Abstract
The CCT (for CONSTANS, CONSTANS-LIKE, TOC1) domain is found in 45 Arabidopsis thaliana proteins involved in processes such as photoperiodic flowering, light signaling, and regulation of circadian rhythms. We show that this domain exhibits similarities to yeast HEME ACTIVATOR PROTEIN2 (HAP2), which is a subunit of the HAP2/HAP3/HAP5 trimeric complex that binds to CCAAT boxes in eukaryotic promoters. Moreover, we demonstrate that CONSTANS (CO), which promotes Arabidopsis flowering, interacts with At HAP3 and At HAP5 in yeast, in vitro, and in planta. Mutations in CO that delay flowering affect residues highly conserved between CCT and the DNA binding domain of HAP2. Taken together, these data suggest that CO might replace At HAP2 in the HAP complex to form a trimeric CO/At HAP3/At HAP5 complex. Flowering was delayed by overexpression of At HAP2 or At HAP3 throughout the plant or in phloem companion cells, where CO is expressed. This phenotype was correlated with reduced abundance of FLOWERING LOCUS T (FT) mRNA and no change in CO mRNA levels. At HAP2 or At HAP3 overexpression may therefore impair formation of a CO/At HAP3/At HAP5 complex leading to reduced expression of FT. During plant evolution, the number of genes encoding HAP proteins was greatly amplified, and these proteins may have acquired novel functions, such as mediating the effect of CCT domain proteins on gene expression.
INTRODUCTION
Plant growth and development continually respond to environmental cues such as light, gravity, and temperature. These responses involve signaling cascades that are specific to plants. CCT domain–containing proteins represent one of the classes of proteins involved in several of these pathways. This domain was originally described as a 43–amino acid region of homology found in the Arabidopsis thaliana proteins CONSTANS (CO), CO-LIKE, and TIMING OF CAB1 (TOC1) (Putterill et al., 1995; Strayer et al., 2000; Robson et al., 2001). In Arabidopsis, members of this family of proteins have been implicated in processes such as photoperiodic flowering (Putterill et al., 1995), regulation of circadian rhythms (Strayer et al., 2000; Nakamichi et al., 2005; Salome et al., 2006), light signaling (Kaczorowski and Quail, 2003; Zobell et al., 2005), and gene expression in response to sugars (Masaki et al., 2005). These proteins are found in a wide range of flowering plants as well as mosses (Shimizu et al., 2004; Zobell et al., 2005) and Chlamydomonas but have not been described in animals or yeast. CCT domain proteins with important functions in other flowering plants include the VERNALIZATION2 (VRN2) protein that confers a vernalization response in wheat (Triticum aestivum) (Yan et al., 2004) and proteins involved in the photoperiodic responses of rice (Oryza sativa) (Yano et al., 2000), barley (Hordeum vulgare) (Turner et al., 2005), and other plants (Liu et al., 2001). Taken together, these observations suggest that the CCT domain evolved in the plant lineage and was amplified in proteins associated with environmental responses to light or temperature or in the circadian clock.
Here, we have studied the function of CO, one of the founding members of the CCT protein family. This protein plays a central role in a pathway in Arabidopsis that controls flowering in response to seasonal changes in daylength. CO encodes a nuclear protein containing B-box zinc fingers as well as the CCT domain. The abundance of CO mRNA is regulated by the circadian clock and by light so that it accumulates at the end of the day under long days (LDs) and during the night under short days (SDs) (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002; Imaizumi et al., 2003, 2005). The coincidence of CO mRNA expression and exposure of plants to light under LDs stabilizes the CO protein in the nucleus, causing activation of FLOWERING LOCUS T (FT) transcription (Valverde et al., 2004), and FT is a potent activator of flowering (Kardailsky et al., 1999; Kobayashi et al., 1999; Abe et al., 2005; Wigge et al., 2005).
Although much is known of the regulation of CO expression, the biochemical activity of CO protein is not well understood. Mutational analysis defined the B-boxes and CCT domain as regions of the protein required for the promotion of flowering (Robson et al., 2001). The B-boxes are closely related in sequence to the B-boxes of animal proteins, which are often involved in protein–protein interactions. Despite its presence in several proteins regulating important traits, the biochemical function of the CCT domain is not known. However, the functional importance of the domain was indicated by the isolation of loss of function alleles of CO (Putterill et al., 1995; Robson et al., 2001) and TOC1 (Strayer et al., 2000) in Arabidopsis and VRN2 in wheat (Yan et al., 2004) as well as natural allelic variation in PPD-H1 in barley (Turner et al., 2005). The CCT domain of CO was sufficient to localize CCT:green fluorescent protein fusions to the nucleus, but some CCT domain mutations that disrupt CO function do not prevent nuclear localization, suggesting that it must have additional functions (Robson et al., 2001). In addition, ABSCISIC ACID INSENSITIVE3, a transcription factor involved in regulating responses to abscisic acid, interacts with the CCT domains of TOC1 and CO (Kurup et al., 2000). Although the biological significance of this interaction has not been defined, it suggests that an additional role of the CCT domain may be in the interaction with other proteins.
We present evidence for a functional relationship between the CCT domain and the heterotrimeric Heme Activator Protein (HAP) complex, a DNA binding transcription factor. Furthermore, we demonstrate a role for the HAP complex in mediating the effect of CO on flowering time, extending the recent observation that a CO homolog from tomato (Solanum lycopersicum) interacts with a tomato HAP5 protein (Ben-Naim et al., 2006). The HAP complex binds to CCAAT boxes, cis-acting elements present in ∼25% of eukaryotic promoters (Gelinas et al., 1985; Bucher, 1990). The HAP complex was independently identified by different groups and is also known as CCAAT box factor (CBF) or nuclear factor Y (NF-Y) (reviewed in Mantovani, 1999). The HAP complex consists of the three subunits: HAP2 (NF-YA; CBF-B), HAP3 (NF-YB; CBF-A), and HAP5 (NF-YC; CBF-C). In mammals, the NF-YA and NF-YC subunits, which belong to the class of histone-fold proteins, first dimerize, and then NF-YA, which contains the sequence-specific DNA binding domain, associates prior to DNA binding (Sinha et al., 1995, 1996; Kim et al., 1996). The yeast HAP complex forms in a single-step mechanism and requires the presence of all three subunits (Mantovani, 1999; McNabb and Pinto, 2005). In the yeast Saccharomyces cerevisiae, the trimeric HAP complex has no transcriptional activation potential, and HAP4, a protein providing an acidic activation domain, associates with the DNA-bound HAP2/HAP3/HAP5 complex to mediate transcriptional activation (Forsburg and Guarente, 1989; McNabb et al., 1997). The mammalian NF-YA and NF-YC subunits contain Gln-rich stretches that can activate transcription (Coustry et al., 1995, 1996). In yeast and mammals, each of the HAP subunits is encoded by a single gene (Mantovani, 1999), which contrasts with the findings in plants where all of the subunits are encoded by gene families (Gusmaroli et al., 2001, 2002). The Arabidopsis genome encodes 10 At HAP2, 13 At HAP3, and 13 At HAP5 subunits but no HAP4 homologs (Gusmaroli et al., 2002). The first plant orthologs were identified by functional complementation of yeast hap3 mutants (Edwards et al., 1998), indicating that plant HAP proteins are functional. The presence of many genes encoding HAP subunits suggests a high degree of genetic redundancy among these proteins in plants. Nevertheless, two mutations in HAP proteins were identified and related to defects in development, suggesting that particular plant HAP proteins may have specialized functions (Lotan et al., 1998; Kwong et al., 2003; Lee et al., 2003).
Here, we present evidence for a functional and structural relationship between the CCT domain and the HAP complex. We demonstrate that CO and the related protein COL15 interact with several At HAP3 and At HAP5 subunits and, therefore, that the interaction between CCT and HAP proteins is much more extensive than found previously (Ben-Naim et al., 2006). We also describe a region in the CCT domain that is conserved with regions in HAP2 involved in protein complex formation and DNA binding. Furthermore, we show that increased expression of HAP complex subunits affects flowering time of Arabidopsis and that this effect is associated with reduced activation of FT by CO. We propose that CCT domain proteins replace At HAP2 subunits to form a trimeric CO/At HAP3/At HAP5 complex that regulates gene expression.
