Abstract
CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 define the boundary domain around organs in the Arabidopsis thaliana meristem. CUC1 and CUC2 transcripts are targeted by a microRNA (miRNA), miR164, encoded by MIR164A, B, and C. We show that each MIR164 is transcribed to generate a large population of primary miRNAs of variable size with a locally conserved secondary structure around the pre-miRNA. We identified mutations in the MIR164A gene that deepen serration of the leaf margin. By contrast, leaves of plants overexpressing miR164 have smooth margins. Enhanced leaf serration was observed following the expression of an miR164-resistant CUC2 but not of an miR164-resistant CUC1. Furthermore, CUC2 inactivation abolished serration in mir164a mutants and the wild type, whereas CUC1 inactivation did not. Thus, CUC2 specifically controls leaf margin development. CUC2 and MIR164A are transcribed in overlapping domains at the margins of young leaf primordia, with transcription gradually restricted to the sinus, where the leaf margins become serrated. We suggest that leaf margin development is controlled by a two-step process in Arabidopsis. The pattern of serration is determined first, independently of CUC2 and miR164. The balance between coexpressed CUC2 and MIR164A then determines the extent of serration.
INTRODUCTION
Plant leaves vary in size, shape, and position on the stem. Two major classes of leaf can be defined on the basis of complexity: simple leaves, like those of Arabidopsis thaliana and tobacco (Nicotiana tabacum), which have a single blade, and compound leaves, like those of tomato (Solanum lycopersicum) or pea (Pisum sativum), in which several blade units called leaflets are attached to one or several axes (Champagne and Sinha, 2004). The margins of the leaf and leaflet blades may be smooth (entire leaves), have small tooth-like indentations (serrated leaves), or large outgrowths (lobed leaves). Leaf shape is regulated by genetic, developmental, and environmental factors (Tsukaya, 2005). Considerable progress has been made toward understanding the basis of leaf patterning and growth control, but the molecular mechanisms underlying leaf shape acquisition remain unclear.
The leaves of dicotyledonous plants are initiated on the flanks of a small group of self-maintaining totipotent stem cells, the meristem (Fleming, 2005). The initially symmetric young leaf initium rapidly becomes flattened on the side facing the meristem. Early in leaf development, three main axes are established, and these axes are maintained throughout the developmental process. The differentiation of an upper part of the leaf specializing in photosynthesis and of a lower part of the leaf specializing in gaseous exchange reflects polarization along an adaxial-abaxial (or dorso-ventral) axis. The formation of the leaf petiole and lamina reflects the existence of a proximo-distal axis. Finally, the midvein and the lamina define a medio-lateral axis.
Leaf patterning and development involve complex crosstalk between hormone signaling and a genetic network involving transcription factors (Hay et al., 2004; Byrne, 2005; Fleming, 2005). Auxin gradients control the position of primordium initiation sites in the meristem (Reinhardt et al., 2003) and are involved in primordium patterning. The response to auxin is mediated by auxin response factors (ARFs), a class of transcription factors that bind to the promoters of auxin response genes in response to auxin activation. The ARF3/ETT and ARF4 proteins, the activities of which are modulated by auxin gradients, facilitate specification of the adaxial and abaxial domains (Pekker et al., 2005), leading to the polarized expression of the genes determining these domains (Heisler et al., 2005). Once established, adaxial-abaxial polarization is maintained via the antagonistic relationship between factors specifying these two identities (Bowman et al., 2002). The juxtaposition of the adaxial and abaxial domains is required for the formation of a leaf blade (Waites and Hudson, 1995; Eshed et al., 2004). Blade outgrowth is associated with the formation of a short-lived marginal meristem along the adaxial-abaxial primordium margin, where cells actively divide (Donnelly et al., 1999). Once the leaf blade has started to grow, auxin is thought to be produced from its margins, generating gradients that coordinate several aspects of leaf development, including blade expansion and vascular differentiation (Mattsson et al., 1999, 2003; Aloni et al., 2003; Zgurski et al., 2005). Cell proliferation activity, which was initially concentrated in the marginal meristem, becomes more broadly distributed along the medio-lateral axis. Finally, cells progressively cease to divide and start to enlarge along a longitudinal gradient from the tip to the base of the leaf (Donnelly et al., 1999).
The mechanisms described above can account for the acquisition of general leaf shape, but other mechanisms must be involved in fine-tuning the shape of the leaf and the specific morphology of the margin in particular. It is clear that margin development is particularly sensitive to the characteristics of leaf growth and differentiation, as the misexpression of a number of genes results in leaf margin defects, ranging from more pronounced serration to highly lobed leaves (Lagarias et al., 1997; Nemeth et al., 1998; Rupp et al., 1999; Ori et al., 2000; Prigge and Wagner, 2001; Tantikanjana et al., 2001; Tian and Chen, 2001; Hugouvieux et al., 2002; Morel et al., 2002; Takahashi et al., 2002). Some of these genes have been linked more directly with leaf margin development. In jagged mutants, the leaves are serrated and the distal part of the petals is missing (Dinneny et al., 2004; Ohno et al., 2004). These defects have been correlated with a reduction in cell cycle activity in the distal part of the petal, as shown by the pattern of histone H4 expression. JAGGED may therefore maintain the cells in a state of active proliferation. Antirrhinum majus cincinnata (cin) mutants have rumpled leaf margins due to the persistence of cell division for longer at the leaf margins than in the central regions (Nath et al., 2003). A similar leaf phenotype is observed in Arabidopsis plants overexpressing miR-JAW, a microRNA (miRNA) targeting several transcripts of the TCP family of transcription factors related to CIN in Antirrhinum (Palatnik et al., 2003). These observations clearly show that the fine regulation of cell proliferation is required for the development of normal leaf margins. However, the molecular mechanisms leading to the formation of entire or serrated leaves have yet to be elucidated. In Arabidopsis, leaf serration is regulated developmentally, as the rosette leaves that form in early development are less serrated than the leaves forming later in development (Tsukaya and Uchimiya, 1997), and different accessions display considerable variability in the extent of leaf serration (Perez-Perez et al., 2002). The tips of the teeth forming the serrated leaf margins are associated with hydathodes (Tsukaya and Uchimiya, 1997) and with local peaks in auxin response, as shown by expression of the auxin-responsive reporter DR5:β-glucuronidase (GUS) (Aloni et al., 2003; Zgurski et al., 2005). We show here that a regulatory module including the CUP-SHAPED COTYLEDON2 (CUC2) gene and the miRNA miR164 plays a central role during the development of serrated leaf margins in Arabidopsis.
The CUC2 gene and two other members of the NAC family of transcription factors, CUC1 and CUC3, are required for embryonic shoot meristem formation and specification of the organ boundary (Aida et al., 1997; Takada et al., 2001; Vroemen et al., 2003). MIR164A, B, and C encode an miRNA, miR164, targeting the transcripts of six NAC genes, including CUC1 and CUC2 but not CUC3 (Rhoades et al., 2002; Kasschau et al., 2003; Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005; Guo et al., 2005; Schwab et al., 2005). The regulation of CUC1 and CUC2 by miR164 is required to prevent organ boundary enlargement during flower development and the formation of extra petals (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). Studies of the early extra petals1 mutant revealed that the MIR164C gene plays a major role in the control of petal numbers (Baker et al., 2005), whereas a mir164b mutant displayed no modification of aerial organ development (Mallory et al., 2004; Baker et al., 2005). We further investigated the role of miR164 in development by isolating a series of MIR164A mutants. These mutants showed enhanced leaf serration that was specifically dependent on CUC2 activity and could be phenocopied by the expression of an miR164-resistant CUC2 gene. We describe here a mechanism for the genetic control of leaf serration and compare the functions of the CUC/MIR164 genes during organ boundary and leaf development.
