Abstract
Regulation by Rho-type small GTPases, such as RAC5, is important for the maintenance of polarity in tobacco (Nicotiana tabacum) pollen tubes. We previously showed that RhoGDI2 is necessary for RAC5 localization. Here, we describe the GTPase activating protein RhoGAP1 that controls the area of RAC5 activity. RhoGAP1 N-terminal and CRIB (for Cdc42/Rac-interactive binding) domains are both necessary for targeting yellow fluorescent protein–RhoGAP1 fusions to the plasma membrane close to, but not in, pollen tube apices. We propose that this localization restricts apical Rho-type GTPase activity from spreading toward the flanks, which ensures the maintenance of RAC signaling at the apex. The CRIB domain is not required but enhances in vitro RhoGAP1 activity toward the pollen tube–specific-RAC5. A mutation reducing GAP activity of RhoGAP1 leads to ballooning pollen tubes resembling those overexpressing RAC5. To ascertain the specific targeting mechanism of RhoGAP1, we isolated a 14-3-3 protein interacting with RhoGAP1. When overexpressed with RhoGAP1, it counteracts the growth-retarding effect of RhoGAP1 overexpression and attenuates RhoGAP1 membrane localization but, overexpressed alone, induces only small architectural changes. We propose that inactivation of RAC5 by the subapically localized RhoGAP1, together with dynamic relocalization of inactivated RAC5 from flanks to tip by RhoGDI2, leads to spatial restriction of RAC5 to pollen tube apices, thereby sustaining polar growth.
INTRODUCTION
Pollen tubes are an ideal model system for the study of rapid polarized growth. They are likely to be comparable to model systems from other species, such as fungal hyphae and neuronal outgrowth in human cells (Lord, 2003). Tobacco (Nicotiana tabacum) pollen tubes can easily grow in vitro at elongation rates reaching up to 100 times their own cell width per hour. This rapid growth also displays an organellar and cell biological organization that allows imaging patterns not usually observable in nonpolar cell growth.
Apart from pollen development, which has been mainly studied using Arabidopsis thaliana mutants (McCormick, 2004), pollen growth has been investigated particularly in respect to self-incompatibility, tube adhesion and allergies in humans, tube guidance, tube development (reviewed in Lord and Russell, 2002; Lord, 2003), and maintenance of polar growth (reviewed in Hepler et al., 2001). Major determinants of maintaining pollen tube polarity were found to be a tip-oriented calcium gradient, thick actin filaments in the body and finer filaments in an actin fringe near the tip of the tube, massive exocytosis at the tip, and consequent membrane recycling by endocytosis (Fu et al., 2001; Hepler et al., 2001; Parton et al., 2003; Lovy-Wheeler et al., 2005).
Many processes regulating cell polarity are probably regulated by small Rho-type GTPases (Jaffe and Hall, 2005). Plant Rho-type GTPases most closely resemble Rac-type proteins and have therefore been alternatively named Rac (Winge et al., 2000) or Rop (rho of plants; Zheng and Yang, 2000). They have been shown to strongly affect pollen tube growth when overexpressed in their wild-type or mutant forms (Kost et al., 1999; Zheng and Yang, 2000). They bind downstream effectors when bound to GTP and are thought to be inactive when the GTP is hydrolyzed to GDP. Mutant versions of these proteins that fail to hydrolyze the GTP and therefore activate downstream signaling proteins constitutively (constitutively active [CA] mutants) have been isolated. Likewise, proteins that do not bind any nucleotide, which are locked in a state that resembles a transition state and therefore bind particularly well to G-nucleotide exchange factors (GEFs) (see below) and block the reactivation of endogenous GTPases (dominant negative [DN]), are also commonly used (Feig, 1999). Downstream targets of these regulators have been linked to calcium regulation and modulation of the actin cytoskeleton (Chen et al., 2003; Gu et al., 2005; Smith and Oppnenheimer, 2005). In N. tabacum pollen tubes, RAC5 is tip localized and leads to ballooning when overexpressed. Even stronger depolarized growth is observed with CA-RAC5, while DN-RAC5 leads to arrest of pollen tube growth (Klahre et al., 2006).
Rho-type small GTPases are regulated by a set of proteins that control the GTP hydrolysis to GDP and GDP-GTP exchange and thereby determine the signaling activity status of these switches (Etienne-Manneville and Hall, 2002). Several of these factors have been identified in plants and were shown to be important for pollen tube growth. Very recently a family of plant-specific GEFs were identified that regulate the replacement of the GDP by GTP (Berken et al., 2005) and are found in the pollen tube tip (Gu et al., 2006). GTPase activating proteins (GAPs) have been identified (Borg et al., 1999; Wu et al., 2000; Baxter-Burrell et al., 2002) and were shown to counteract Rop/Rac overexpression (Fu et al., 2001). Finally, GDP nucleotide dissociation inhibitors (GDIs) were shown to counteract Rop/Rac overexpression but are also necessary for correct GTPase localization in root hairs (Carol et al., 2005) and pollen tubes (Klahre et al., 2006).
In animal cells, GAPs have been shown to regulate neuronal morphogenesis, organ and cell size determination, mitosis, the actin cytoskeleton, endocytosis, and tumor formation (Moon and Zheng, 2003). GAPs exist for basically all GTPases. For the human genome, >160 GAPs for the Ras superfamily have been predicted, of which >60 contain a RhoGAP domain (Bernards and Settleman, 2004). Substantially fewer have been predicted for plants (Wu et al., 2000), although several more may exist (Molendjik et al., 2004). While RhoGAP proteins found in the human genome have a great variety of protein domains, including membrane binding domains (Scheffzek and Ahmadian, 2005), all published plant genes share a similar structure encompassing both a CRIB (for Cdc42/Rac-interactive binding) and a RhoGAP domain. The main difference between those plant genes lies in their N- and C-terminal extensions (Wu et al., 2000). Recently, several more RhoGAP proteins from Arabidopsis have been annotated, which have a PH domain instead of the CRIB domain.