RESULTS
CO Interacts with At HAP3 and At HAP5 Proteins in Yeast
To better understand the biochemical function of CO, a yeast two-hybrid library was screened to identify proteins that interact with the CCT domain (see Methods). Five clones that expressed At HAP5 homologs (At3g48590 [twice], At1g54830 [twice], and At1g56170) and one clone that expressed At HAP3a (At2g38880) were identified. In yeast, HAP2, HAP3, and HAP5 form a complex that interacts with DNA via the HAP2 subunit (Sinha et al., 1995). The observation that CO interacts with At HAP3 and At HAP5 suggested that CO might act as a novel HAP2 subunit and replace At HAP2 in a modified HAP complex.
Twenty-six proteins proposed to be At HAP3 and At HAP5 subunits are encoded in the Arabidopsis genome. In addition, there are 16 additional CO-LIKE (COL) genes of Arabidopsis that all encode related CCT domains, suggesting that these might also interact with the HAP complex. To provide an indication of the full extent of the interactions between the CCT domain proteins and the At HAP proteins, 18 At HAP proteins (nine At HAP3 and nine At HAP5) were tested in yeast for their ability to interact with the CCT domains of CO and a second COL protein, COL15. These experiments demonstrated that CO and COL15 can interact with several At HAP3 and At HAP5 homologs (Table 1). Therefore, the interactions between the At HAP and COL families of proteins in planta could be complex, with multiple At HAP proteins able to interact with several COL proteins.
Table 1.
Interactions of HAP Proteins with the CCT Domains of CO and COL15 Detected Using the Yeast Two-Hybrid System
| HAP3 AGI Code | Other Name | CO | COL15 | HAP5 AGI Code | Other Name | CO | COL15 |
|---|---|---|---|---|---|---|---|
| At2g47810 | NA | + | + | At3g48590 | HAP5a | + | + |
| At1g21970 | LEC1 | + | + | At5g63470 | NA | − | + |
| At5g47670 | L1L | + | − | At1g08970 | HAP5c | + | + |
| At3g53340 | NA | − | + | At1g54830 | NA | + | − |
| At2g38880 | HAP3a | + | + | At1g56170 | HAP5b | + | + |
| At5g47640 | HAP3b | + | + | At5g50490 | NA | + | + |
| At5g50470 | NA | + | + | ||||
| At5g50480 | NA | + | + |
No interactions between the empty bait vector and HAP proteins or between the CCT domains of CO and COL15 with the empty prey vector were detected (data not shown). CO and COL15 were also tested with other HAP3 proteins (At1g09030, At2g13570, and At4g14540) and another HAP5 protein (At3g27910), but no interaction was detected, although expression of the HAP proteins in these strains was not confirmed. +, positive interactions; −, negative interactions; NA, not applicable. AGI, Arabidopsis Genome Initiative.
CO Interacts with At HAP3 and At HAP5 Proteins in Vitro
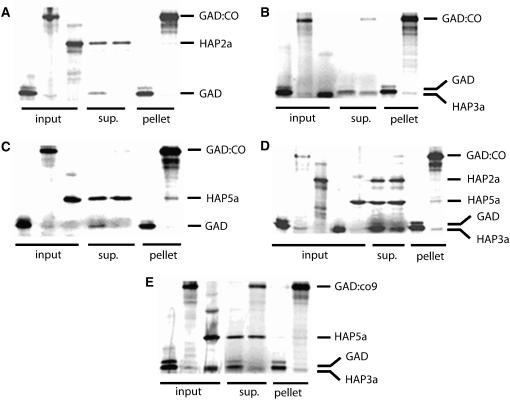
The interactions observed in yeast occurred in the presence of the yeast HAP proteins. Therefore, the ability of CO to interact directly with each of the At HAP subunits was tested in vitro where individual interactions could be assessed. CO was expressed in vitro as a fusion protein with the Gal4 activation domain (GAD). GAD:CO was combined with At HAP2a, At HAP3a, or At HAP5a and precipitated using a GAD-specific antibody (Figures 1A to 1C). At HAP3a (Figure 1B) and At HAP5a (Figure 1C) were coimmunoprecipitated with GAD:CO, indicating that they interact with GAD:CO in this assay. At HAP3a and At HAP5a were also combined simultaneously with GAD:CO, and both proteins were immunoprecipitated with GAD:CO. This experiment indicated that At HAP3a and At HAP5a do not compete for the same binding site on GAD:CO and that CO may bind the At HAP3/At HAP5 dimer in a similar way to HAP2. By contrast, coimmunoprecipitation of At HAP2a and GAD:CO was barely detectable (Figure 1A), suggesting that At HAP2a did not interact with GAD:CO or did so to a much lesser extent than At HAP3a and At HAP5a. Furthermore, when At HAP2a, At HAP3a, and At HAP5a were added simultaneously to GAD:CO in the immunoprecipitation reaction, At HAP2a was coprecipitated to a much lesser extent than At HAP3a and At HAP5a, indicating that GAD:CO does not bind to the trimeric HAP complex (Figure 1D). Mutations that impair the function of the CO CCT domain were identified because they delay flowering (Figure 2A), and one of these, co-9, was tested for its effect on coimmunoprecipitation of the At HAP5a and At HAP3a proteins. GAD:co-9 coprecipitated At HAP5a and At HAP3a to a similar extent to GAD:CO (Figure 1E), indicating that the co-9 mutation does not abolish interaction with these proteins in this assay, although it does strongly affect CO function in planta.
Figure 1.
In Vitro Analysis of the Protein–Protein Interactions Observed in Yeast.
In vitro binding experiments performed with GAD:CO or GAD and At HAP2a (A), At HAP3a (B), At HAP5a (C), At HAP2a/At HAP3a/At HAP5a (D), and with GAD:co9 and At HAP3a/At HAP5a (E). Protein complexes were immunoprecipitated using an anti-GAD antibody (Santa Cruz). Input, aliquots (from left to right) of GAD, GAD:CO, or GAD:co-9 and prey in vitro–produced proteins that were used for coimmunoprecipitations; sup., supernatant fraction containing unbound proteins (from left to right) GAD plus prey and GAD:CO or GAD:co-9 plus prey; pellet, fraction after precipitation of GAD or GAD:CO or GAD:co-9.
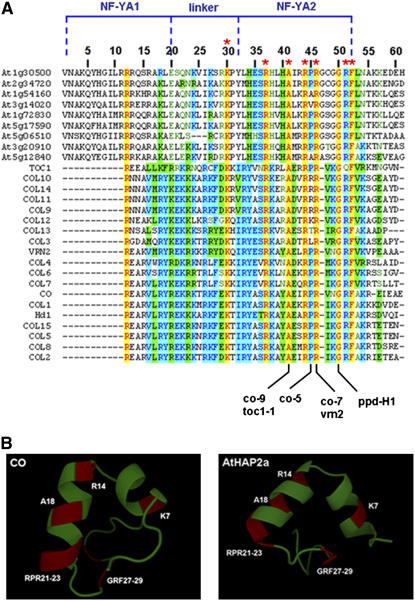
Figure 2.
Sequence Alignment of the Conserved Domain of All Arabidopsis COL Proteins, Rice Hd1, Wheat VRN2, Arabidopsis TOC1, and All Arabidopsis HAP2.
(A) Mutations affecting the residues that impair the function of CO, TOC1, VRN2, and PPD-H1 are indicated. Yellow, conserved residues; blue, identical residues; green, similar residues. Residues marked with an asterisk are of importance in yeast HAP2 proteins. The NF-YA1/linker/NF-YA2 domain is marked in blue according to Romier et al. (2003).
(B) Ab initio prediction of protein structures of the NF-YA2 domain of CO and At HAP2a using the Rosetta program.