RESULTS
mir164a Mutants Display a Higher Level of Leaf Serration Than the Wild Type
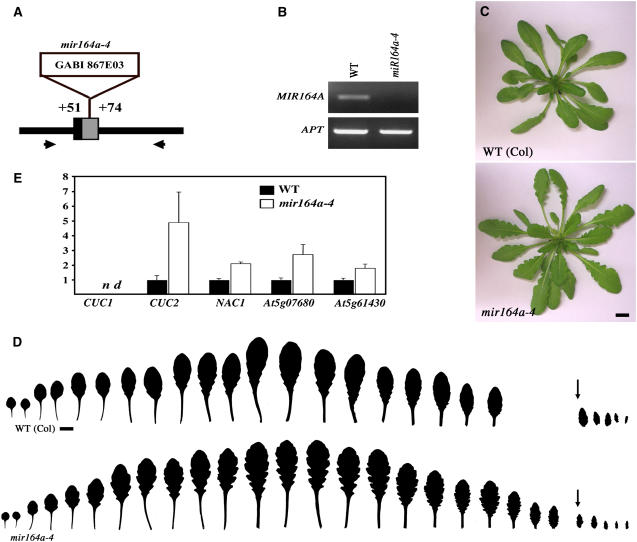
The MIR164C gene controls petal number, whereas MIR164B has no specific role in development of the aerial parts of the plant (Mallory et al., 2004; Baker et al., 2005). We investigated the function of the MIR164A gene in the aerial organ development by analyzing mir164a-4, a new knockout mutant from the GABI collection (Rosso et al., 2003). In this line, a T-DNA is inserted in the MIR164A gene, leading to the deletion of nucleotides 51 to 74 downstream from the miRNA (Figure 1A). We detected no expression on RT-PCR, suggesting that mir164a-4 is probably a null allele of MIR164A (Figure 1B).
Figure 1.
mir164a Mutants Show an Increase in Leaf Margin Serration.
(A) Schematic representation of the T-DNA insertion in the mir164a-4 mutant line. The mature miR164 sequence is shown in black, and the predicted pre-miRNA (Reinhart et al., 2002) is shown in gray. Positions are indicated relative to the first nucleotide of the mature miRNA.
(B) No pri-miR164As expression is detected in the mir164a-4 mutant line by RT-PCR. The primers used to amplify the pri-miR164As are indicated by arrows in (A). APT is used here as a control.
(C) Phenotype at bolting of the wild type and the mir164a-4 mutant.
(D) Leaf series of the wild type and the mir164a-4 mutant. The arrows indicate the first cauline leaves. Leaf serration is more pronounced in the mutant but follows the same pattern as in the wild type.
(E) Real-time PCR quantification of the expression levels of CUC1, CUC2, NAC1, At5g07680, and At5g61430 in wild-type and mir164a-4 leaves. Expression levels are normalized with respect to Elongation Factor1 expression levels (n = 4; bars = se).
Bars = 1 cm in (C) and (D).
The mir164a-4 mutant line had a specific leaf phenotype, with mutant leaves having deeper serrations than wild-type leaves (Figures 1C and 1D). In the wild type, successive rosette and cauline leaves showed a typical pattern of serration. The overall level of serration increased between successive leaves, and the proximal part of each blade was more deeply serrated than the distal part (Figure 1D). This serration pattern was not affected by the mir164a-4 mutation, which merely increased the depth of the groove between the teeth (Figure 1D). The total numbers of rosette and cauline leaves, bolting time, and the distribution of trichomes on the abaxial and adaxial surfaces of the leaf were unchanged in the mutant line (Table 1). The mir164a-4 mutant therefore differs from heterochronic mutants, such as hasty and zippy, which display early leaf serration due to accelerated phase changes (Bollman et al., 2003; Hunter et al., 2003). Flower organ numbers were not significantly modified in the mir164a-4 mutant (Table 2).
Table 1.
Effects of mir164a-4 on Leaf Identity and Flowering Time
| Days to Flowering | Rosette Leaves | Cauline Leaves | Leaves without Abaxial Trichomes | |
|---|---|---|---|---|
| Wild type (Col) | 32.5 ± 0.5 | 16.8 ± 0.4 | 4.1 ± 0.2 | 6.5 ± 0.3 |
| mir164a-4 | 32.9 ± 0.4 | 16.6 ± 0.3 | 3.8 ± 0.2 | 6.4 ± 0.2 |
Plants were grown under long-day conditions, and values are means ± se (20 plants counted).
Table 2.
Floral Organ Numbers in the Wild Type, mir164a-4 Mutants, and CUC2g-m4 Transgenic Plants
| Flower Position | Sepals | Petals | Stamens | Carpels | Carpel Fusion Defects | |
|---|---|---|---|---|---|---|
| Wild type (Col) | 1–5 | 4.10 ± 0.05 | 4.02 ± 0.03 | 5.24 ± 0.11 | 2.00 ± 0.00 | 0% |
| 16–20 | 4.00 ± 0.00 | 4.00 ± 0.00 | 5.76 ± 0.07 | 2.00 ± 0.00 | 0% | |
| mir164a-4 | 1–5 | 4.22 ± 0.08 | 4.02 ± 0.05 | 5.26 ± 0.12 | 2.00 ± 0.00 | 0% |
| 16–20 | 4.02 ± 0.03 | 4.00 ± 0.00 | 5.60 ± 0.09 | 2.00 ± 0.00 | 0% | |
| CUC2g-m4 | 1–5 | 4.50 ± 0.10 (P = 0.0003) | 4.18 ± 0.07 (P = 0.03) | 5.56 ± 0.12 (P = 0.04) | 2.00 ± 0.00 | 100% (P = 0) |
| 16–20 | 4.06 ± 0.05 | 3.98 ± 0.03 | 5.92 ± 0.04 (P = 0.05) | 2.00 ± 0.00 | 100% (P = 0) |
Values are mean ± se, 50 flowers counted. The P values are indicated when the mean values in the mutant or transgenic plants are significantly different from those for the wild-type (t test; P < 0.05)
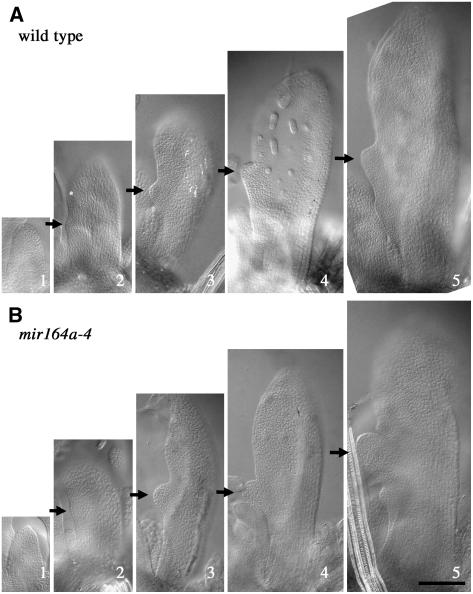
We then investigated the origin of this more marked serration during leaf development. We compared early stages of the development of leaves 11 to 15 in partially dissected and cleared wild-type and mir164a-4 mutant plants (Figure 2). In the wild type, the young leaf primordium had smooth margins until it reached ∼250 μm in length (Figure 2A, panel 1). The first pair of teeth was initiated at the leaf margin at this stage (Figure 2A, panel 2) and gradually became clearer, being much more marked in 300- to 400-μm-long primordia (Figure 2A, panels 3 and 4). The teeth were initiated approximately midway between the base and the tip of the primordium. In some cases, the two teeth were initiated at slightly different positions or differed in size, consistent with the asymmetry of fully grown leaf margins. Additional teeth became visible when the leaf reached 500 to 600 μm in length (Figure 2A, panel 5). No differences were observed between the mir164a-4 mutant and the wild type in the early stages of leaf development: young primordia had smooth margins (Figure 2B, panel 1), and the first pair of teeth was initiated in primordia of ∼250 μm in length (Figure 2B, panel 2). However, the teeth rapidly became larger and rounder in the mutant, with a more pronounced sinus (for example, compare Figures 2A, panel 4, and 2B, panel 4). These findings suggest that the early phases of leaf serration are not affected by the mir164a-4 mutation but that the teeth grow out faster due to a higher growth rate of the teeth, stronger growth inhibition in the sinus, or both.
Figure 2.
Early Stages of Leaf Development in the Wild Type and in mir164a-4 Mutants.
Four-week-old plants were partly dissected, with leaves 11 to 15 removed. Arrows indicate tooth initiation at the leaf margin. Bar = 100 μm.
(A) Developmental sequence of wild-type leaves.
(B) Developmental sequence of mir164a-4 mutant leaves.