In plants, RhoGAPs have been identified by yeast two-hybrid interaction (Borg et al., 1999) and by homology search of the Arabidopsis genome (Wu et al., 2000), and activity toward the respective Racs was demonstrated. One RhoGAP was functionally characterized in an Arabidopsis mutant and shown to be important for oxygen metabolism and H2O2 signaling (Baxter-Burrell et al., 2002). In tobacco pollen tubes, overexpression of Arabidopsis RhoGAP leads to growth arrest, similar to the overexpression of DN Rac/Rop, and counteracts GEF-induced apical swelling (Fu et al., 2001; Gu et al., 2003, 2006).
CRIB domains are thought to specifically bind activated GTPases and are found in all eukaryotes, although many more CRIB domain–containing proteins are found in humans than in lower eukaryotes, such as yeast (Pirone et al., 2001). In plants, two groups of proteins contain CRIB domains, namely, RhoGAPs and a novel class of effectors termed RICs (for Rop interactors with CRIB motif; Wu et al., 2001). For RhoGAPs, the function of the CRIB domain is not yet clear, as the RhoGAP domain itself already should bind the GTPase. Although the CRIB domain is necessary for AtRopGAP1 activity on AtRop1, it inhibits human Cdc42 GTPase activation by the same protein (Wu et al., 2000).
Here, we present the characterization of a tobacco GAP, RhoGAP1, which is localized to spatially restrict the action of small Rho-type GTPases to the tip of pollen tubes. The function of a GAP in delimiting GTPase activity has, to our knowledge, not been described before. We show that the CRIB domain is necessary to confer specificity to this protein, both for activity and localization. The overexpression of a mutant with lower GAP activity induces ballooning of the pollen tube, which may be explained by its higher affinity to RAC5. We propose that a 14-3-3 protein that is highly expressed in pollen tubes contributes to the correct, subapical localization of RhoGAP1 and thereby maintains the restricted activity of Rho GTPases to the tip and, hence, polarity of the cell. The 14-3-3 proteins are small proteins found in all eukaryotes and have been ascribed a variety of functions related to their phosphorylation-dependent interaction with other proteins in animals and plants (Aitken, 2002; Ferl, 2004).
RESULTS
Isolation of a cDNA Encoding N. tabacum RhoGAP1
In a yeast two-hybrid screen with the CA mutant of RAC5, similar to one we previously described (Klahre et al., 2006), we isolated several proteins that are putative effectors or regulators of the pollen tube–expressed GTPase. Among these, we identified a partial clone that showed strong homology to known GAPs from plants and animals. Using the clone from the yeast two-hybrid screen, we designed oligos to clone the 5′ end of RhoGAP1. Through nested amplifications we obtained an amplified clone that most likely encodes full-length RhoGAP1. Stop codons in all three reading frames upstream of the ATG, the approximate length of the mRNA determined by RNA gel blotting (see below), and a good match of the predicted protein with that from the Arabidopsis gene At5g22400 justify this statement.
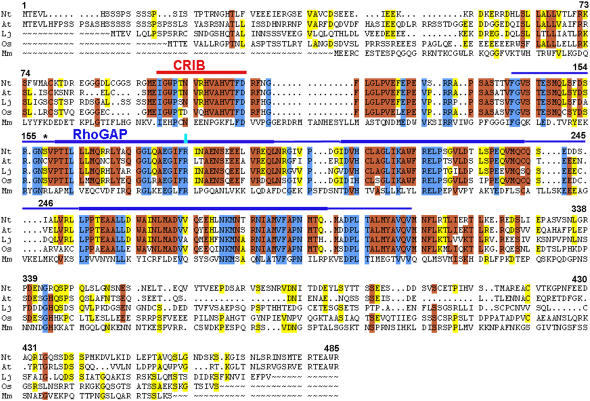
RhoGAP1 shows a high degree of sequence similarity to several sequences available in databases (Figure 1). It is closely related to proteins from Arabidopsis (At5g22400; 72% similarity, 65% identity), Lotus japonicus (AAC62624; 62% similarity, 55% identity; Borg et al., 1999), and rice (Oryza sativa; AAX95236; 63% similarity, 57% identity), which are all structurally similar. Arabidopsis At5g22400 has been previously described as At GAP1 (Wu et al., 2000). GAPs from vertebrates (mouse NP_083546; 33% similarity, 25% identity) or arthropods (Drosophila AAO39441; 33% similarity, 26% identity) show much lower similarities and are also structurally divergent. While the mouse protein has a PH domain (Pleckstrin homology; Lemmon et al., 1995), the Drosophila GAP is much larger and contains a MyTH4 domain present in myosin and kinesin tails (myosin tail homology region 4; Chen et al., 1996). No significant homology is found with GAPs that are specific to other classes of small GTPases, such as ARF or RAB, which have their own class of activating proteins (Scheffzek and Ahmadian, 2005). Although the N terminus of the mouse GAP encompasses a PH domain where plant proteins have a CRIB domain, the conservation of several amino acids in the CRIB core region between these proteins seems remarkable (Figure 1).
Figure 1.
Sequence Comparison of RhoGAPs from Plants and Mammals.
Alignment of RhoGAP1 sequences from plants and the most similar protein from animals. Sequences were aligned using the GAP program of GCG (see Methods). Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Lj, Lotus japonicus; Os, Oryza sativa; Mm, Mus musculus. Shading represents identical amino acids in three, four, or five of the sequences. The central part of the CRIB domain has a line above it, as does the RhoGAP domain. Thick lines represent the core domains (for RhoGAP box 1, 2, and 3), and thinner lines represent less well-conserved areas. Domains were marked as defined by Wu et al. (2001) and by SMART domain definition for RhoGAP domains (SM00324, http://smart.embl.heidelberg.de/index2.cgi). The conserved Arg (in RhoGAP1 R183) is highlighted in the overlined area of box 1. The mutated Ser at position 158 is indicated by an asterisk. Numbers correspond to amino acids of RhoGAP1.