The CCT Domain Contains a Functionally Important Region of Homology with the NF-YA1/2 Subdomains of HAP2
The yeast two-hybrid and in vitro experiments suggested that CO interacts with At HAP3/At HAP5 in an analogous way to the At HAP2 subunit. This hypothesis was supported by detailed comparisons of HAP2 and COL protein sequences that identified a domain of homology that is shared by these proteins and was not previously detected. The highly conserved core region of the HAP2 protein contains two functionally important subdomains named NF-YA1 and NF-YA2 that are connected by a small linker sequence (Romier et al., 2003). The NF-YA1 subdomain forms an α helix proposed to interact with the HAP3/HAP5 dimer, whereas NF-YA2 was modeled to interact with the DNA of the CCAAT box (Romier et al., 2003). The CCT domain contains a region of striking similarity to NF-YA2 as well as a lower degree of conservation to the linker domain and NF-YA1 (Figure 2A). Mutational analysis of the NF-YA1 domain of yeast HAP2 identified several residues that are required for the formation of the trimeric complex. One of the residues that strongly affected complex assembly was the Arg residue Arg-12 (Figure 2). The R12G mutation abolished HAP complex formation, resulting in loss of DNA binding (Xing et al., 1994). Interestingly, Arg-12 is conserved among all HAP and CCT domain proteins and defines the start of the CCT domain (Robson et al., 2001). Furthermore, in a 23–amino acid segment of the NF-YA2 domain, seven amino acids are identical in all CCT domain and At HAP2 proteins (Figure 2A). Six of these seven residues are important for DNA binding of NF-YA2, and four are altered by mutations that impair the function of the CCT domain in different proteins. Residues important for CCT function and conserved with residues important for DNA binding of HAP2 are the Ala affected in toc1-1 (Strayer et al., 2000) and co-9 and the Gly affected in ppdH-1 (Turner et al., 2005). The Arg affected in co-7 (Robson et al., 2001) and vrn2 (Yan et al., 2004) is absolutely conserved in all of the CCT domain and At HAP2 proteins and is proposed to be involved in DNA binding of HAP2 (Mantovani et al., 1994). Similarly, mutations in HAP2 that alter the Pro analogous to the Pro affected in the co-5 mutation (Robson et al., 2001) do not affect DNA binding (Mantovani et al., 1994). This residue is not conserved in all of the NF-YA2 domains of HAP2 proteins but is present in 18 of the 28 NF-YA2 sequences shown in Figure 2A. The mutation causes a weak delay in flowering, suggesting that it is not as essential as the absolutely conserved residues identified as mutations (Robson et al., 2001). These results indicate that functionally important domains within HAP2 and the CCT domain of CO share homology and that the most conserved residues are important for the function of the CCT domain.
To determine whether the NF-YA2 subdomain of HAP2 and the related region of the CCT domain of CO are predicted to form similar three-dimensional structures, the Rosetta program was employed (see Methods). This analysis predicted that the NF-YA2 subdomain of HAP2 forms two short α helices, as previously proposed (Romier et al., 2003; Figure 2B). The homologous region of CO is also predicted to form two α helices. Furthermore, in HAP2 three of the residues predicted to interact with DNA (Arg-14, Ala-18, and Arg-21) are on the same side of one of the α helices, and the analogous residues in CO are similarly positioned (Figure 2B). These observations suggest that this region of CO would form a similar structure to NF-YA2; therefore, CO might interact with DNA in a similar way as proposed for HAP2 (Romier et al., 2003).
CO Interacts with At HAP3 and At HAP5 in Planta
To test the significance of the similarities between CO and HAP2 in plants, we first tested whether CO interacts with At HAP3a and At HAP5a in plant cells as shown in yeast and in vitro. Yellow fluorescent protein (YFP):CO protein and cyan fluorescent protein (CFP) fusion proteins with At HAP3a or At HAP5a were expressed in Arabidopsis leaf epidermal cells after bombardment and detected using a confocal microscope. YFP:CO and the CFP:At HAP proteins colocalized in the nuclei of Arabidopsis epidermal cells (Figure 3A).
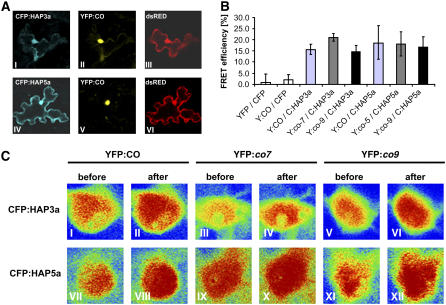
Figure 3.
CO Interacts with At HAP3a and At HAP5a in Plant Cells.
(A) Transient coexpression of 35S:YFP:CO and 35S:CFP:HAP constructs. A 35S:dsRED construct was cotransformed to highlight the whole cell. (I) to (III) Colocalization of (I) CFP:At HAP3a, (II)YFP:CO, and (III) dsRED. (IV) to (VI). Colocalization studies of (IV) CFP:At HAP5a, (V)YFP:CO, and (VI) dsRED. These studies demonstrated nuclear colocalization for CO and HAP3a/HAP5a.
(B) Quantification of FRET efficiencies after acceptor photobleaching. FRET efficiencies of negative controls, such as YFP/CFP, YFP:CO, co-7, or co-9/CFP, were in the range of 0 to 3%.
(C) In vivo FRET analysis showing that CO interacts with the HAP3a and HAP5a subunits as an increase in the CFP fluorescence was detected after photobleaching. The co-7 and co-9 mutant forms of CO have no effect on the interaction with At HAP3a and At HAP5a, since an increase in intensity of the CFP fluorescence was observed after photobleaching. YFP:CO tested with HAP3a (I and II) and HAP5a (VII and VIII); YFP:co7 tested with HAP3a (III and IV) and HAP5a (IX and X); and YFP:co9 tested with HAP3a (V and VI) and HAP5a (XI and XII) is shown. Images display the CFP channel in false colors before and after photobleaching.
Whether the protein interactions observed in yeast and in vitro also occur in planta was determined by fluorescence resonance energy transfer (FRET). FRET was detected between YFP:CO and each of the CFP-labeled At HAP subunits (Figures 3B and 3C), indicating that CO is likely to interact with At HAP3a/At HAP5a in planta. Much weaker FRET signals were detected in negative control experiments employing YFP and CFP alone, or YFP:CO and CFP (Figure 3B). Whether known mutations in the CCT domain that caused delayed flowering in Arabidopsis (co-7 and co-9) also caused a decrease in FRET efficiency was then tested. YFP:co-7 and YFP:co-9 both produced strong FRET signals with CFP:At HAP3a and CFP:At HAP5a (Figures 3B and 3C). These results indicate that co-7 and co-9 do not prevent interaction with At HAP3a and At HAP5a in plant cells as observed in vitro for co-9.
HAP3a mRNA Abundance Exhibits a Diurnal Rhythm and Is Regulated by the Flowering-Time Gene GIGANTEA
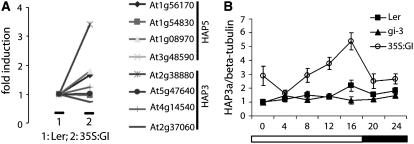
CO mRNA abundance is regulated by GIGANTEA (GI) (Suarez-Lopez et al., 2001; Mizoguchi et al., 2005). To test whether any of the At HAP genes were also regulated by GI, Affymetrix arrays were hybridized with probes derived from 35S:GI and wild-type plants. CO mRNA levels are increased in 35S:GI plants (Mizoguchi et al., 2005), and the array data were analyzed to determine whether any At HAP genes were also increased in expression in these plants. The mRNA of the At HAP3a (At2g38880) gene was more than threefold more abundant than in wild-type plants, suggesting it was also regulated by GI (Figure 4A). The levels of At HAP3a mRNA were then tested by RT-PCR in RNA extracted from LD-grown Landsberg erecta, gi-3, and 35S:GI seedlings at 4-h intervals over 24 h. In wild-type plants, At HAP3a mRNA levels showed a diurnal rhythm, with a peak at 16 h after dawn (Figure 4B). This peak was absent in gi-3 mutant plants. In plants overexpressing GI from the 35S promoter, expression levels of At HAP3a mRNA were significantly higher at all times of the day but showed highest levels at 16 h after dawn. Therefore, At HAP3a mRNA peaks in abundance ∼16 h after dawn, a similar time in the diurnal cycle to the first peak in the biphasic pattern of CO mRNA accumulation, and both genes are regulated by GI.