MIR164 Genes Encode Variable Primary miRNAs with a Locally Conserved Secondary Structure
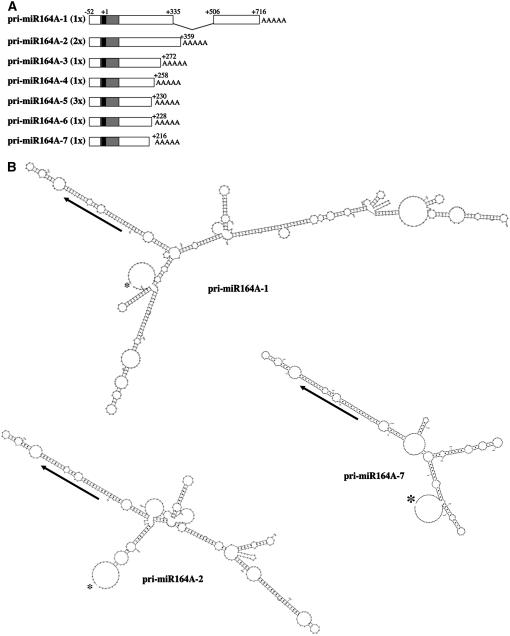
As a first step toward functional characterization of the MIR164A gene, we isolated the primary transcript by 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR. A single transcription start site was identified for the pri-miR164A (a primary miRNA [pri-miRNA]) 52 nucleotides upstream from the start of the mature miRNA (Figure 3A), as previously reported (Xie et al., 2005). This transcription site was confirmed by in silico promoter prediction and was downstream from a putative TATA box. The 3′ end was extremely variable. Five of the seven different 3′ ends identified were cloned only once, with the other two being cloned two or three times (Figure 3A). This 3′ end variability resulted in variation in the size of the pri-miR164As, from 268 to 597 nucleotides, corresponding to one or two exons. To determine whether this variability was specific to pri-miR164A or more general, we isolated the primary transcripts of the MIR164B and C genes. We identified a single transcription start site 85 nucleotides upstream from the miRNA for the MIR164B gene, whereas MIR164C transcription began 193 or 201 nucleotides upstream from the miRNA (see Supplemental Figure 1 online). As for the pri-miR164A, termination sites were highly variable for pri-miR164B and C (see Supplemental Figure 1 online).
Figure 3.
MIR164A Encodes pri-miRNA Transcripts with a Conserved Secondary Structure in the miRNA Region Despite Large Differences in Size.
(A) Schematic representation of the pri-miR164A identified by 5′ and 3′ RACE PCR. The number of times each 3′ end was cloned is indicated in parentheses. The mature miR164 sequence is shown in black, and the predicted pre-miRNA (Reinhart et al., 2002) is shown in gray. Positions are indicated relative to the first nucleotide of the mature miRNA (+1).
(B) Secondary structure of the pri-miR164A-1, pri-miR164A-2, and pri-miR164A-7 transcripts, as predicted by Mfold (Zuker, 2003). Asterisks indicate the beginning of the transcript, and arrows show the miRNA. The sequences of the pri-miR164B and C and the predicted secondary structures of the other pri-miR164s are available in Supplemental Figure 1 online.
The processing of pri-miRNAs involves recognition of a stem loop structure, and it has been suggested that the structure of the precursor is more important than its sequence for processing and that the pre-miRNA is sufficient for miRNA maturation (Parizotto et al., 2004; Guo et al., 2005). We therefore analyzed the predicted secondary structures of the pri-miR164s encoded by the three MIR164 genes. The secondary structure around the mature miRNA corresponding to the pre-miRNA (Reinhart et al., 2002; Wang et al., 2004), predicted by the Mfold program (Mathews et al., 1999; Zuker, 2003), was conserved in all the pri-miRNAs (Figure 3B; see Supplemental Figure 1 online). The rest of the transcript could adopt a variety of structures. The conservation of the pre-miRNA structure suggested that the various pri-miR164s may all be processed to generate functional miR164. Thus, all of the MIR164 genes are transcribed to generate a large population of pri-miRNAs. Despite their different sizes, all these pri-miRNAs can fold into a conserved structure and therefore probably give rise to functional miRNAs.
Functional Characterization of the MIR164A Locus
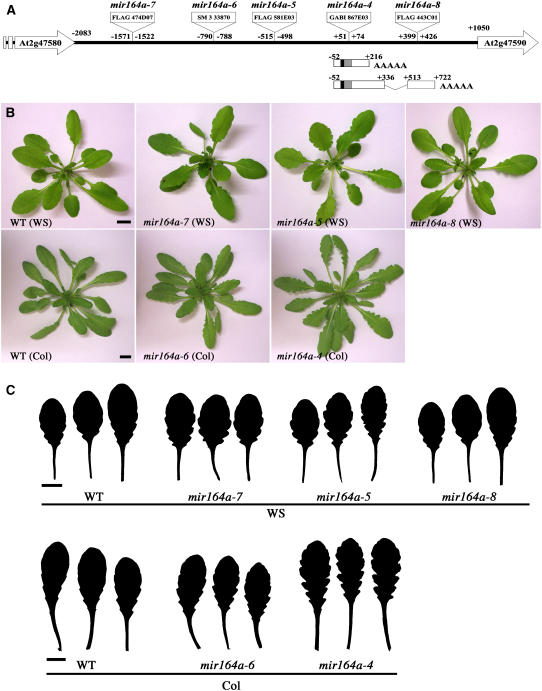
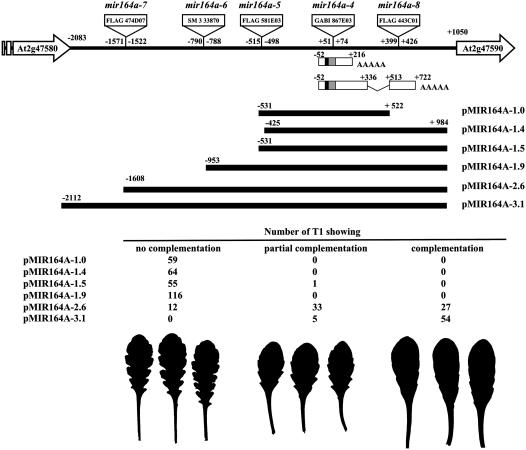
We characterized the MIR164A locus by analyzing four additional T-DNA insertion lines with insertions scattered throughout the region between the neighboring annotated genes At2g47580 and At2g47590 (Figure 4A). One insertion (FLAG 443C01, Versailles T-DNA collection, mir164a-8) was located in the 3′ region of the longest pri-miR164A-1 transcript but downstream from the 3′ end of the shorter pri-miR164As forms. Serration was unaffected in this line (Figures 4B and 4C; see Supplemental Figure 2 online), suggesting that the region disrupted by the T-DNA in this line was not essential for MIR164A function. By contrast, three insertions located within the promoter, up to 1.5 kb upstream from the transcription initiation site, led to deeper leaf serration (Figure 4). One insertion (line SM 3 33870, mir164a-6) was isolated in the Columbia (Col) background and could be compared with the knockout mir164a-4 mutant, which is in the same background. The increase in serration was less marked in the mir164a-6 line than in the mir164a-4 mutant. The other two insertion lines (FLAG 474D07 and FLAG 581E03, mir164a-7 and mir164a-5, respectively) were isolated from the Versailles T-DNA collection in the Wassilewskija (Ws) background. Both mutants showed a mild increase in serration. Thus, MIR164A was required to limit the extent of leaf serration in two different ecotypes, and a large promoter region was required for full activity.
Figure 4.
Allelic Series of the MIR164A Gene.
(A) Insertion positions of an allelic series along the MIR164A locus. The MIR164A locus is represented with the two neighboring genes. The longest and shortest pri-miR164As identified by RACE PCR are indicated.
(B) Phenotype at bolting of the mutants.
(C) Detail of the three largest rosette leaves (the entire leaf series is available in Supplemental Figure 2 online).
We assessed the ability of various genomic clones to complement the leaf phenotype of mir164a-4 (Figure 5) to confirm that a large promoter region was required for full MIR164A function. We tested six different genomic clones. Only the largest, pMIR164A-3.1, containing 2.1 kb of promoter sequence, efficiently complemented the mir164a-4 mutant (normal leaf phenotype was restored in 92% of the transformants [n = 59]). The pMIR164A-2.6 fragment, containing 1.6 kb of promoter, also complemented the mutant, but less efficiently (37% of the transformants [n = 72] had a fully restored leaf phenotype). Smaller genomic fragments were unable to complement the leaf phenotype of the mir164a-4 mutant.
Figure 5.
Complementation of the mir164a-4 Mutant.
The MIR164A locus is represented with the two neighboring genes. The longest and shortest pri-miR164As identified by RACE PCR are indicated. For each genomic construct, the numbers of independent transgenic lines showing no complementation, partial complementation, or full complementation of the mir164a-4 leaf phenotype are indicated. Below are representatives of the largest rosette leaves of noncomplemented, partially complemented, and fully complemented mir164a-4 mutants.
