Li, Y.F., and Zhou, D.X. (2004). Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett. 562 147–154. [DOI] [PubMed] [Google Scholar]
- Berger, S.L. (2002). Histone modification in transcriptional regulation. Curr. Opin. Genet. Dev. 12 142–148. [DOI] [PubMed] [Google Scholar]
- Bertrand, C., Benhamed, M., Li, Y.F., Ayadi, M., Lemonnier, G., Renou, J.P., Delarue, M., and Zhou, D.X. (2005). Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280 1465–1473. [DOI] [PubMed] [Google Scholar]
- Bertrand, C., Bergounioux, C., Domenichini, S., Delarue, M., and Zhou, D.X. (2003). Arabidopsis histone acetyltransferase GCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278 28246–28251. [DOI] [PubMed] [Google Scholar]
- Carrozza, M.J., Utley, R.T., Workman, J.L., and Côté, J. (2003). The diverse function of histone acetyltransferase complexes. Trends Genet. 19 321–329. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Watson, L.A., and Gray, J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M.F., Altschuler, S.J., Wu, L.F., and Rando, O.J. (2005). Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S.R., Schroeder, M., Fernandez, P., Taubert, S., and Amati, B. (2001). Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Duggan L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., Berger, S.L., and Workman, J.L. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11 1640–1650. [DOI] [PubMed] [Google Scholar]
- Horn, P.J., and Peterson, C.L. (2002). Chromatin higher order folding: Wrapping up transcription. Science 297 1824–1827. [DOI] [PubMed] [Google Scholar]
- Huisinga, K.L., and Pugh, B.F. (2004). A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 12 573–583. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Hyun, Y., Park, J.Y., Park, M.J., Park, M.K., Kim, M.D., Kim, H.J., Lee, M.H., Moon, J., Lee, I., and Kim, J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36 167–171. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., Causton, H.C., Holstege, F.C.P., Shen, W.C., Hannett, N., Jennings, E.G., Winston, F., Green, M.R., and Young, R.A. (2000). Redundant roles for TFIID and SAGA complexes in global transcription. Nature 405 701–704. [DOI] [PubMed] [Google Scholar]
- Loidl, P. (2004). A plant dialect of the histone language. Trends Plant Sci. 9 84–90. [DOI] [PubMed] [Google Scholar]
- Lusser, A. (2002). Acetylated, methylated, remodeled: Chromatin states for gene regulation. Curr. Opin. Plant Biol. 5 437–443. [DOI] [PubMed] [Google Scholar]
- Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277 88–91. [DOI] [PubMed] [Google Scholar]
- Marmorstein, R., and Roth, S.Y. (2001). Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genet. Dev. 11 155–161. [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez, A., Lopez-Ochoa, L., Arguello-Astorga, G., and Herrera-Estrella, L. (2002). Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 128 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, B.B., Andersson, C.R., Poole, D.S., Kay, S.A., and Chory, J. (2003). HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiol. 133 1565–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R., Muller, A., Napoli, C.A., Selinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.E., and Berger, S.L. (2000). Acetylation of histone and transcription-related factors. Microbiol. Mol. Biol. Rev. 64 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Fong, M.P., Wang, J.J., Wei, N.E., Jiang, H., Doerge, R.W., and Chen, Z.J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Wang, J., Fong, M.P., Chen, M., Cao, H., Gelvin, S.B., and Chen, Z.J. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (HD1) expression. Genetics 165 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto, S., Locatelli, S., Canova, S., Pipal, A., Motto, M., and Rossi, V. (2003). Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 133 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios, K.E., Thomashow, M.F., and Triezenberg, S.J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000). Functional analysis of a RPD3 histone deacetylase homolog in Arabidopsis thaliana. Plant Mol. Biol. 44 167–176. [DOI] [PubMed] [Google Scholar]
- Yadav, V., Kundu, S., Chattopadhyay, D., Negi, P., Wei, N., Deng, X.W., and Chattopadhyay, S. (2002). Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 31 741–753. [DOI] [PubMed] [Google Scholar]
- Yadav, V., Mallappa, C., Gangappa, S.N., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light–mediated photomorphogenic growth. Plant Cell 17 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]