Figure 4.
GI Regulates the mRNA of HAP3a.
(A) GI regulates the mRNA abundance of HAP3a. Data extracted from microarray analysis (Affymetrix 8k chip) comparing the expression levels of several HAP genes between Landsberg erecta (Ler) and 35S:GI.
(B) Quantitative RT-PCR analysis of the HAP3a expression pattern in a 24-h LD time course in Ler, gi-3, and 35S:GI. Expression levels were quantified against β-tubulin. Error bars represent standard deviations.
Flowering Time of Transgenic Plants Overexpressing At HAP2, At HAP3a, and FLAG:At HAP3a
The protein–protein interaction data described above suggest that CO participates in a protein complex that includes At HAP3 and At HAP5. To test the significance of this protein complex in flowering-time regulation, loss-of-function T-DNA mutations in At HAP3a, which is regulated by GI, were characterized. Plants carrying a T-DNA insertion allele of At HAP3a flowered at the same time as wild-type plants (see Methods; GABI-KAT line 769B07 carrying an insertion in the coding sequence). However, the large number of genes encoding At HAP3 and At HAP5 subunits in Arabidopsis that are coexpressed with CO in the phloem (Zhao et al., 2005) and the ability of CO to interact with several At HAP3 and At HAP5 subunits in the yeast two-hybrid system (Table 1) suggested that there may be genetic redundancy between these genes in the regulation of flowering time.
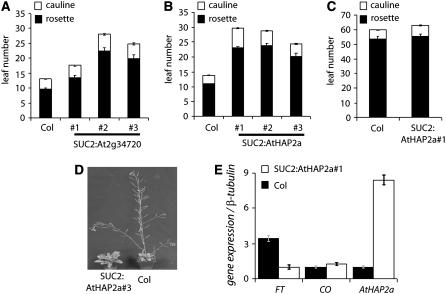
The likely genetic redundancy between genes encoding subunits of At HAP3 or At HAP5 suggested an alternative approach to test the requirement of these subunits for CO function. We reasoned that since At HAP2 also interacts with At HAP3 and At HAP5, then At HAP2 may compete with CO for binding of the At HAP3 and At HAP5 subunits. CO is expressed in the phloem companion cells; therefore, two At HAP2 genes (At5g12840 [AtHAP2a] and At2g34720) were fused to the SUC2 promoter to drive their expression at high levels in these cells. The At HAP2a mRNA contains a binding site for microRNA169 in the 3′ untranslated region (Kidner and Martienssen, 2005). This binding site was removed to allow expression of high levels of At HAP2a in the phloem by the SUC2:At HAP2a fusion. Transformants containing either construct flowered late under LDs (Figures 5A and 5B) SUC2:At HAP2a delayed flowering more severely and in a higher proportion of transformants than SUC2:At2g34720 (see Supplemental Figure 1 online). This might be due to the striking sequence similarity between this specific At HAP2 gene and CO, which extends outside of the NF-YA2 domain (Figure 2A). The involvement of At HAP2a in causing this late-flowering phenotype was supported by the correlation between the abundance of At HAP2a transcript and the severity of the phenotype observed in individual lines. Furthermore, FT transcript levels in a late-flowering line homozygous for the SUC2:At HAP2a construct were fourfold to fivefold lower compared with the wild type, whereas CO mRNA levels were not changed (Figure 5E). Under SDs SUC2:HAP2a#1 plants were not later flowering than wild-type plants, although they were strongly delayed under LDs (Figures 5B and 5C). Therefore, SUC2:At HAP2a impairs flowering under the conditions in which CO acts to promote flowering. These observations suggest that SUC2:At HAP2a plants are delayed in flowering under LDs because CO activation of FT transcription is impaired.
Figure 5.
SUC2:AtHAP2a Delays Flowering and Impairs CO Function.
(A) and (B) Comparison of the late-flowering phenotypes of SUC2:At3g34720 (A) and SUC2:AtHAP2a (B) transgenic plants compared with the wild type (Col) grown under LDs. Representative independent transgenic lines are shown. When comparing all transgenic lines in the experiments, those containing the SUC2:At HAP2a transgene were much later flowering (see Supplemental Figure 1 online).
(C) Flowering times of the SUC2:HAP2a#1 plants grown under SDs compared with wild-type plants. Bars in (A) to (C) show standard error of the mean of at least eight plants.
(D) Image of a representative late-flowering SUC2:At HAP2a#3 plant 5 weeks old under LDs compared with a wild-type Col plant of the same age.
(E) Quantification of FT, CO, and At HAP2a mRNA levels in the late-flowering homozygous SUC2:AtHAP2a#1 line compared with wild-type Col plants of the same age. RNA samples were collected from 9-d-old seedlings grown under LDs. Samples were collected 16 h after dawn before the end of each day. Expression was measured by quantitative real-time RT-PCR (see Methods). Expression of each gene was calculated relative to β-tubulin (At5g62690). Bars show standard error of the mean of three biological repeats. Similar results were obtained in separate experiments.
Whether reducing the levels of At HAP2 subunits accelerates flowering by reducing competition with CO for At HAP3/At HAP5 was also tested. Lines containing T-DNA insertions in At HAP2a and At2g34720 (Alonso et al., 2003; Tzafrir et al., 2003) were characterized for alteration in flowering time under LDs. Homozygous plants containing a T-DNA insertion within an exon of At2g34720 (Salk line N644313, T-DNA insertion in exon 6) showed no detectable accumulation of the At2g34720 transcript (data not shown) but did not display an obvious flowering-time phenotype. A previously characterized T-DNA insertion within the 5′ untranslated region of At HAP2a (EMB2220; http://www.seedgenes.org) causes female gametophyte lethality; therefore, the flowering phenotype caused by a homozygous loss-of-function mutation in this gene could not be tested.
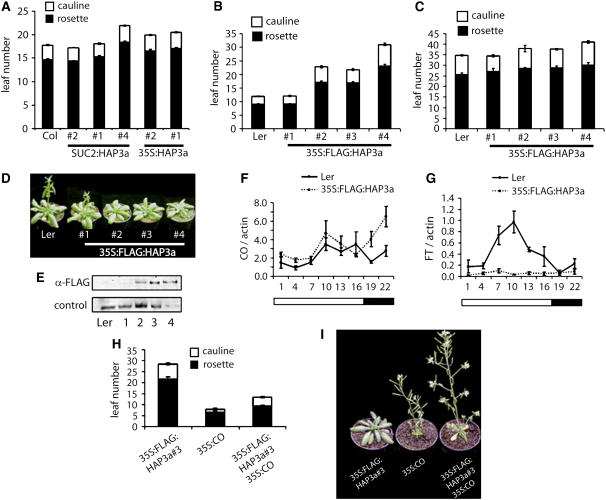
To determine whether elevating the expression levels of At HAP3a would cause early flowering, as observed for 35S:CO plants, the At HAP3 subunit was overexpressed from the 35S promoter of Cauliflower mosaic virus (CaMV35S) and in the phloem from the SUC2 promoter. In addition, a FLAG:At HAP3a fusion protein was expressed from the CaMV35S promoter. Unexpectedly, overexpression of both At HAP3a and FLAG:At HAP3a caused late flowering under LDs (Figures 6A and 6B). Two of six 35S:At HAP3a transgenic lines flowered significantly later than wild-type plants under LDs, as did one of four SUC2:At HAP3a lines. Approximately 63% of the 35S:FLAG:At HAP3a T1 plants (n = 107) were late flowering under LDs. In addition, ∼20% of 35S:FLAG:At HAP3a T1 lines showed other phenotypes, such as growth arrest of the shoot apical meristem and the formation of club-shaped siliques from which new inflorescences developed (data not shown). Under SDs, only a slight delay in flowering was observed for the most severely delayed transformants under LDs. Five representative late-flowering lines in which the transgene was present at a single locus were selected for molecular analysis. The involvement of FLAG:At HAP3a in causing the late-flowering phenotype was supported by a correlation between the abundance of FLAG:At HAP3a protein and the severity of the phenotype in individual lines (Figures 6D and 6E).