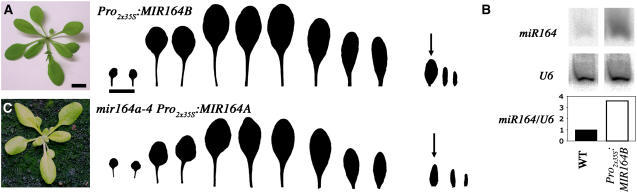
miR164 Overexpression Decreases Leaf Serration
We investigated whether an increase in miR164 levels could abolish leaf serration by examining Ws transgenic lines carrying the Pro2x35S:MIR164A, Pro2x35S:MIR164B, or Pro2x35S:MIR164C constructs (see Methods; Laufs et al., 2004), which led to the overaccumulation of miR164 (Figure 6B). Indeed, these transgenic plants had low levels of serration and formed leaves with smooth margins (Figure 6A), which was not observed in control plants carrying a Pro2x35S:erGFP (erGFP for endoplasmic reticulum–targeted green fluorescent protein) construct. We also checked the ability of the Pro2x35S:MIR164A, Pro2x35S:MIR164B, and Pro2x35S:MIR164C constructs to abolish serration in mir164a-4 mutants. In these experiments, we transformed homozygous mir164a-4 plants with these constructs. The leaves of the transformants did not have the strongly serrated phenotype of the mutant. Instead, they formed leaves with smooth margins, like those of Ws wild-type overexpressing miR164 (Figure 6C).
Figure 6.
Overexpression of miR164 Abolishes Leaf Serration in the Wild Type and mir164a-4 Mutants.
(A) Phenotype at bolting and series of leaves of a Pro2x35S:MIR164B transgenic plant forming leaves with a smooth margin.
(B) Quantification of miR164 in wild-type and Pro2x35S:MIR164B leaves.
(C) Phenotype at bolting and series of leaves from a mir164a-4 mutant expressing the Pro2x35S:MIR164A construct and forming leaves with a smooth margin.
The arrows indicate the first cauline leaf. Bars = 1 cm.
Expression of an miR164-Resistant CUC2 Gene, but Not of an miR164-Resistant CUC1 Gene, Phenocopies mir164a Mutant Leaves
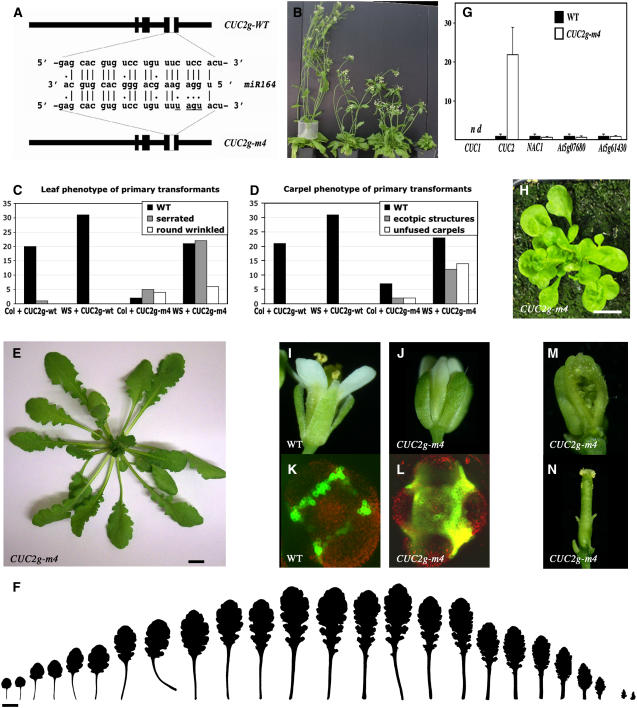
We investigated the molecular basis of the mir164a-4 leaf phenotype. Overexpression of miR164 showed that this miRNA posttranscriptionally regulates the expression of six members of the NAC gene family: CUC1, CUC2, NAC1, At5g07680, At5g39610, and At5g61430 (Laufs et al., 2004; Mallory et al., 2004; Guo et al., 2005; Schwab et al., 2005). We compared the expression levels of five of these target genes in wild-type and mir164a-4 mutant leaves. In mir164a-4 leaves, NAC1, At5g07680, and At5g61430 transcript levels were two to three times higher than those in the wild type, whereas CUC2 transcript levels were about five times higher than those in the wild type (Figure 1E). CUC1 was undetectable in both the wild type and the mutant. As the overexpression of CUC1 or CUC2 affected leaf development (Takada et al., 2001; Hibara et al., 2003; Laufs et al., 2004; Mallory et al., 2004), we hypothesized that the leaf phenotype of the mir164a mutants might be due to the defective miR164-mediated regulation of CUC1 and/or CUC2. Mallory et al. (2004) reported that the expression of an miR164-resistant CUC1 gene led to shorter leaf petioles and a broader leaf shape but described no increase in leaf serration. We generated 23 new miR164-resistant CUC1 transgenic lines in the Ws background and 54 such lines in the Col background using the miR164-resistant CUC1 gene (5mCUC1) described by Mallory et al. (2004). None of these lines showed deeper leaf serration, suggesting that the serrated leaf phenotype of mir164a mutants does not result from the absence of CUC1 regulation by miR164.
We investigated whether the serrated phenotype of mir164a mutants resulted from defects in the miR164-mediated regulation of CUC2 by generating transgenic lines expressing an miR164-resistant CUC2 gene. We used a 6-kb genomic fragment of CUC2 that was sufficient to complement the cuc1 cuc2 double mutant (Taoka et al., 2004) and that therefore contains all the information required for correct expression of the CUC2 gene. We generated the miR164-resistant CUC2 gene (CUC2g-m4) by introducing four nucleotide changes into the CUC2 gene, resulting in the abolition of miR164 function without changing the sequence of the CUC2 protein (Laufs et al., 2004) (Figure 7A). This miR164-resistant CUC2 gene (CUC2g-m4) or the wild-type CUC2 (CUC2g-wt) gene was introduced into plants of the Ws or Col ecotype. Twenty-seven of the 60 CUC2g-m4 lines formed leaves with deeper serration, resembling those of the mir164a-4 mutant (Figures 7C, 7E, and 7F). This phenotype was observed in only one of the 62 CUC2g-wt lines. As expected, CUC2g-m4 was epistatic to miR164 overexpression, as plants carrying both the CUC2g-m4 and Pro2x35S:MIR164A constructs formed strongly serrated leaves similar to those of CUC2g-m4 plants (see Supplemental Figure 3 online). We quantified the expression of five target genes of miR164 in CUC2g-m4 and wild-type plants. Only one of the five genes, CUC2, showed an increase in mRNA levels in the serrated leaves of CUC2g-m4 plants (Figure 7G). In particular, no CUC1 expression was detected, suggesting that the increase in CUC2 expression was sufficient to enhance the serration of CUC2g-m4 leaves. We conclude from these observations that disruption of the miR164-dependent regulation of CUC2, but not that of CUC1, led to an increase in leaf serration similar to that observed in mir164a mutants. Increased serration or lobing is observed in leaves of plants overexpressing KNOX genes, such as KNAT1 and KNAT2 (e.g., see Lincoln et al., 1994; Ori et al., 2000; Pautot et al., 2001). However, no ectopic KNAT1 and KNAT2 expression was observed in the serrated CUC2g-m4 leaves (see Supplemental Figure 4 online).
Figure 7.
Expression of an miR164-Resistant CUC2 Gene Phenocopies the mir164a Mutant Leaves.
(A) Representation of the wild-type and miR164-resistant CUC2 genes, with detail of the mutations introduced into the miR164 binding site. Thick black lines indicate the exons.
(B) General view of the phenotypic changes observed in CUC2g-m4 plants. In CUC2g-m4 lines with a strong phenotype, the inflorescence was not more than a few centimeters long due to severe internode length reduction (plant on the right). CUC2g-m4 lines with weaker phenotypes had shorter inflorescences, with abnormal flower phyllotaxy (two central plants). No decrease in growth rate was observed in any of the CUC2g-wt lines (plant on the left).
(C) Numbers of CUC2g-wt and CUC2g-m4 transgenic lines showing normal, serrated (as in [E]), or round and wrinkled leaves (as in [H]). Transgenic plants in the Col and Ws backgrounds were analyzed.
(D) Numbers of CUC2g-wt and CUC2g-m4 lines developing normal carpels, carpels with ectopic structures along the replum (as in [N]), or unfused carpels (as in [M]). Transgenic plants in the Col and Ws backgrounds were analyzed.