Unlike vertebrate and fly RhoGAPs, RhoGAP1 contains a CRIB domain, a feature that was found in all RhoGAPs from Arabidopsis published so far (Wu et al., 2000). In RhoGAP1, the CRIB domain ends shortly before the GAP domain. Previous reports have defined CRIB domains as long as 43 amino acids or longer, based on binding studies and sequence homologies (Thompson et al., 1998). The CRIB domain in RhoGAP1, however, seems to be limited to at most 45 amino acids, as the Rho-GAP1 domain begins immediately following this region (Figure 1).
Expression Profile and Interaction with RAC5
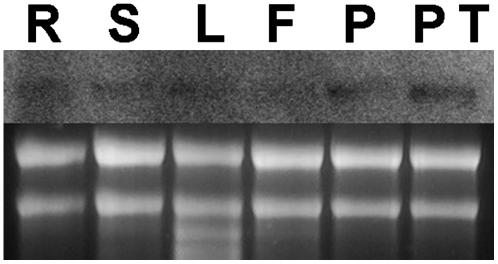
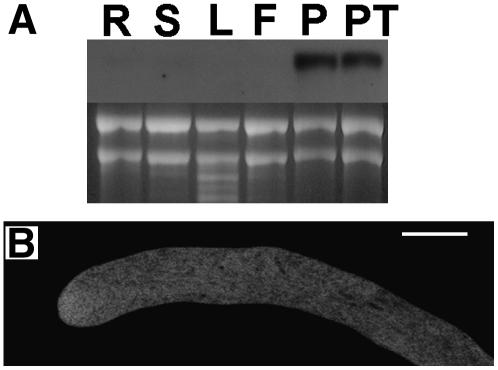
To determine the expression pattern of RhoGAP1, we analyzed RNA samples from different tissues in RNA gel blots. We found that RhoGAP1 is expressed at extremely low levels and is present in all tested tissues. Slightly higher levels were detected in pollen samples, both germinated and nongerminated (Figure 2). However, as the tobacco genome has not been entirely sequenced, we cannot exclude that under the stringent hybridization conditions used, other mRNA species may be detected (see Discussion).
Figure 2.
Expression of RhoGAP1 Is Not Tissue Specific.
The RNA gel blot was loaded with 5 μg of total RNA and probed with a radioactively labeled RhoGAP1 cDNA fragment under stringent conditions. Weak signals were consistently detected from all tissue samples. R, roots; S, stems; L, leaves; F, flowers without anthers; P, pollen grains; PT, pollen tubes germinated for 3 h.
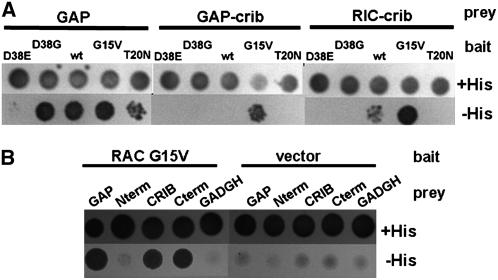
We isolated RhoGAP1 by performing a yeast two-hybrid screen using a CA form of RAC5. To determine the interaction of RhoGAP1 and other forms of RAC5, we tested several combinations of the two proteins in yeast cells. First, we determined whether RhoGAP1 interacts with all forms of RAC5. Because RhoGAP1 should inactivate GTP-bound RAC by increasing the GTPase activity, one would expect that RhoGAP1 preferentially binds to GTP-bound RAC, or in terms of mutants, to the CA mutant form. As seen in the left panel of Figure 3A, interaction of RhoGAP1 and RAC5 was strongest when RAC5 was mutated to the CA G15V mutant that characteristically has much lower GTPase activity than wild-type protein. Lower interaction was observed with the wild-type protein, while RAC5 T20N, a DN mutant, barely interacted with RhoGAP1.
Figure 3.
Yeast Two-Hybrid Interactions of RhoGAP1 and GAP Domains with RAC5.
(A) Plasmids encoding GAL4 activating domain fusion proteins (prey) with RhoGAP1 (GAP), RhoGAP1 CRIB domain (GAP-crib, amino acids 96 to 126), or Nt RIC1 CRIB domain (RIC-crib, amino acids 26 to 57) were cotransformed into yeast HF7c cells together with plasmids encoding the GAL4 DNA binding domain (bait) fused to RAC5 mutant proteins with the indicated point mutations.
(B) Yeast cells were transformed with GAL4-RAC5 G15V or a GBKT7 vector control (bait) and GAL4 activating domain fusions (prey) with the entire GAP protein (GAP), the N terminus (Nterm, amino acids 1 to 95), the CRIB domain (CRIB, amino acids 96 to 126), the C terminus (Cterm, amino acids 127 to 391), or the empty vector (GADGH).
Images were taken 3 d after replating on medium with the lack of amino acids Leu and Trp and the indicated presence or absence of His for selection.
Plant GAPs isolated so far have a CRIB domain N-terminally to the RhoGAP domain (Wu et al., 2000). As one would expect that both CRIB domains and RhoGAP domains contribute to RAC binding, we tested yeast two-hybrid interactions of truncated versions of RhoGAP1 with RAC5. Figure 3B shows that the CRIB domain and the C terminus interact with the GTPase and the N terminus not at all. This indicates that the CRIB domain and the RhoGAP domain contribute to GTPase binding (see Discussion).
We also tested whether the CRIB domain alone is conferring specificity to the GAP protein. To test this, we made yeast two-hybrid interaction assays with a prey construct containing only the CRIB domain (amino acids 96 to 126, see also Discussion for the definition of the CRIB domain). This construct was tested for interaction with RAC5 and mutant versions as bait. As a comparison, we used the full-length GAP protein (Figure 3A, left panel) and a CRIB domain from Nt RIC1 (Klahre et al., 2006). The middle panel of Figure 3A shows that the interaction with RAC5 G15V is stronger with the GAP-CRIB domain than with any other RAC5 version tested. Similarly, the CRIB domain of Nt RIC1 (Figure 3A, right panel) interacts more strongly with the G15V mutant but also interacts with the wild-type RAC5. Mutations in RAC5 at position D38 in the effector domain abolish interaction with the CRIB domain constructs, but only RAC5 D38E fails to interact with full-length RhoGAP1. None of the other mutants interacts with the CRIB domains. This indicates that the CRIB domains specifically modulate the interaction with RAC5.