Figure 6.
Overexpression of FLAG:At HAP3a or HAP3a Delays Flowering and Impairs CO Function.
(A) Quantification of flowering time of SUC2:HAP3a and 35S:HAP3a in LD compared with Col-0.
(B) and (C) Quantification of flowering time by counting the number of leaves of 35S:FLAG:HAP3a plants produced at bolting in LD (B) and SD conditions (C). Error bars indicate standard errors of the average of at least nine individuals per line.
(D) Flowering-time phenotypes of four independent homozygous 35S:FLAG:HAP3a transgenic lines (#1 to 4) in comparison with the wild type grown in LD.
(E) Protein gel blot using an α-FLAG antibody. Lanes labeled 1 to 4 show protein extracts from transgenic 35S:FLAG:HAP3a lines. The late-flowering phenotype correlates with FLAG:HAP3a protein levels. Control: histone 3 detected with the rabbit anti-histone 3 antibody.
(F) and (G) Time-course experiment comparing the expression levels of CO (F) and FT mRNA (G) in Ler and 35S:FLAG:HAP3a. Triplicate values were quantified per time point, and the standard error of the average is indicated. The experiment was repeated twice with similar results.
(H) Quantification of flowering time by counting the number of leaves produced at bolting in LD of 35S:FLAG:HAP3a, 35S:CO, and 35S:FLAG:HAP3a 35S:CO. Standard errors of averages were determined and are indicated as above.
(I) Flowering-time phenotypes of 35S:FLAG:HAP3a, 35S:CO, and 35S:FLAG:HAP3a 35S:CO plants grown under LD conditions.
To determine whether FLAG:At HAP3a delays flowering by antagonizing CO function, FT and CO mRNA levels were tested in late-flowering 35S:FLAG:At HAP3a plants. FT mRNA abundance was significantly lower in 35S:FLAG:At HAP3a plants than in the wild type (Figure 6G), but CO mRNA levels were not reduced in 35S:FLAG:At HAP3a plants (Figure 6F). These observations suggest that the late-flowering phenotype of these plants is caused by reduced CO activity leading to reduced FT expression.
Overexpression of At HAP3a might sequester CO into nonfunctional complexes. To test this hypothesis, the abundance of CO relative to FLAG:At HAP3a was increased by constructing 35S:CO 35S:FLAG:At HAP3a plants by crossing. The flowering times of homozygous F3 lines were measured and showed that overexpression of CO can attenuate the late flowering caused by high expression of FLAG:At HAP3a (Figures 6H and 6I), consistent with the idea that 35S:FLAG:At HAP3a delays flowering by sequestering CO into inactive complexes and that increasing the abundance of CO overcomes this effect.
The association of FLAG:At HAP3a with the FT promoter was tested in early- and late-flowering transgenic plants by chromatin immunoprecipitation (ChIP). No association of FLAG:At HAP3a with the FT promoter could be demonstrated in the assay, although positive control reactions were successful (data not shown). These experiments may have failed due to the low abundance of the protein complex or because the complex does not bind directly to the FT promoter.
DISCUSSION
We demonstrated that in yeast the CCT domains from two Arabidopsis proteins interact with multiple homologs of the At HAP3 and At HAP5 subunits of the Arabidopsis HAP complex. Selected At HAP3 and At HAP5 subunits were also shown to interact with full-length CO protein in plant cells and in vitro. Strikingly, the CCT domain shows homology to the NF-YA1/2 domains of HAP2 that mediate HAP complex formation and DNA binding. Furthermore, highly conserved residues in this region are required for both CO and HAP2 function. Redundancy between At HAP subunit homologs prevented the use of loss-of-function mutations to demonstrate their importance in flowering-time control, but the relevance of the interaction between CO and the At HAP proteins was strengthened by the observation that overexpression of At HAP2a or At HAP3a impaired CO function and delayed flowering. We propose that one function of the CCT domain is to mediate formation of a protein complex with the HAP proteins At HAP3 and At HAP5 and that this complex has a role in the regulation of gene expression. Specifically, in the case of CO, these proteins are involved in the regulation of FT mRNA expression during the promotion of flowering.
Significance of Interactions between CO and At HAP3a or At HAP5a
The interactions between the CCT domain of CO and At HAP3a or At HAP5a were detected using three independent methods. These results extend the recent observation that a tomato homolog of CO and Arabidopsis CO interact with a tomato HAP5 subunit in yeast and in vitro (Ben-Naim et al., 2006). We did not detect a convincing interaction between CO and At HAP2 in vitro, suggesting that these two proteins do not interact and that the At HAP2 subunit is not required for CO function. The finding that At HAP3 and At HAP5 subunits interact with CO suggested that CO might function in an analogous manner to At HAP2.
CFP:At HAP3a and CFP:At HAP5a interacted with YFP:co-9 and YFP:co-7 in plant cells, as detected by FRET, suggesting that these mutations do not affect complex assembly but impair another aspect of CO function. Since the residues altered by these mutations are conserved in the DNA binding domain of HAP2, they might impair the capacity of the complex to access DNA. By contrast, Ben-Naim et al. (2006) showed that the co-7 mutation affects the interaction with tomato HAP5a in yeast. These results could be reconciled if the yeast system is more quantitative than the in vitro immunoprecipitation and FRET experiments we performed and therefore might have detected a reduction in the efficiency of the interaction that was not detected in our experiments. Alternatively, GAD:co-7 might not have accumulated in yeast, preventing detection of an interaction, or mutations such as co-7 may cause an overall structural change in this region of the protein that affects both DNA binding and complex assembly. Further mutational and functional analysis of the CCT domain will be required to define the roles of individual residues in complex assembly and DNA binding as was performed for HAP2 in yeast (Olesen and Guarente, 1990; Maity and de Crombrugghe, 1992; Xing et al., 1993; Xing et al., 1994).
Significance of the Region of Homology between HAP2 and CCT Domain Proteins
A 23–amino acid region of the CCT domain shows striking homology to a subdomain called NF-YA2 within the core homologous region of HAP2. This region of HAP2 is involved in DNA binding as defined by functional analysis of mutants (Olesen and Guarente, 1990; Maity and de Crombrugghe, 1992; Xing et al., 1993, 1994) and a model of the three-dimensional structure based on the determined structure of the HAP3/HAP5 heterodimer (Romier et al., 2003). The residues of HAP2 analogous to those affected by the toc1-1/co-9 mutations and the ppdH-1 recessive allele are important for DNA binding of HAP2 (Maity and de Crombrugghe, 1992; Xing et al., 1993). Similarly, residues such as Arg-37, Ala-41, Arg-44, Arg-46, and Phe-52 (Figure 2A) are important for DNA binding of HAP2 and are absolutely conserved between all HAP2 isoforms and all CCT domains presented (except TOC1 for Arg-51). Furthermore, the CCT domain is predicted to fold in a similar way to the subdomain of HAP2, and in each case, critical residues that interact with DNA are all on the same side of an α helix. These observations are consistent with CO binding to DNA directly in a manner analogous to HAP2. However, three His residues (His-34, His-38, and His-40) conserved in all HAP2 proteins and required for DNA binding (Xing et al., 1993; Romier et al., 2003) are not conserved in the CCT domain, making a precise functional analogy less likely (Figure 2A).
The similarity in structure of the HAP2 NF-YA2 subdomain with the CCT domain could indicate that CO and other CCT-containing proteins bind directly to DNA. However, HAP2 requires interaction with HAP3/HAP5 to stabilize DNA binding (Romier et al., 2003). Therefore, an attractive possibility is that CCT domain proteins replace At HAP2 in the CCAAT binding factor so that At HAP3/At HAP5 stabilize the interaction of CO with DNA. This model is consistent with the observed interactions between CO and At HAP3/At HAP5a. Since part of the NF-YA1 domain is also conserved in the CCT domain, the At HAP3/At HAP5 interaction might be through the NF-YA1/linker domain forming a similar structure to that of HAP2. An alternative model consistent with our data is that an unidentified transcription factor stabilizes the interaction between CO and DNA and that CO then interacts with the HAP complex bound at adjacent sites on promoters.