(E) Just after bolting, CUC2g-m4 plants have leaves with exaggerated serration, similar to the leaves of the mir164a-4 mutant.
(F) Series of leaves from CUC2g-m4 plants.
(G) Real-time PCR quantification of the levels of expression of CUC1, CUC2, NAC1, At5g07680, and At5g61430 in the wild type and in serrated leaves from CUC2g-m4 plants (n = 4; bars = se).
(H) CUC2g-m4 plants with a strong phenotype form small rosettes with round, wrinkled leaves.
(I) and (J) Wild-type sepals are joined from the base, whereas the sepals of CUC2g-m4 flowers are separated by a gap.
(K) and (L) Expression of the boundary marker ProSTM:ALCR-ProalcA:erGFP (Laufs et al., 2004), showing that the boundary domain is larger in CUC2g-m4 flowers than in the wild type.
(M) Dissected pistil of a CUC2g-m4 line with a strong phenotype showing carpel fusion defects
(N) Dissected pistil of a CUC2g-m4 line with a weak phenotype showing ectopic structures along the replum.
Bars = 1 cm.
CUC2g-m4 plants displayed other developmental abnormalities in addition to the increase in leaf serration. Ten of the 60 CUC2g-m4 lines had small, wrinkled leaves (Figures 7C and 7H). In lines with strong phenotypes, the inflorescence was very small due to a large decrease in internode elongation (Figure 7B). In lines with weaker phenotypes, inflorescence elongation was affected only slightly, but flower phyllotaxy was strongly perturbed (the phyllotaxy defects will be described elsewhere; A. Peaucelle, H. Morin, J. Traas, and P. Laufs, unpublished data). Early flowers formed more sepals, petals, and stamens than the wild type, whereas only the number of stamens was higher in later flowers (Table 2). The spacing between sepals was also greater in CUC2g-m4 plants with a strong phenotype (Figures 7I and 7J). Studies of a boundary GFP reporter (ProSTM:ALCR-ProalcA:erGFP; Laufs et al., 2004) further confirmed the enlargement of boundaries. CUC2g-m4 lines with a strong phenotype had carpel fusion defects, whereas CUC2g-m4 lines with weaker phenotypes developed ectopic structures along the replum (Figures 7M and 7N, Table 2), resembling those observed in organ polarity mutants (Eshed et al., 1999).
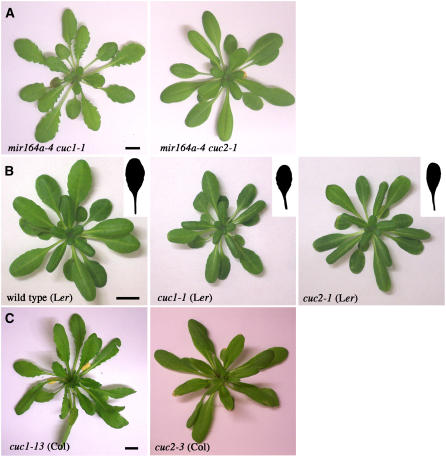
CUC2 Is Required for Leaf Serration in mir164a Mutants and the Wild Type, whereas CUC1 Is Not
The similarity between the serrated leaf phenotypes observed in CUC2g-m4 transgenic and mir164a mutant plants suggests that the serrated leaf phenotype of mir164a is due to an increase in CUC2 activity resulting from defective miR164 regulation. To assess the respective contribution of CUC1 and CUC2 genes to the serrated leaf phenotype of mir164a mutants, we constructed the mir164a-4 cuc1-1 and mir164a-4 cuc2-1 double mutants. Double mutants were identified by genotyping in a segregating F2 population, and their phenotype was confirmed in the F3 generation. The leaves of mir164a-4 cuc1-1 double mutants were deeply serrated and resembled those of the mir164a-4 single mutant, whereas mir164a-4 cuc2-1 double mutants developed leaves with smooth margins (Figure 8A). We therefore conclude that CUC2 is required for the serrated leaf phenotype of the mir164a-4 mutant, whereas CUC1 is not.
Figure 8.
CUC2 Is Required for Leaf Serration in mir164 Mutants and the Wild Type, whereas CUC1 Is Not.
(A) Phenotype, at bolting, of mir164a-4 cuc1-1 and mir164a-4 cuc2-1 double mutants.
(B) Phenotype of control wild-type Ler, cuc1-1, and cuc2-1 mutants at 8 weeks of short days. Details of the largest leaf are shown.
(C) Phenotype, at bolting, of cuc1-13 and cuc2-3 mutants in the Col background.
Bars = 1 cm.
We investigated whether the CUC2 gene played a similar role in leaf serration in the wild type. The cuc1-1 and cuc2-1 mutant alleles are found in the Landsberg erecta (Ler) background (Aida et al., 1997), an ecotype displaying only mild leaf serration. As serration increases in successive leaves, we assessed the contribution of the CUC1 and CUC2 genes to leaf serration in plants kept in short-day conditions to ensure that they remained in the vegetative phase. Unlike wild-type Ler and cuc1-1 mutants, which have slightly serrated leaves, cuc2-1 mutants produced leaves with smooth margins, displaying a total absence of serration (Figure 8B). For confirmation of this observation, we analyzed the leaf phenotype associated with the recently identified strong cuc1-13 and cuc2-3 alleles (Hibara et al., 2006) in Col, an ecotype with more strongly serrated leaves than Ler. Examination of these mutants showed that the level of leaf serration of the cuc1-13 mutant was similar to that in the wild type, whereas the cuc2-3 mutant formed leaves with smooth margins (Figures 1C, for the wild type, and 8C). We therefore conclude that CUC2 is required for the serration of wild-type Ler and Col leaves and that the absence of MIR164A-mediated CUC2 regulation leads to the exaggerated serrated phenotype of mir164a mutant leaves.
CUC2 Is Expressed in the Leaf Sinus
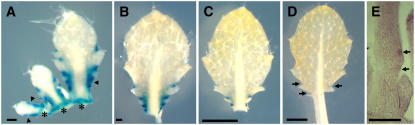
As our genetic observations had revealed a new role for CUC2 in leaf development, we investigated the pattern of expression of CUC2 in the leaves using a ProCUC2:GUS reporter. This reporter includes 3.1 kb of CUC2 promoter sequences. A 5.4-kb genomic clone of CUC2 including the same promoter can complement the embryo phenotype of cuc1 cuc2 double mutants, suggesting that this promoter is sufficient for normal CUC2 expression. During leaf development, GUS activity was observed in the margins of the developing primordium (Figure 9). This activity was initially restricted to a stretch of the two margins of the proximal part of the primordium (smallest primordium in Figure 9A). GUS activity then ceased in the outgrowing teeth and was restricted to the sinus region (Figures 9A to 9C). Several small domains in which outgrowth was not yet visible but GUS staining was already weaker were observed (Figure 9A, arrowheads), suggesting that CUC2 repression precedes tooth outgrowth. In larger leaves, GUS staining was restricted to the proximal part of the leaves, in which the teeth were still forming (Figures 9B and 9C), later becoming limited to small domains in the sinus of the proximal part of the leaves (Figure 9D). In addition to this expression in the leaf margin, GUS staining was observed early in development at the base of the primordium at its junction with the meristem (Figure 9A, asterisks).
Figure 9.
CUC2 Is Expressed in the Sinus of the Leaves.
(A) to (D) ProCUC2:GUS expression in rosette leaves. Arrowheads in (A) indicate regions with weaker GUS staining in which no outgrowth is yet visible, and asterisks indicate the insertion point of the primordium on the apex. Arrows in (D) indicate GUS expression restricted to the sinus.
(E) In situ hybridization of CUC2 mRNA in a longitudinal leaf section. Arrows point to cells expressing CUC2.
Bars = 100 μm in (A), (B), and (E) and 1 mm in (C) and (D).
The results of the ProCUC2:GUS reporter were confirmed by CUC2 mRNA in situ hybridization. Cells located at the base of outgrowing teeth showed weak levels of CUC2 mRNA (Figure 9E).
MIR164A Is Expressed in the Leaves, and Its Expression Domain Largely Overlaps with That of CUC2
We characterized the pattern of expression of MIR164A by generating three GUS reporter constructs driven by MIR164A promoter sequences. ProMIR164A-2.1:GUS and ProMIR164A-1.6:GUS included the promoters from the genomic clones pMIR164A-3.1 and pMIR164A-2.6, respectively, which complemented the mir164a-4 mutant. ProMIR164A-0.9:GUS contained the promoter from the genomic clone pMIR164A-1.9, which did not complement the mutant.