RhoGAP Activity Is Enhanced by the CRIB Domain
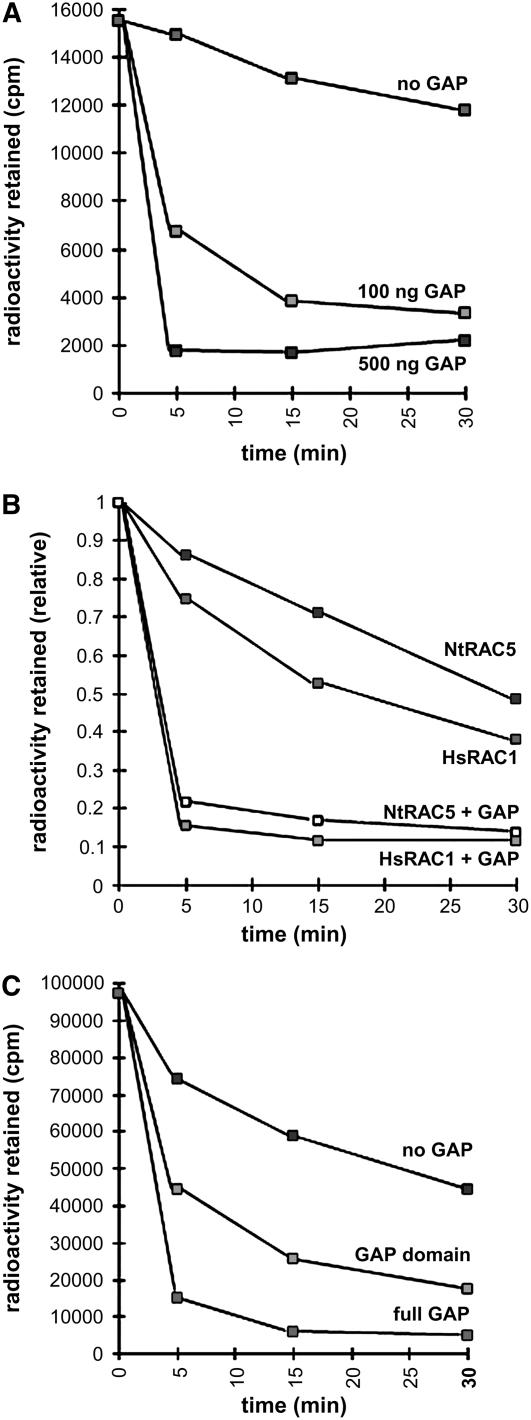
To establish whether the RhoGAP domain alone is necessary and sufficient for the GAP activity of RhoGAP1, we performed GAP assays in vitro. First, we measured the activity of RhoGAP1 on RAC5 using the whole RhoGAP1 protein. Different amounts of RhoGAP1 were added to a GTPase assay as described using [γ-32P]GTP (Self and Hall, 1995). As shown in Figure 4A, RhoGAP1 accelerates RAC5 GTPase activity dramatically. Even low amounts of RhoGAP1 strongly enhance RAC5 activity, suggesting that a rapid catalytic activation, followed by the dissociation from the GTPase, takes place. To make sure that the decrease of GTP bound to the GTPase is not caused by dissociation of the entire nucleotide, we also performed the above experiment with GTP labeled radioactively at the α-phosphate position. These experiments showed very little loss of GTPase-bound radioactivity (data not shown).
Figure 4.
GAP Accelerates GTPase Activity of RAC5.
(A) RhoGAP1 accelerates GTPase activity of RAC5 in a dose-dependent manner. Approximately 100 or 500 ng of GST:RhoGAP1 protein was added to the reactions, and the amount of radioactivity retained on filters indicating the amount of GTP-bound RAC5 measured after the indicated time.
(B) RhoGAP1 accelerates GTPase activity of RAC5 and Hs RAC1. Approximately 0.5 μg of RhoGAP1 protein was used. Otherwise, experimental conditions were similar to those described in (A).
(C) Both full-length and the C-terminal domain containing the RhoGAP domain of RhoGAP1 accelerate RAC5 GTPase activity. Approximately 0.2 μg each of GST:RhoGAP1 and GST:RhoGAP1 C-terminal protein (amino acids 142 to end) were used to activate RAC5. Measurements were made as in (A).
Assays are representative examples of experiments repeated at least three times, with similar results.
We also tested whether RhoGAP1 was active on nonplant GTPases, such as Hs RAC1. When equal amounts of protein were used, GTPase activity of both human and plant RACs was activated. Interestingly, both activities were increased by approximately the same degree (Figure 4B).
To investigate whether the CRIB domain is necessary for the GAP activity of RhoGAP1, we constructed a glutathione S-transferase (GST) fusion protein that does not contain the N-terminal domain and the CRIB domain, starting only at the beginning of the RhoGAP domain. This protein showed significant but lower activity toward RAC5 (Figure 4C), indicating that the CRIB domain may not be strictly required for activity but may rather confer isoform specificity to the protein.
We also determined whether RhoGAP1 showed activity toward the CA mutant (G15V) of RAC5. As seen for other systems, no activity was observed (data not shown), even though this RAC5 mutant interacts strongly with RhoGAP1 (Figure 3). It appears that the interaction of the two proteins is stronger in the GTP-bound conformation and may be disrupted when the RAC5-bound GTP is hydrolyzed (see also Discussion).