Relationship between At HAP3a, At HAP2a, and CO-Mediated Control of Flowering Time
At HAP3a and CO are both regulated by GI, and CO is expressed in the phloem, where it acts to promote flowering through activation of FT transcription (An et al., 2004; Ayre and Turgeon, 2004). Affymetrix microarray analysis of phloem cells indicated that At HAP3a is expressed in the phloem, although it is not enriched in comparison to other cell types (Zhao et al., 2005). The coregulation of CO and At HAP3a and their spatial expression in the same cells is consistent with these proteins interacting in vivo. A functional relationship between At HAP3a and flowering-time regulation is also suggested by the late flowering of plants overexpressing At HAP3a from the CaMV35S or SUC2 promoters or FLAG:At HAP3a from the CaMV35S promoter. The late-flowering phenotype does not correlate with a reduction in CO mRNA abundance but with a severe decrease in abundance of the mRNA of FT, a gene activated by CO (Samach et al., 2000; Abe et al., 2005; Wigge et al., 2005), suggesting that the basis of the effect of FLAG:At HAP3a is in antagonism of CO function. The impairment of CO activity is further implicated by showing that the delay in flowering does not occur under SDs for most transformants, and overexpression of CO partially suppresses the late-flowering phenotype. At HAP3a and FLAG:At HAP3a might impair CO activity by sequestering CO or other components of a complex into inactive complexes and therefore reduce FT expression. CO protein is expressed at very low levels (Valverde et al., 2004) and is likely to be the limiting component in a CO/At HAP3/At HAP5 complex. This conclusion is emphasized by the observation that reducing CO levels by half in heterozygous plants delays flowering (Robson et al., 2001). The HAP complex of mammals forms by the HAP3 subunit interacting with the HAP5 subunit to form a heterodimer, which then binds the third subunit HAP2 (Kato, 2005). If a complex involving CO must also form in a defined series of steps, then overexpression of At HAP3a could alter the stoichiometry of components and recruit CO or one of the other components into unproductive complexes. Overexpression of At HAP5 subunits would also be of interest to determine whether this has a similar effect on flowering to overexpression of At HAP3. Overexpression of a heterologous tomato HAP5 gene caused slightly early flowering of Arabidopsis (Ben-Naim et al., 2006). Future analysis of the role of At HAP3 and At HAP5 forms in flowering control will require detailed analysis of loss-of-function alleles, but likely redundancy between forms will probably make it necessary to combine mutations to detect phenotypic effects. This suggestion is supported by our observation that mutations in the At HAP3a gene did not delay flowering. However, the roles of the HAP complex in regulating expression of many genes may lead to highly pleiotropic phenotypes when mutations in different subunits are combined, and this would complicate analysis of the flowering phenotype.
Ectopic expression of At HAP2a in the phloem also caused a delay in flowering. At HAP2a interacts with At HAP3 and At HAP5 subunits; therefore, overexpression of At HAP2a was predicted to compete with CO for binding of the other At HAP subunits. The microRNA binding site in the At HAP2a mRNA was removed from the SUC2:At HAP2a construct, so that overexpression of At HAP2a was as effective as possible. The high levels of transcription from the SUC2 promoter plus removal of microRNA-based posttranscriptional regulation may have resulted in such effective overexpression of At HAP2a that levels of free At HAP3 and/or At HAP5 became limiting on CO activity. As shown for plants overexpressing FLAG:At HAP3a, the observation that FT mRNA levels but not CO mRNA levels are reduced in the SUC2:At HAP2a lines supports the suggestion that delayed flowering in these plants is due to impaired CO function. Similarly, these plants were delayed in flowering only under LDs where CO promotes flowering.
In yeast and humans, functional CCAAT boxes are typically found 80 to 300 bp upstream of the transcriptional start site (Chodosh et al., 1988a, 1988b; Hatamochi et al., 1988). FT transcription is regulated by CO, but the FT promoter region contains no CCAAT boxes in the proximal promoter region, although it does contain three CAAT sequences within 250 bp 5′ of the ATG. An interaction of CBF with CAAT elements of the tobacco (Nicotiana tabacum) AtpC gene was previously demonstrated (Kusnetsov et al., 1999). Interestingly, the CAAT element located 43 bp upstream of the ATG in FT is conserved in rice Hd3a, the FT homolog (Kojima et al., 2002). However, relevant CAAT/CCAAT sequences might be located further from the start codon, since recent analysis of human genes by ChIP demonstrated binding of the complex at distal promoter locations (Testa et al., 2005). Alternatively, if CO replaces At HAP2 in the HAP complex, then the resulting complex might bind to different DNA sequences. Extensive efforts to demonstrate binding of CO or FLAG:At HAP3 to the FT promoter by ChIP failed, perhaps because of the extreme low abundance of the CO complex in planta (Valverde et al., 2004) or because the CO/At HAP3/At HAP5 complex does not bind directly to FT DNA. However, the recent observation that a CO-related protein from tomato could be immunoprecipitated with tomato HAP5a from CBF target genes in yeast supports the idea that such a complex binds DNA (Ben-Naim et al., 2006).
Conclusion
We propose that the CCT domain acts by a mechanism related to that of the HAP complex. Our data suggest that CCT domain proteins act by interacting with the At HAP3 and At HAP5 subunits to form a higher-order complex that regulates gene expression and that residues in the CCT domain are conserved with those in HAP2 that interact with DNA. The gene family encoding the HAP proteins greatly expanded in the plant lineage, and they may have been recruited in plants to a wider range of functions than in animals and yeast, which contain a single gene encoding each subunit. This was previously suggested by a specific At HAP3 subunit having a defined function in inducing embryo development (Lee et al., 2003) and by interactions between a rice HAP3 and a MADS box transcription factor (Masiero et al., 2002). We propose that a further function of the HAP complex in plants is to enable gene activation by CCT domain proteins that play many important roles in environmental responses, and since we observed that CO and COL15 both interact with a large number of HAP subunits, the matrix of interactions between 45 CCT domain–containing proteins and the 26 proteins that represent At HAP3 and At HAP5 subunits is likely to be complex. These interactions will require further detailed analysis combining loss-of-function mutations in At HAP genes.
METHODS
Yeast Two-Hybrid Screening
The yeast two-hybrid screen that identified At HAP5a interacting with CO was performed using the CCT domain (amino acids 306 to 373) of CO cloned into pAS2.1, yielding an in-frame fusion with the Gal4-DNA binding domain. The construct was transformed into yeast (PJ694A) and mated with Y187 harboring two different cDNA libraries cloned into pGAD-T7 (shoot apex and total plant libraries; kind gift from Hans Sommer, Max Planck Institute für Züchtungsforschung, Cologne, Germany).
Which HAP protein interacts with the CCT domains of CO and COL15 was tested using the PROQUEST system (Invitrogen). The CCT domains of CO (amino acids 306 to 348) and COL15 (amino acids 281 to 323) were recombined into the pDEST32 vector using the LR recombinase mix from Invitrogen. The HAP proteins were taken as entry clones from the REGIA transcription factor library (in pDONR201) and recombined into pDEST22 using LR recombinase. Baits were transformed in the MAV203 yeast strain and tested for autoactivation before transformation of the HAP plasmids. The screen was performed on SD medium lacking His, Leu, and Trp plus 30 mM 3-aminotriazole.
In Vitro Coimmunoprecipitations
The cDNAs of HAP proteins were recombined into pTNT-GW, and the CO cDNA was introduced into the pTNTGAD-GW vector, providing an in-frame fusion to the GAD domain using the Gateway recombination system (Invitrogen). Prey and bait proteins were synthesized in the presence of 35S-Met using the TNT Quick Coupled Transcription/Translation system (Promega). Coimmunoprecipitations were performed as described by Laubinger and Hoecker (2003).