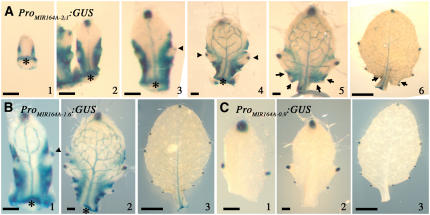
GUS staining was observed at the point of insertion of the primordium into the meristem in plants carrying the ProMIR164A-2.1:GUS construct (Figure 10A, asterisks). In small primordia, GUS activity was observed at the margins (Figure 10A, panel 1) and later appeared at the tip of the organ and in the vasculature (Figure 10A, panels 2 to 6). Once the teeth had started to develop, GUS staining was excluded from the outgrowing region (Figure 10A, panels 2 to 5). A focus of GUS expression then appeared at the tip of each tooth in the hydathodes (Figure 10A, panels 3 and 4, arrowheads). As the teeth grew out, GUS staining became restricted to a small domain at the sinus (Figure 10A, panels 5 and 6, arrows). This pattern of expression in the leaf margin largely overlapped with the domain of CUC2 expression (Figure 9). Interestingly, we observed no increase in GUS staining at any position at which a tooth was expected, suggesting that CUC2 downregulation and initial tooth outgrowth are not mediated by a local increase in MIR164A expression.
Figure 10.
Expression of MIR164A in the Sinus of the Leaves.
(A) Expression of ProMIR164A-2.1:GUS in developing rosette leaves.
(B) Expression of ProMIR164A-1.6:GUS in developing rosette leaves.
(C) Expression of ProMIR164A-0.9:GUS in developing rosette leaves.
Arrowheads indicate GUS expression in the hydathodes, and arrows indicate GUS expression in the sinus. Asterisks indicate the insertion point of the primordium on the apex. Bars = 100 μm in (A1) to (A5), (B1), (B2), (C1), and (C2) and 1 mm in (A6), (B3), and (C3).
Plants transformed with the medium-sized promoter construct ProMIR164A-1.6:GUS displayed a similar pattern of expression during leaf development (Figure 10B).
By contrast, plants carrying the construct including the smallest promoter, ProMIR164A-0.9:GUS, displayed GUS staining only in the tips of the leaf and teeth (Figure 10C, panel 3). The lack of complementation observed when MIR164A was expressed only in the tips of the teeth or leaf (construct pMIR164A-1.9 containing the small 0.9-kb promoter; Figure 5) suggests that miR164 has only very short-range action, consistent with other observations of the short-range action of miRNAs (Alvarez et al., 2006; Schwab et al., 2006). It is therefore unlikely that the complementation observed with the medium or large promoter resulted from miR164 expression in the vasculature. Thus, we conclude that coexpression of MIR164A with its target CUC2 in the leaf margin is essential for MIR164A function during leaf development.
DISCUSSION
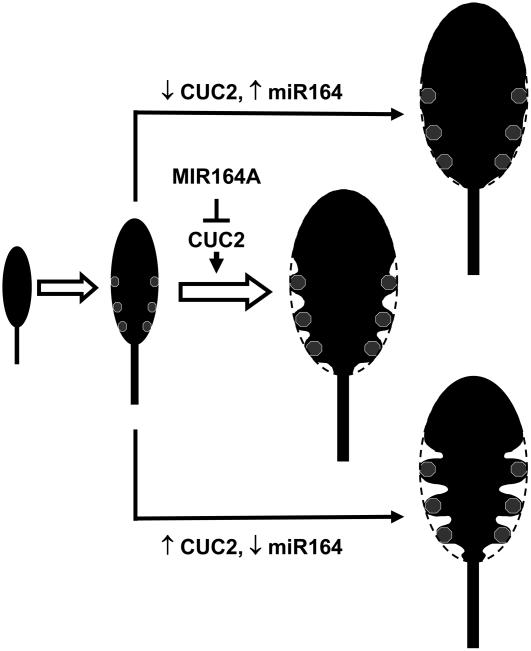
Based on our results, we propose a two-step model for the control of leaf serration development in Arabidopsis (Figure 11). In the first step, the number and positions of the teeth are determined (Figure 11, gray dots). This step is independent of MIR164A, as the serration pattern is not affected by mir164a mutations (Figure 1D). Tooth formation is made possible by local repression of the CUC2 gene (Figure 9) and does not require MIR164A activity because the early events of teeth formation are unaffected in the mir164a-4 mutant (Figure 2). In the second step, the balance between the coexpressed MIR164A and CUC2 genes controls the depth of serration. An increase in CUC2 expression in the sinus resulting from a decrease in MIR164A activity (Figures 1 and 4) or the resistance of CUC2 to miR164 (Figure 7) strengths or prolongs growth repression, resulting in more pronounced serration. Alternatively, low levels of CUC2 activity, in the cuc2 mutant (Figure 8) or in plants overexpressing miR164 (Figure 6), leads to a less marked sinus and the formation of leaves with smooth margins.
Figure 11.
Model of the Control of Leaf Margin Development by CUC2 and MIR164A.
We propose a two-step model of leaf margin development. In the first step, the tips of serration teeth are defined (gray dots). In the second step, CUC2-mediated growth repression controlled by miR164 activity generates the sinus. See text for further details.
This work also provides new insight into the basic mechanisms of miRNA gene expression and their effects on target gene expression.
Termination of MIR164 Gene Transcription Is Not Strictly Controlled
Characterization of the primary transcripts produced by the MIR164A, B, and C genes revealed unexpectedly high levels of variability in terms of the termination site. This variability is unlikely to result from the cloning procedure, as it was not observed with a non-miRNA transcript amplified in the same way (K. Nikovics and P. Laufs, unpublished data), and all the amplified 3′ regions contained a poly(A) tail. Several observations are consistent with the hypothesis that the 3′ termination site of the transcript is not critical for miR164 processing. First, modeling showed that the various pri-miR164s conserve the secondary structure of the pre-miRNA. Second, disruption of the 3′ region in the mir164a-8 mutant has no effect on development. Finally, we have characterized an insertion in the MIR164C gene, located in the pri-miR164C, 24 nucleotides downstream from the predicted pre-miRNA (data not shown). The corresponding plant line did not display the flower phenotype reported for a partial loss of function of MIR164C (Baker et al., 2005), suggesting that MIR164C function was not strongly affected in this line. These findings suggest that miRNA processing is dependent on the hairpin structure of the pre-miRNA and extends to miRNAs expressed under control of their own promoter observations made previously for endogenous or artificial miRNAs expressed under the control of strong promoters, such as the 35S promoter (Parizotto et al., 2004; Guo et al., 2005; Alvarez et al., 2006; Schwab et al., 2006).
Modular Organization of the MIR164A Promoter
We show that the MIR164A promoter displays the modular organization typical of genes encoding proteins (Watanabe and Okada, 2003; Baurle and Laux, 2005). Expression analyses with the GUS reporter and complementation assays led to the identification of three distinct domains in this promoter. The proximal domain is sufficient for expression in the hydathodes but does not restore MIR164A function during leaf development. The central domain confers expression in leaf margin and vasculature. The activity of this domain is sufficient for partial MIR164A function during leaf development. Full MIR164A function requires the distal part of the promoter. The lack of any clear difference in the pattern of expression according to the presence or absence of the distal fragment suggests that this domain mainly increases promoter activity without changing the pattern of expression.
Quantitative Interaction between miR164 and CUC2
It has been suggested that miRNAs facilitate rapid changes in cell differentiation programs by clearing the cells of inappropriate transcripts (Rhoades et al., 2002). This model assumes that the miRNA and its targets are expressed in largely complementary domains and is supported by experimental data (Chen, 2004; Kidner and Martienssen, 2004; Axtell and Bartel, 2005). However, we show here that the patterns of transcription of the MIR164A and CUC2 genes overlap at the leaf margin. Deletions of the MIR164A promoter demonstrated that this coexpression was required for MIR164A activity. These observations are consistent with an alternative model of miRNA function, in which the miRNA is expressed in a domain overlapping with that of its target (Baker et al., 2005; Vaucheret et al., 2006). Our observations build on this model. The mir164a mutant series and complementation experiments showed that miR164 acts in a quantitative manner to control the intensity of leaf serration. The quantity of target transcript present in a cell is therefore controlled by the balance between target gene transcription and endonucleotide cleavage activity, which is in turn controlled by the miRNA level.