Phenotype of RhoGAP1 Overexpression
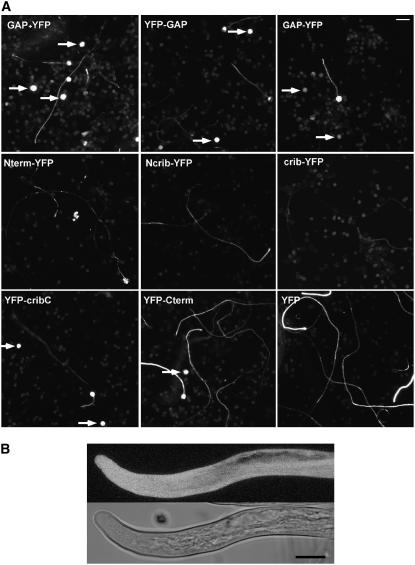
One major difference between plant and nonplant GAPs is that plant proteins contain a CRIB domain. Because this domain is thought to bind to active forms of GTPases, one would expect that in plant GAPs this domain is required for normal activity. As we have shown above, however, the CRIB domain of RhoGAP1 is not strictly required for the activation of GTPase activity in vitro (Figure 4C). To establish whether the CRIB domain has more of an impact on RhoGAP1 function in vivo, we tested the effect of RhoGAP1 and truncated versions fused to yellow fluorescent protein (YFP) when overexpressed in pollen tubes (Figure 5A). When full-length RhoGAP1 was expressed transiently under the strong promoter of LAT52 (see Methods), a large proportion of the pollen failed to even germinate. The germinating pollen tubes were usually short and had a characteristically narrow tip (Figure 5B). This phenotype was expected, as the overexpression of a GAP leads to hydrolysis of the GTP bound to RAC and thereby the inactivation of the signal transduction cascade initiated by RAC5 (see Discussion).
Figure 5.
Overexpression of GAP Leads to Arrest of Germination and Growth.
(A) Overview picture of pollen tubes bombarded with LAT52-driven RhoGAP1 cDNA, together with YFP to visualize transfected pollen and pollen tubes, and YFP fusions of full-length RhoGAP1 proteins (top row). Bottom panels show images of pollen tubes transformed with YFP fusions of RhoGAP1 truncations: Nterm, N terminus of RhoGAP1 (amino acids 1 to 95); Ncrib, N terminus and CRIB domain (amino acids 1 to 143); crib, CRIB domain (amino acids 96 to 126); cribC, CRIB domain and C terminus (amino acids 96 to 391); Cterm, C terminus (amino acids 127 to 391). All truncated proteins were expressed with YFP attached to the N or C terminus, as indicated in the figure. Pictures were taken 20 h after bombardment. Arrows indicate nongerminating pollen expressing the respective YFP fusion protein. Background fluorescence is derived from nontransfected pollen grains. Bar = 100 μm.
(B) High magnification of a pollen tube overexpressing RhoGAP1 and YFP to visualize the tube, both under the LAT52 promoter. The top panel shows a projection of a confocal stack, and the bottom panel shows a differential interference contrast image of the same tube. Bar = 10 μm.
To determine what the single domains of RhoGAP1 contribute to the growth-retarding phenotype of RhoGAP1, we constructed YFP fusions of the N terminus alone, the N terminus together with the CRIB domain, the CRIB domain alone, the CRIB domain and the C terminus, and the C terminus alone. As these constructs express only parts of RhoGAP1, we reasoned that adding the YFP to the truncated end would make the proteins more stable and thereby more comparable and would allow monitoring of expression levels.
Overexpression of the N terminus lead to only a small effect on pollen tube length and germination, although tubes often aborted after 20 h and spilled their cytoplasm (Figure 5A). When the CRIB domain was added to the overexpressed protein, no strong phenotype was observed either, but fewer aborted tubes were found. Similarly, the overexpression of the CRIB domain alone was not accompanied by a strong reduction in tube length, and expression levels seemed somewhat lower, possibly due to a lower stability of the protein. However, a substantial reduction in tube length and several ungerminated pollen were observed when the C-terminal portion of RhoGAP1, which contains the RhoGAP domain, was overexpressed. Interestingly, the combination of the CRIB domain with the C terminus showed a phenotype similar to the overexpression of the full-length protein, demonstrating that in vivo the CRIB domain appears to be necessary for full GAP activity, probably through its binding capacity to active GTPase.
Subcellular Localization of RhoGAP1
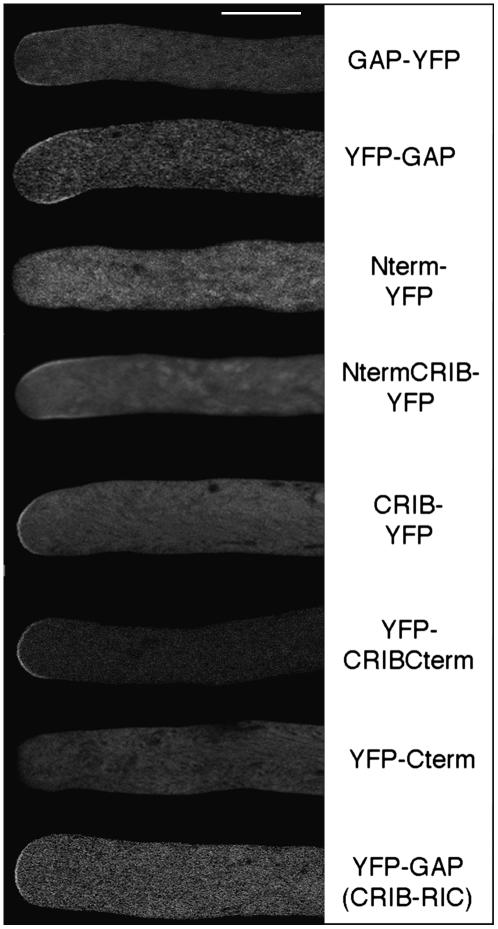
Next, we wanted to correlate the activity of RhoGAP1 with its localization within the pollen tube. Therefore, we expressed proteins tagged either at the N or C terminus with YFP in pollen tubes. We always measure the growth rate of pollen tubes we use for YFP fusion protein localization to ensure that there is no mislocalization of the fusion protein due to a phenotypic change caused by the overexpression. All pollen tubes shown in images of confocal sections had growth rates of at least 3 μm per minute and are therefore comparable to wild-type tubes, which grow at a rate of ∼5 μm per minute in our in vitro system (see also Parton et al., 2003). Great care was also taken to ensure a medial section through tubes, discarding images with slanted section planes or tubes obstructed by other tubes, which might equivocate the localization of YFP fusion proteins.