Protein Extraction, ChIP Assays, and Protein Gel Blotting
Protein extraction and ChIP assays were performed as described (Searle et al., 2006). For protein gel blotting, 5 μg of nuclear extract was blotted to polyvinylidene difluoride membranes after SDS-PAGE and probed with mouse monoclonal anti-FLAG antibodies (Sigma-Aldrich F3165) and rabbit anti-histone 3 (Upstate 050499) simultaneously. Proteins were visualized and quantified after incubation with anti-rabbit IRDye800 (Rockland 611-132-122) and anti-mouse AlexaFluor680 (Invitrogen AL1055) secondary antibodies in an Odyssey infrared scanner. All antibodies were used as 1:5000 dilutions.
Transient Expression of Fluorescent Proteins, Confocal Microscopy, and FRET Analysis
To generate fluorescent proteins, the pENSG-YFP:GW and pENSG-CFP:GW vectors were used, yielding N-terminal fusions of YFP/CFP driven by the 35S promoter (kind gift from Nieves Medina-Escobar, Max Planck Institute für Züchtungsforschung). Transformation, subcellular localization, and FRET experiments were performed as described by Feys et al. (2005).
Plant Material and Growth Conditions
Plants used for gene expression and ChIP analysis were grown on germination medium plates containing 1% sucrose. After ethanol sterilization, seeds were spread on plates, stratified for 3 d at 4°C, and finally transferred to growth chambers with LD (16 h/8 h dark) or SD (8 h light/16 h dark) regimes at 22°C.
The mutation described in the text as co-9 was identified as a mutation in a 35S∷CO transgene as part of a screen for suppressors of the 35S∷CO phenotype (R. Hayama and G. Coupland, unpublished data).
To generate transgenic plants overexpressing the cDNAs encoding At HAP3a and At HAP2a, the Gateway recombination system was used. The open reading frame for the At HAP3a entry clone was recombined into the pLeela binary vector and into pJAN33 to create the fusion to the FLAG tag. The SUC2∷AtHAP2a fusion was constructed by recombining the At HAP2a open reading frame into a modified pGREEN vector in which the SUC2 promoter was inserted upstream of the GATEWAY recombination site (L. Gissot and G. Coupland, unpublished). These constructs were introduced into Agrobacterium tumefaciens GV3101. Transgenic Arabidopsis thaliana plants were made using the floral dip method. T1 transgenic plants were isolated after selection with BASTA and lines that contained the T-DNA at a single locus confirmed by following the segregation ratio in the T2 generation. Flowering times were scored either in homozygous T3 families or by scoring BASTA-resistant T2 plants.
Flowering-Time Experiments
For flowering-time experiments, seeds were sown on soil, cold-treated for 3 d at 4°C, transferred to Percival growth chambers, and grown in the desired light regime (SD or LD) at 20°C. Flowering time was determined by counting the number of rosette leaves at bolting and the number of cauline leaves after bolting.
Gene Expression Analysis Using Quantitative RT-PCR
In all experiments except the one shown in Figure 5E, plant material grown on plates was harvested 12 d after transfer to the growth chambers, and RNA was extracted using the RNAeasy kit (Qiagen). The isolated RNA was DNaseI treated and phenol/chlorophorm extracted. RNA was reverse transcribed using oligo(dT) primers and Superscript II reverse transcriptase (Invitrogen). Real-time quantitative PCR reactions were performed on an iQ5 real-time PCR detection system (Bio-Rad Laboratories), and data analysis was performed using Bio-Rad iQ5 software.
The following primers were used for real-time quantitative PCR: β-tubulin forward, 5′-ACACCAGACATAGTAGCAGAAATCAAG-3′; β-tubulin reverse, 5′-ACTCGTTGGGAGGAGGAACT-3′; CO forward, 5′-TAAGGATGCCAAGGAGGTTG-3′; CO reverse, 5′-CCCTGAGGAGCCATATTTGA-3′; FT forward, 5′-CGAGTAACGAACGGTGATGA-3′; FT reverse, 5′-CGCATCACACACTATATAAGTAAAACA-3′; HAP3a forward, 5′-GCGTTGCCTCCTAATGGTAA-3′; HAP3a reverse, 5′-ACCCTCCAACTCCCTGTACC-3′.
In the experiment presented in Figure 5E, cDNA was synthesized from poly(A)+ RNA according to Teper-Bamnolker and Samach (2005). Real-time quantitative PCR reactions were performed using kits from Abgene UK: ABsolute SYBR Green Mix kit (AB-1162) for reactions with Syber-Green (β-tubulin and At HAP2a) and ABsolute QPCR mix (AB-1138A) for reactions with Taqman probes (FT and CO). Reactions were run on a Corbett Research Rotor-Gene 2000 cycler. Quantification of each gene was performed using Corbett Research Rotor-Gene software as described by Teper-Bamnolker and Samach (2005). The forward primer used for β-tubulin (At5G62690) gene expression was 5′-AAACTCACTACCCCCAGCTTTG-3′, and the reverse primer was 5′-CACCAGACATAGTAGCAGAAATCAAGT-3′. The forward primer used for At HAP2a (At5g12840) was 5′-GGAAAGTCATCCGGGACAGAAAGC-3′, and the reverse primer was 5′-TTTCTTCGCAAACCGGCCTCCA-3′. The Taqman probe used for FT expression was 5′-CCTTTGGCAATGAGATTGTGTGTTACGAAAAT-3′, the forward primer was 5′-GATATCCCTGCTACAACTGGAACA-3′, and the reverse primer was 5′-GAATTCCTGCAGTGGGACTTG-3′. The Taqman probe used for CO expression was 5′-TTCCATTAACCATAACGCATACATTTCATCCATG-3′, the forward primer was 5′-CAGGGACTCACTACAACGACAATG-3′, and the reverse primer was 5′-TCCGGCACAACACCAGTTT-3′.
Protein Structure Predictions
Protein structures were ab initio predicted using the Rosetta program (Simons et al., 1997, 1999; Bonneau et al., 2001, 2002) and the resulting pdb-files analyzed using PYMOL.
Accession Numbers
Sequence data for the proteins mentioned in this article are as follows: Arabidopsis Genome Initiative code (name, if applicable/GenBank accession number). At2G47810 (AAC63635); At1g21970 (LEC1/NP_173616); At5g47670 (L1L/AAN15924); At3g53340 (NP_190902); At2g38880 (HAP3a/NP_850305); At5g47640 (HAP3b/NP_199575); At3g48590 (HAP5a/NP_190428); At5g63470 (NP_201152); At1g08970 (HAP5c/NP_973796); At1g54830 (NP_974030); At1g56170 (HAP5b/NP_176013); At5g50490 (NP_199860); At5g50470 (NP_199858); At5g50480 (NP_199859); At5G15840 (CO/NP_001031887); AT1G28050 (COL15/Q9C7E8); At1g65480 (FT/NP_176726); At1g22770 (GI/NP_564180).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Comparison of the Late-Flowering Phenotypes of SUC2∷At3g34720 and SUC2∷At HAP2a Transgenic Plants Compared with the Wild Type Grown under LDs.