A Conserved Regulatory Module Acting during Meristem and Leaf Development
We show here that the balance between the coexpressed MIR164A and CUC2 genes controls the development of leaf margins in Arabidopsis. CUC2 and miR164 have well-established roles in organ initiation at the shoot apical and floral meristems. CUC2, CUC1, and CUC3 repress growth in the boundary domain, facilitating the separation of adjacent organs (Aida et al., 1997, 1999; Takada et al., 2001; Vroemen et al., 2003). miR164 stabilizes the boundary domain by preventing its enlargement and/or by modulating the level of expression of the CUC1 and CUC2 genes within boundary cells (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). Our results indicate that the miR164/CUC regulatory module operating during the initial phase of organ initiation at the meristem is also involved at a later stage, when the final shape of the organ is being determined. The groove separating the primordium from the meristem and the sinuses between adjacent teeth at the leaf margin therefore form in a similar way. This similarity extends even beyond the miR164/CUC balance, as both the primordium and the tip of the teeth display an enhanced auxin response, as shown with the auxin-sensitive reporter element DR5 (Heisler et al., 2005; Zgurski et al., 2005). Indeed, it has been suggested that high auxin levels may inhibit CUC2 expression (Vernoux et al., 2000; Heisler et al., 2005). The downregulation of CUC2 at the site of tooth outgrowth may therefore be a similar response to high auxin levels. The role of miR164 in this model involving auxin is unclear. A transient increase in miR164 levels was observed in young seedlings treated with auxin, suggesting that MIR164 genes may be induced by auxin (Guo et al., 2005). However, our data indicate that although peaks in auxin levels may be associated with tooth outgrowth, they do not induce a local increase in MIR164A expression. The interplay between auxin, miR164, and the CUC genes therefore remains unclear.
Specific Role of CUC2 in Leaf Development
The miR164/CUC balance controls both formation of the primordium boundary domain and leaf margin development, but there is an important difference in CUC involvement between these two processes. Both CUC1 and CUC2 are involved in primordium development, whereas we demonstrate that only CUC2 is involved in leaf margin development. This difference may reflect differences in the functions of the CUC1 and CUC2 proteins. For example, the specificity of these transcription factors may result from their sets of target genes being only partially overlapping (Taoka et al., 2004). However, we favor an alternative, nonexclusive hypothesis. In RT-PCR experiments, CUC2 was found to be expressed, whereas CUC1 mRNA levels were below the detection limit. The differences in the contributions of the CUC1 and CUC2 genes during leaf development may therefore result from differences in the regulatory sequences of these two genes, leading to the expression of CUC2, but not CUC1, in leaves.
A Conserved Role for the CUC Genes during Leaf Development in Different Species?
The CUC genes belong to the NAC gene family, one of the largest families of plant-specific transcription factors, with 105 members in Arabidopsis and 75 members in rice (Oryza sativa; Ooka et al., 2003). The Arabidopsis CUC1, CUC2, and CUC3 genes are involved in establishment of the embryonic meristem and determination of the boundary domain (Aida et al., 1997; Takada et al., 2001; Vroemen et al., 2003). These functions are conserved for the petunia (Petunia hybrida) NAM and Antirrhinum CUP homologues (Souer et al., 1996; Weir et al., 2004). Indeed, conservation of the roles of the CUC genes in meristem and boundary function may extend to monocotyledonous plants, as suggested by the recently described patterns of expression of maize (Zea mays) homologues (Zimmermann and Werr, 2005). Phylogenetic analysis based on the conserved NAC domain has shown that CUC proteins cluster into two clades, which probably separated before the divergence of monocotyledons and dicotyledons (Zimmermann and Werr, 2005). The first clade contains CUC3-related sequences from both monocotyledons and dicotyledons. The second clade, the NAM clade, can be further subdivided into a monocotyledon-specific branch and a dicotyledon-specific branch containing CUC1, CUC2, NAM, and CUP. Arabidopsis is the only dicotyledonous species for which two CUC genes have been identified in the NAM clade. No expression of the known dicotyledon or monocotyledon homologues has been reported in lateral organs. Furthermore, the two species for which loss-of-function mutations of the CUC homologues are available form simple leaves with entire margins, in which the CUC genes are likely to play only a minor role. This is also the case for the Ler ecotype of Arabidopsis, which has only slightly serrated leaves. The contribution of CUC genes to leaf development may have remained unnoticed for these reasons. Alternatively, the role of the CUC genes in leaf development may be limited to certain species, including Arabidopsis. It has been suggested that CUC1 and CUC2 result from a gene duplication event specific to Arabidopsis (Zimmermann and Werr, 2005). A new role for CUC2 in leaf margin development may therefore have arisen following this gene duplication in Arabidopsis.
METHODS
Plasmid Construction
The Pro2x35S:MIR164A and Pro2x35S:MIR164B constructs have been described elsewhere (Laufs et al., 2004). For Pro2x35S:MIR164C, 1 kb of genomic sequence centered on the miRNA was amplified using miR164C-1 and miR164C-5 primers (primer sequences available in Supplemental Table 1 online) and cloned under control of the 2x35S promoter, as previously described (Laufs et al., 2004).
The miR164-resistant CUC1 gene (5mCUC1; Mallory et al., 2004) was kindly provided by A.C. Mallory. CUC2g-m4 transgenic plants were generated using a 6-kb EcoRI-EcoRV genomic fragment of CUC2 that complements the cuc1 cuc2 double mutant (Taoka et al., 2004). This fragment was introduced into the binary vector pGreenII0229 (Hellens et al., 2000) to generate pGreenII0229-CUC2g-wt. The four previously reported point mutations in the miR164 binding site (Laufs et al., 2004) were introduced by PCR. The wild-type fragment surrounding the miR164 binding site in pGreenII0229-CUC2g-wt was replaced by the mutated form, as a PmlI-EcoRI fragment, to generate pGreenII0229-CUC2g-m4.
For the ProCUC2:GUS reporter, an EcoRV-BglII fragment (3.7 kb) of the CUC2 promoter was fused to the BamHI-EcoRI fragment of GUS-ter derived from pBI101, in pBS+. A SalI-EcoRI cassette (consisting of the 3.1-kb promoter fragment, GUS, and nos terminator) was excised from this plasmid and inserted between the SalI and EcoRI sites of pBI101.
For the ProMIR164A:GUS reporters, the small, medium, and large promoter regions were amplified using the mir164A-8 primer, together with miR164A-10, miR164A-26, and miR164A-24 primers, respectively. The PCR products were inserted into pGEM-T, and their sequence was checked. The promoters were excised from pGEM-T by NotI digestion and introduced into the XmaI site of the pBI101.3 GUS binary vector after a partial fill-in reaction.
The various constructs used to complement the mir164a-4 mutant were generated by amplifying several fragments of the MIR164A gene by PCR and reconstructing the gene in the pGreenII0229 binary vector (Hellens et al., 2000). The central region of the MIR164A gene was amplified with the mir164A-2 and mir164A-3 primers, inserted into pGEM-T (pGEM-T MIR164A2-3), excised by NotI digestion, and ligated into the NotI site of pGreenII0229 to generate the pMIR164A-1.0 construct. The 3′ region of MIR164A was amplified with the mir164A-1 and mir164A-20 primers, inserted into the pCRII-Topo plasmid, excised by SacI digestion, and inserted into pGreenII0229. The HindIII restriction site was removed from this vector by HindIII digestion, blunt-ending, and self-ligation. The central part of MIR164A was added to this vector as a NotI-NdeI fragment from pGEM-T MIR164A2-3 to generate the pMIR164A-1.5 construct. The promoter region was extended by amplification with the mir164A-1 and mir164A-10 primers. It was inserted into pGEM-T, excised as a HindIII-NotI fragment, and ligated into pMIR164A-1.5, generating the pMIR164A1.9 construct. Additional promoter sequences were added by amplification with mir164A-8 and mir164A-26 or mir164A-8 and mir164A-24, insertion into pGEM-T, excision by digestion with XmnI and NotI, and insertion into pMIR164A-1.9 to generate pMIR164A-2.6 and pMIR164A-3.1, respectively. Finally, pMIR164A-1.4 was generated by replacing the promoter region of pMIR164A-1.5 by the region amplified with the mir164A-8 and mir164A-25 primers via NotI-HindIII digestion.
Plant Material and Growth
The mir164a insertion lines were identified in the GABI insertion collection (Rosso et al., 2003), the Salk collection (Alonso et al., 2003), the Exotic collection, or the Versailles collection (Bechtold et al., 1993; Bouchez et al., 1993). Plant transformation and transgenic plant selection have been described elsewhere (Deveaux et al., 2003). The controlled long-day growth conditions consisted of 16 h of light at 23°C and 8 h of darkness at 15°C.