Interestingly, both fusion proteins with full-length RhoGAP1 were found at the plasma membrane close to the apex of pollen tubes but slightly removed from the very tip of the tube, leading to what could be described as subapical labeling. The attachment of the reporter protein at the N or C terminus did not significantly change this labeling, although the YFP-RhoGAP1 protein was consistently more removed from the very tip and distributed more to the flanks than the RhoGAP1-YFP fusion (Figure 6, top two panels). Both images had to be taken of tubes expressing low levels of YFP fusion protein, as higher levels are toxic to pollen tubes and result in membrane labeling at the very tip.
Figure 6.
GAP Is Localized at the Flanks of Pollen Tubes, and Localization Is Dependent on Both the N Terminus and the CRIB Domain.
Pollen tubes were transiently transformed by bombardment with plasmids expressing the indicated YFP fusion proteins and imaged 5 to 7 h later using a confocal laser scanning microscope. Great care was taken to make medial sections through pollen tubes that were growing normally. Constructs expressing partial proteins of RhoGAP1 were as described in the legend to Figure 5. The construct in which the CRIB domain was exchanged with that of Nt RIC1 had amino acids 96 to 126 replaced with the corresponding stretch of Nt RIC1 (YFP-GAP [CRIB-RIC]).
As RhoGAP1 itself has no obvious membrane localization domain, we wanted to find out which domains are responsible and necessary for the correct localization. YFP fusion proteins containing the CRIB domain were found to be membrane localized, while fusions to both the N and C terminus alone were not found at the membranes, but rather diffusely in the cytoplasm (Figure 6). This shows that the CRIB domain is necessary and sufficient for membrane localization. However, the CRIB domain alone showed labeling at the very apex of the pollen tube, consistent with the hypothesis that CRIB domains bind mainly to GTP-bound, active GTPase (Hwang et al., 2005). Only when the N terminus was included in the fusion protein was subapical labeling observed. The combination of the CRIB domain with the C-terminal part showed also strong membrane labeling, more extended than that found with the CRIB domain alone, but in contrast with RhoGAP1 localization, this protein was also found at the very apex. This set of experiments establishes that for membrane association, the CRIB domain is essential, while for subapical labeling, both the N terminus and the CRIB domain are required. On the other hand, the C terminus containing the RhoGAP domain has only a minor effect on localization.
To test whether the CRIB domain of RhoGAP1 exemplifies a module necessary for GTPase-dependent membrane labeling that could be replaced by any other CRIB domain, we replaced the RhoGAP1 CRIB domain with the CRIB domain of Nt RIC1. Surprisingly, the chimeric protein localized to the very tip of pollen tubes, more or less like the RhoGAP1 CRIB domain alone, albeit with substantially lower intensity (Figure 6). Together with the somewhat stronger binding of the CRIB domain of Nt RIC1 to RAC5 (Figure 3), this indicates that a precise affinity of the two proteins is crucial for the localization of RhoGAP1.
RhoGAP1 Mutant with Reduced GAP Activity
To check what effect the reduction of GAP activity would have on RhoGAP1 localization and on the RhoGAP1 overexpression phenotype, we constructed a R183A mutant in which the Arg found to be essential for GAP activity in mammalian proteins containing a RhoGAP domain (Leonard et al., 1998; Graham et al., 1999,) was changed to an Ala. To verify the lack of activity of this mutant, we compared equivalent amounts of the wild-type and mutant protein in GAP assays and found that the mutant has an ∼5- to 10-fold lower activity (data not shown). The mutated bacterial protein was also somewhat more stable than the highly unstable wild-type protein (data not shown).
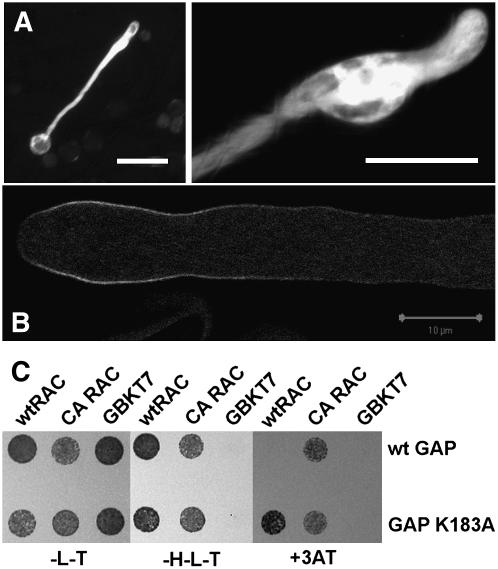
Pollen tubes overexpressing this RhoGAP1 R183A construct showed a very different phenotype from those overexpressing wild-type RhoGAP1 (Figure 7A). Pollen tubes were short as well, but instead of showing a narrow tip, they were bulged, reminiscent of RAC5 overexpression (Klahre et al., 2006). Also, the failure of pollen to germinate was reduced. Probably overexpression of a GAP with reduced activity leads to a DN effect on resident, endogenous RhoGAP1 (see below and Discussion).
Figure 7.
Overexpression and Localization of the R183A Mutant of RhoGAP1.
(A) Overexpression of YFP-R183A leads to bulged pollen tubes reminiscent of RAC5 overexpression. Tobacco pollen tubes were transiently transfected with a plasmid encoding RhoGAP1 R183A and a plasmid expressing YFP. Bars = 100 μm.
(B) Confocal localization of transient expression of YFP-RhoGAP1 R183A 6 h after bombardment. YFP localization was found at the subapex of swollen, but growing, pollen tubes but extended to the flanks compared with wild-type YFP-RhoGAP1. Bar = 10 μm.