Supplementary Material
Acknowledgments
We thank Jo Putterill and Nabil Elrouby for helpful suggestions on the manuscript and Elmon Schmelzer for assistance with the confocal microscope. We also thank Ryosuke Hayama for his unpublished information on the co-9 mutation, Eliezer Lifschitz and Orna Ben-Naim for providing data on tomato HAP5 and COLs prior to publication, and Heiko Schoof and Seth Davis for help with the structural predictions. We thank Nieves Medina-Escobar and Hans Sommer for providing the FRET vectors and yeast two-hybrid libraries, respectively. We also thank Ehud Katz for help initiating experiments on the T-DNA insertions of At HAP2 genes and Lior Seri and Elke Kemper for their excellent technical help. This work was supported by a joint German-Israel Foundation grant to A.S. and G.C. and by a core grant from the Max Planck Society to the laboratory of G.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: George Coupland (coupland@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- An, H.L., Roussot, C., Suarez-Lopez, P., Corbesler, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ayre, B.G., and Turgeon, R. (2004). Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 135 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim, O., Eshed, R., Parnis, A., Teper-Bamnolker, P., Shalit, A., Coupland, G., Samach, A., and Lifschitz, E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46 462–476. [DOI] [PubMed] [Google Scholar]
- Bonneau, R., Strauss, C.E.M., Rohl, C.A., Chivian, D., Bradley, P., Malmstrom, L., Robertson, T., and Baker, D. (2002). De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322 65–78. [DOI] [PubMed] [Google Scholar]
- Bonneau, R., Tsai, J., Ruczinski, I., Chivian, D., Rohl, C., Strauss, C.E.M., and Baker, D. (2001). Rosetta in CASP4: Progress in ab initio protein structure prediction. Proteins 5 (suppl.), 119–126. [DOI] [PubMed] [Google Scholar]
- Bucher, P. (1990). Weight matrix descriptions of 4 eukaryotic RNA Polymerase-II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212 563–578. [DOI] [PubMed] [Google Scholar]
- Chodosh, L.A., Baldwin, A.S., Carthew, R.W., and Sharp, P.A. (1988. a). Human CCAAT-binding proteins have heterologous subunits. Cell 53 11–24. [DOI] [PubMed] [Google Scholar]
- Chodosh, L.A., Olesen, J., Hahn, S., Baldwin, A.S., Guarente, L., and Sharp, P.A. (1988. b). A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell 53 25–35. [DOI] [PubMed] [Google Scholar]
- Coustry, F., Maity, S.N., and Decrombrugghe, B. (1995). Studies on transcription activation by the multimeric CCAAT-binding Factor CBF. J. Biol. Chem. 270 468–475. [DOI] [PubMed] [Google Scholar]
- Coustry, F., Maity, S.N., Sinha, S., and deCrombrugghe, B. (1996). The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J. Biol. Chem. 271 14485–14491. [DOI] [PubMed] [Google Scholar]
- Edwards, D., Murray, J.A.H., and Smith, A.G. (1998). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S.L., and Guarente, L. (1989). Identification and characterization of HAP4 - A 3rd component of the CCAAT-bound HAP2 HAP3 heteromer. Genes Dev. 3 1166–1178. [DOI] [PubMed] [Google Scholar]
- Gelinas, R., Endlich, B., Pfeiffer, C., Yagi, M., and Stamatoyannopoulos, G. (1985). G-substitution to a-substitution in the distal CCAAT box of the a-gamma-globin gene in greek hereditary persistence of fetal hemoglobin. Nature 313 323–325. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Tonelli, C., and Mantovani, R. (2001). Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264 173–185. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Tonelli, C., and Mantovani, R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283 41–48. [DOI] [PubMed] [Google Scholar]
- Hatamochi, A., Golumbek, P.T., Vanschaftingen, E., and Decrombrugghe, B. (1988). A CCAAT DNA-binding factor consisting of 2 different components that are both required for DNA-binding. J. Biol. Chem. 263 5940–5947. [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-BOX protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306. [DOI] [PubMed] [Google Scholar]
- Kaczorowski, K.A., and Quail, P.H. (2003). Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15 2654–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kato, M. (2005). An overview of the CCAAT-box binding factor in filamentous fungi: Assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 69 663–672. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8 38–44. [DOI] [PubMed] [Google Scholar]
- Kim, I.S., Sinha, S., deCrombrugghe, B., and Maity, S.N. (1996). Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16 4003–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Kurup, S., Jones, H.D., and Holdsworth, M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21 143–155. [DOI] [PubMed] [Google Scholar]
- Kusnetsov, V., Landsberger, M., Meurer, J., and Oelmueller, R. (1999). The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin and the stage of the plastids. J. Biol. Chem. 274 36009–36014. [DOI] [PubMed] [Google Scholar]
- Kwong, R.W., Bui, A.Q., Lee, H., Kwong, L.W., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S., and Hoecker, U. (2003). The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 35 373–385. [DOI] [PubMed] [Google Scholar]
- Lee, H.S., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 100 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Y., Yu, J.P., McIntosh, L., Kende, H., and Zeevaart, J.A.D. (2001). Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 125 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A.L., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Maity, S.N., and de Crombrugghe, B. (1992). Biochemical analysis of the B subunit of the heteromeric CCAAT-binding factor. A DNA-binding domain and a subunit interaction domain are specified by two separate segments. J. Biol. Chem. 267 8286–8292. [PubMed] [Google Scholar]
- Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239 15–27. [DOI] [PubMed] [Google Scholar]
- Mantovani, R., Lio, X.Y., Pessara, U., Vanhuisjduijnen, R.H., Benoist, C., and Mathis, D. (1994). Dominant-negative analogs of NF-YA. J. Biol. Chem. 269 20340–20346. [PubMed] [Google Scholar]
- Masaki, T., Tsukagoshi, H., Mitsui, N., Nishii, T., Hattori, T., Morikami, A., and Nakamura, K. (2005). Activation tagging of a gene for a protein with novel class of CCT-domain activates expression of a subset of sugar-inducible genes in Arabidopsis thaliana. Plant J. 43 142–152. [DOI] [PubMed] [Google Scholar]
- Masiero, S., Imbriano, C., Ravasio, F., Favaro, R., Pelucchi, N., Gorla, M.S., Mantovani, R., Colombo, L., and Kater, M.M. (2002). Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J. Biol. Chem. 277 26429–26435. [DOI] [PubMed] [Google Scholar]
- McNabb, D.S., and Pinto, I. (2005). Assembly of the HAP2p/HAP3p/HAP4p/HAP5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 4 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb, D.S., Tseng, K.A.S., and Guarente, L. (1997). The Saccharomyces cerevisiae HAP5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 17 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46 609–619. [DOI] [PubMed] [Google Scholar]
- Olesen, J.T., and Guarente, L. (1990). The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition - Model for the HAP2/3/4 complex. Genes Dev. 4 1714–1729. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Robson, F., Costa, M.M.R., Hepworth, S.R., Vizir, I., Pineiro, M., Reeves, P.H., Putterill, J., and Coupland, G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28 619–631. [DOI] [PubMed] [Google Scholar]
- Romier, C., Cocchiarella, F., Mantovani, R., and Moras, D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278 1336–1345. [DOI] [PubMed] [Google Scholar]
- Salome, P.A., To, J.P.C., Kieber, J.J., and McClung, C.R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., Amasino, R.A., and Coupland, G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, M., Ichikawa, K., and Aoki, S. (2004). Photoperiod-regulated expression of the PpCOL1 gene encoding a homolog of CO/COL proteins in the moss Physcomitrella patens. Biochem. Biophys. Res. Commun. 324 1296–1301. [DOI] [PubMed] [Google Scholar]
- Simons, K.T., Bonneau, R., Ruczinski, I., and Baker, D. (1999). Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins 3 (suppl.), 171–176. [DOI] [PubMed] [Google Scholar]
- Simons, K.T., Kooperberg, C., Huang, E., and Baker, D. (1997). Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268 209–225. [DOI] [PubMed] [Google Scholar]
- Sinha, S., Kim, I.S., Sohn, K.Y., DeCrombrugghe, B., and Maity, S.N. (1996). Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 16 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S., Maity, S.N., Lu, J.F., and Decrombrugghe, B. (1995). Recombinant rat CBF-C, the 3rd subunit of CBF/NF-Y, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. USA 92 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa, A., Donati, G., Yan, P., Romani, F., Huang, T.H.-M., Vigano, M.A., and Mantovani, R. (2005). Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J. Biol. Chem. 280 13606–13615. [DOI] [PubMed] [Google Scholar]
- Turner, A., Beales, J., Faure, S., Dunford, R.P., and Laurie, D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., Dickerman, A., Brazhnik, O., Nguyen, Q., McElver, J., Frye, C., Patton, D., and Meinke, D. (2003). The Arabidopsis SeedGenes Project. Nucleic Acids Res. 31 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xing, Y.Y., Fikes, J.D., and Guarente, L. (1993). Mutations in Yeast Hap2/Hap3 define a hybrid CCAAT box-binding domain. EMBO J. 12 4647–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y.Y., Zhang, S.U., Olesen, J.T., Rich, A., and Guarente, L. (1994). Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 91 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L.L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., Bennetzen, J.L., Echenique, V., and Dubcovsky, J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell, O., Coupland, G., and Reiss, B. (2005). The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 7 266–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.