Expression Analysis
Transcripts were quantified by fluorescence-based real-time RT-PCR. Total RNA was extracted using the RNeasy plant mini kit (Qiagen) according to the manufacturer's recommendations. Contaminating DNA was removed by DNaseI treatment (Invitrogen). The SuperScript II first-strand cDNA synthesis kit (Invitrogen) was used to generate cDNA from 5 μg of RNA in a reaction volume of 20 μL. Each cDNA sample was diluted 1:5 in water, and 5 μL of this dilution was used as a template for quantitative PCR. Reactions were performed with the LightCycler FastStart DNA master SYBR Green I kit (Roche) on a Roche LightCycler real-time PCR machine according to the manufacturer's instructions. The gene-specific primers used to detect uncleaved transcripts of CUC1 and CUC2 were as previously described (Mallory et al., 2004). Expression levels were normalized with respect to Elongation Factor1 expression levels and were averaged over at least two replicates from two independent biological samples.
Small RNAs were RNA gel blotted as previously described (Laufs et al., 2004), except that we used U6 as a standard. For the RT-PCR detection of pri-miR164A, we used the primers mir164A-13 and miR164A-28. GUS staining was performed as previously described (Sessions et al., 1999).
For the mRNA in situ hybridization, an antisense probe of the full-length open reading frame of CUC2 was synthesized in vitro and labeled by DIG-UTP using a gel-purified PCR product including the T7 RNA polymerase binding site as template. Tissue fixation, embedding, sectioning, and in situ hybridization was done as described before (Laufs et al., 1998), with the following changes. After dehydration of the tissues on the slide, an additional prehybridization step was performed (2 h at 45°C in 50% formamide, 5× SSC, 100 μg·mL−1 tRNA, 50 μg·mL−1 heparin, and 0.1% Tween). Hybridization was done overnight at 45°C using DakoCytomation mRNA in situ hybridization solution. Washing was performed as described earlier except that a first washing (0.1× SSC, 0.5% SDS; 30', 45°C) and a second washing (2× SSC, 50% formamide; 1 h, 45°C) were included. The specificity of the probe was checked on the cuc2.1 mutant.
Analysis of the pri-miRNAs
We performed 3′ and 5′ RACE for each gene with the GeneRacer cDNA amplification kit (Invitrogen). We used two or three rounds of nested PCR to amplify the 3′ and 5′ ends of the three pri-miR164s. The miR164A-15, miR164A-17, and miR164A-18 primers were used for the 3′ end of pri-miR164A, and miR164A-16 and miR164A-7 were used for the 5′ end. The miR164B-13, miR164B-15, and miR164B-16 primers were used for the 3′ end of pri-miR164B, and miR164B-5 and miR164B-14 were used for the 5′ end. We used miR164A-15 and miR164C-1 for the 3′ end of pri-miR164A and miR164C-3 and miR164C-5 for the 5′ end. Following gel electrophoresis, RACE reaction products were inserted into the pGEM-T Easy vector (Promega), and the two strands were sequenced. Two to four independent 5′ clones and 5 to 10 independent 3′ clones were analyzed for each gene. Promoter and transcription start sites were predicted by the TSSP program (http://www.softberry.com/berry.phtml?topic=tsspandgroup=programsandsubgroup=promoter). The pri-miRNA secondary structure was determined with the Mfold program (Mathews et al., 1999; Zuker, 2003) (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CUC1 (At3g15170), CUC2 (At5g53950), and MIR164A (At2g47585).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation, Sequence, and Secondary Structure of the pri-MIR164s.
Supplemental Figure 2. Full Leaf Series of the mir164a Mutants.
Supplemental Figure 3. Epistasis of CUC2g-m4 to miR164 Overexpression.
Supplemental Figure 4. No ectopic KNAT1 and KNAT2 Expression Is Observed in the Serrated CUC2g-m4 Leaves.
Supplemental Table 1. Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank R. Hellens, P. Mullineaux, F. Divol, the GABI and Exotic consortia, the SALK Institute, and the Institut National de la Recherche Agronomique for materials used in this work. We thank A.C. Mallory for the cuc1-13 insertion line and the 5mCUC1 construct. We thank B. Dubreucq for advice on real-time RT-PCR and B. Letarnec for plant care. We thank N. Da Silveira, K. Deschesne, and A. Duflot for skillful technical assistance, V. Pautot, J.D. Faure, and J.C. Palauqui for helpful discussions, and J. Traas for critical reading of the manuscript. This work was partly supported by an Action Concertée Incitative Jeunes Chercheurs award to P.L. and by the trilateral Genoplante GENOSOME program TRIL-046.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Patrick Laufs (laufs@versailles.inra.fr).
Online version contains Web-only data.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alvarez, J.P., Pekker, I., Goldshmidt, A., Blum, E., Amsellem, Z., and Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Bartel, D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.C., Sieber, P., Wellmer, F., and Meyerowitz, E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15 303–315. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Laux, T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Life Sci. 316 1194–1199. [Google Scholar]
- Bollman, K.M., Aukerman, M.J., Park, M.Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130 1493–1504. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camillieri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Life Sci. 316 1188–1193. [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E. (2005). Networks in leaf development. Curr. Opin. Plant Biol. 8 59–66. [DOI] [PubMed] [Google Scholar]
- Champagne, C., and Sinha, N. (2004). Compound leaves: Equal to the sum of their parts? Development 131 4401–4412. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux, Y., Peaucelle, A., Roberts, G.R., Coen, E., Simon, R., Mizukami, Y., Traas, J., Murray, J.A., Doonan, J.H., and Laufs, P. (2003). The ethanol switch: A tool for tissue-specific gene induction during plant development. Plant J. 36 918–930. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131 1101–1110. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215 407–419. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fleming, A.J. (2005). The control of leaf development. New Phytol. 166 9–20. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., Xie, Q., Fei, J.F., and Chua, N.H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Craft, J., and Tsiantis, M. (2004). Plant hormones and homeoboxes: Bridging the gap? Bioessays 26 395–404. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hibara, K., Karim, M.R., Takada, S., Taoka, K., Furutani, M., Aida, M., and Tasaka, M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara, K., Takada, S., and Tasaka, M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36 687–696. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Murata, Y., Young, J.J., Kwak, J.M., Mackesy, D.Z., and Schroeder, J.I. (2002). Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 130 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C., Sun, H., and Poethig, R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Lagarias, D.M., Crepeau, M.W., Maines, M.D., and Lagarias, J.C. (1997). Regulation of photomorphogenesis by expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Cell 9 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125 1253–1260. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. (1999). Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288 911–940. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, U., Crawford, B.C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299 1404–1407. [DOI] [PubMed] [Google Scholar]
- Nemeth, K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, C.K., Reddy, G.V., Heisler, M.G., and Meyerowitz, E.M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131 1111–1122. [DOI] [PubMed] [Google Scholar]
- Ooka, H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10 239–247. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., and Traas, J. (2001). KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 13 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker, I., Alvarez, J.P., and Eshed, Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez, J.M., Serrano-Cartagena, J., and Micol, J.L. (2002). Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162 893–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rupp, H.M., Frank, M., Werner, T., Strnad, M., and Schmulling, T. (1999). Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 18 557–563. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20 259–263. [DOI] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Matsuhara, S., Abe, M., and Komeda, Y. (2002). Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana, T., Yong, J.W., Letham, D.S., Griffith, M., Hussain, M., Ljung, K., Sandberg, G., and Sundaresan, V. (2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 15 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka, K., Yanagimoto, Y., Daimon, Y., Hibara, K., Aida, M., and Tasaka, M. (2004). The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 40 462–473. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2005). Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 49 547–555. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256 231–238. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Mallory, A.C., and Bartel, D.P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127 5157–5165. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., and Gaasterland, T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5 R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., and Okada, K. (2003). Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, I., Lu, J., Cook, H., Causier, B., Schwarz-Sommer, Z., and Davies, B. (2004). CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131 915–922. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A., and Carrington, J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurski, J.M., Sharma, R., Bolokoski, D.A., and Schultz, E.A. (2005). Asymmetric auxin response precedes asymmetric growth and differentiation of asymmetric leaf1 and asymmetric leaf2 Arabidopsis Leaves. Plant Cell 17 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, R., and Werr, W. (2005). Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 58 669–685. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.