(C) Yeast two-hybrid interaction of the wild type and R183A mutant of RhoGAP1 with the wild type and CA-G15V mutant of RAC5. To roughly estimate the interaction strength, yeast was plated on permissive (-T-L) and selective medium (-H-L-T) containing 1 mM inhibitor 3-amino-1,2,5-triazole.
To examine the effect of the reduced RhoGAP1 catalytic activity on the localization of the protein, we compared confocal images of wild-type and R183A mutant proteins fused to YFP. Both proteins localized to the subapical area of pollen tubes, but the mutant protein showed a clearly extended labeling along the flanks (cf. Figures 6, top panel, and 7B), indicating that a slightly abnormal localization is caused by the mutation.
We also measured the interaction of RhoGAP1 R183A with wild-type and CA-G15V mutant RAC5 in yeast two-hybrid assays. As shown in Figure 7C, the RhoGAP1 R183A mutant interacts slightly more strongly with RAC5 than the wild-type protein does. This may partially be due to the lower GAP activity of the protein leading to slower release of the two proteins (see Discussion).
Involvement of a 14-3-3 Protein in RhoGAP1 Activity and Localization
As we have shown above, the N-terminal domain is necessary for the correct localization of RhoGAP1, while it is itself not localized at the membrane. As membrane labeling is conferred by the CRIB domain (Figure 6), there are several possibilities how the N terminus may be responsible for subapical labeling. First, it may bind to membrane components (lipids or proteins) that are localized to this area of the membrane. Second, it may be necessary for recycling of RhoGAP1 to its specific localization, as has been observed, for example, in auxin transporters (Grebe et al., 2002). Third, the protein may be removed from areas at the apex and flanks by proteins that associate with it in a pH-, ion-, or lipid-dependent fashion.
All these possibilities probably include interaction with proteins or membrane components through the N terminus of RhoGAP1. We therefore performed a yeast two-hybrid screen with the N terminus of RhoGAP1 as bait. Among several interacting proteins was a 14-3-3 protein, belonging to a family of proteins for which several functions have been proposed so far (Sorrell et al., 2003; Ferl, 2004; Paul et al., 2005). The N-terminally truncated protein (missing 43 amino acids) we isolated was identical to the previously deposited sequence for N. tabacum 14-3-3 b-1 (GenBank accession number BAD12169). This protein interacted well with the N terminus and even more strongly with the full-length RhoGAP1 protein in yeast two-hybrid interaction tests (data not shown). RNA gel blots probed with the 14-3-3 cDNA showed high expression of the protein in pollen and pollen tubes (Figure 8A). Full-length 14-3-3 b-1 fusion proteins with GAL4 domains were less effective in binding and showed higher background in yeast two-hybrid assays, probably due to the sensitive N-terminal area responsible for protein dimerization (Aitken, 2002). Overexpression of YFP-14-3-3 in pollen tubes showed cytoplasmic localization (Figure 8B). If any membrane association occurred through association with RhoGAP1, it was too weak to detect.
Figure 8.
14-3-3 b-1 Is Expressed in the Cytoplasm of Pollen Tubes.
(A) RNA gel blot probed with 14-3-3 b-1. RNA (5 μg) was loaded per lane and probed with a DIG-labeled probe encompassing the 14-3-3 b-1 coding region. R, roots; S, stems; L, leaves; F, flowers without anthers; P, pollen grains; PT, pollen tubes germinated for 3 h.
(B) Localization of YFP-14-3-3 b-1 in pollen tubes. Pollen tubes were bombarded with plasmids encoding YFP-14-3-3 b-1 and photographed 6 h after bombardment using a confocal microscope. Bar = 10 μm.
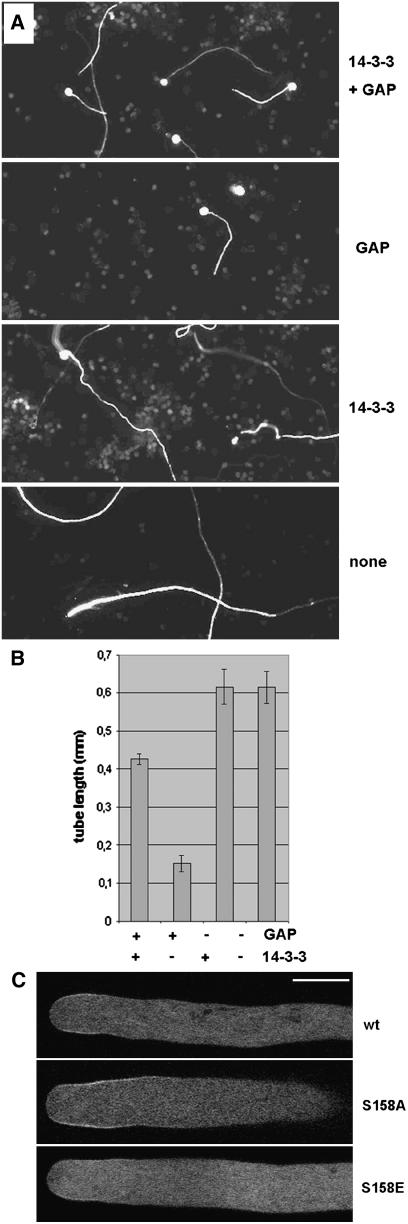
If 14-3-3 b-1 is indeed involved in localizing the RhoGAP1 protein, we would expect that coexpression of this protein together with RhoGAP1 would have an effect on the localization and the overexpression phenotype of RhoGAP1. First we tested what would happen when we coexpressed 14-3-3 b-1 together with RhoGAP1 and compared it to overexpression of either protein alone. Figure 9A shows that while pollen tubes expressing only RhoGAP1 are substantially shorter than wild-type tubes after 24 h, overexpression of 14-3-3 b-1 shows basically no effect on tube length but only leads to the formation of somewhat less straight tubes. However, when both proteins were expressed together, pollen tubes showed a strong suppression of the RhoGAP1 phenotype (Figure 9A). When tubes were measured after 8 h, they were substantially longer than tubes expressing RhoGAP1 alone. They were much longer than RhoGAP1 tubes expressing a correspondingly high amount of RhoGAP1 only (Figure 9B) and as long as wild-type tubes when smaller amounts of plasmid were bombarded (data not shown). No complementation effect was seen when RhoGAP1 was expressed together with β-glucuronidase (GUS) protein under the control of the same promoter (see Methods). These results suggest that 14-3-3 b-1 plays a role in regulating RhoGAP1 localization or activity.
Figure 9.
14-3-3 b-1 Alters the GAP Overexpression Phenotype.
(A) Phenotype of pollen tubes expressing 14-3-3 b-1, RhoGAP1, or a combination of both. Pollen grains were bombarded with equal amounts of plasmids expressing 14-3-3 b-1 or RhoGAP1, and in the cases where only one plasmid was bombarded, the total amount of plasmid was equalized with a plasmid expressing GUS under the same promoter.
(B) Quantification of pollen tubes expressing 14-3-3 b-1, RhoGAP1, or a combination of both. Four micrograms of plasmid DNA was bombarded, and tubes were measured 6 h after bombardment. Measurements are means of at least 30 tubes. Error bars represent sd.
(C) Membrane localization of pollen tubes expressing YFP fusions of RhoGAP1 and mutant GAPs as described. Pollen tubes were imaged ∼6 h after bombardment. All pollen tubes grew at a minimal rate of 3 μm/min.
To find out whether RhoGAP1 localization is affected by coexpression of 14-3-3 b-1, we expressed YFP-RhoGAP1 alone or together with 14-3-3 b-1. We consistently saw weaker subapical labeling of RhoGAP1 when 14-3-3 b-1 was coexpressed (Figure 9C), while some tubes showed absolutely no membrane labeling (data not shown). To further investigate the specificity of the interaction between RhoGAP1 and 14-3-3 b-1, we mutated the conserved motif found in RhoGAP1 (amino acids 155 to 160, RGNSVP, consensus R/KxxpS/TxP) that is a putative phosphorylation-sensitive binding site for 14-3-3 proteins (Ferl, 2004). Mutations S158A and S158E may mimic the nonphosphorylated and phosphorylated protein, thus acting as a weaker and stronger substrate, respectively, for 14-3-3 b-1 than wild-type RhoGAP1. We do not expect that these mutations would completely abolish binding, as the N terminus of RhoGAP1 clearly contributes to the interaction. To test for in vivo effects of the mutations, we examined these proteins for membrane localization in pollen tubes and found that while RhoGAP1 S158A seemed to be localized more strongly to the membrane than the wild type, RhoGAP1 S158E was seen substantially less at the membrane (Figure 9C). This may mean that the interaction between the two proteins indeed depends on the phosphorylation status of RhoGAP1 and that the interaction between the proteins is specific. Unfortunately, yeast two-hybrid interaction assays and attempts to demonstrate in vitro interaction differences could not confirm these data (see Discussion).
Apart from regulating RhoGAP1 localization, 14-3-3 b-1 might also influence RhoGAP1 activity, as was seen in animal cells, where a GAP for a heterotrimeric G-protein reduced GTPase activity (Benzing et al., 2000). To test this, we made in vitro GAP activity assays on RAC5 in the presence and absence of 14-3-3 b-1. However, even at high concentrations of 14-3-3 b-1, we could not detect any effect on GTPase activity (data not shown).
DISCUSSION
We have presented here the isolation of a novel tobacco GAP and its characterization. We detected a subapical membrane localization of a GAP and its thereby implied function as a spatially limiting factor for GTPase signaling. To our knowledge, a spatial control of GTPase signaling by a GAP has not been described before. Furthermore, we propose the role of a 14-3-3 protein in the control of this localization.
RhoGAP1 Sequence and Expression Compared with Other Known RhoGAPs
Considering the important role of Rho-type small GTPases in pollen tube growth (Kost et al., 1999; Li et al., 1999; Zheng and Yang, 2000; Cheung et al., 2003), the presence of a RhoGAP in pollen is not surprising. It is unexpected, though, to find this GAP expressed at such low levels in pollen tubes and other tissues. Naturally, extrapolating from the situation in Arabidopsis and other organisms, many more RhoGAPs are expected to be expressed in tobacco. As the genome of tobacco has not been sequenced, it remains to be seen whether RhoGAP1 is the major RhoGAP in pollen tubes, but we have not identified any other RhoGAP in our yeast two-hybrid screens. More than one RhoGAP may be important for pollen tube growth, as T-DNA insertion lines of the most similar GAP in Arabidopsis (At5g22400) showed no altered transmission through the pollen (our unpublished data). The low expression levels may be explained by the fact that RhoGAP1 seems to be highly processive (i.e., little GAP is needed due to a rapid turnover on the GTPase) (Figure 4).
RhoGAP1 expression was found to occur in all tissues tested. Considering that Arabidopsis contains six genes with significant homology to RhoGAPs, it is surprising that RhoGAP1 is expressed more or less ubiquitously, with slightly higher expression levels in pollen (Figure 2). Unfortunately, little is known about tissue-specific expression of RhoGAPs in plants. Chip data (www.genevestigator.ethz.ch) show background level expression for the Arabidopsis genes related to RhoGAP1, with no elevated levels for pollen.
RhoGAP1 Interaction with RAC5, and GAP activity
Yeast two-hybrid interactions with Rho-type small GTPases are often found to be weak, as is the case for RhoGAP1 (see above). This is not surprising considering that RAC5 is usually prenylated and membrane associated. For the yeast system used in this work, the prenylation site was mutated to prevent membrane association to facilitate transport to the nucleus. Furthermore, as the RhoGAP1 interaction with RAC5 may be transient, as expected for a catalytically enhancing protein, the interaction in yeast may seem less strong but can be stabilized by locking the GTPase in the GTP-bound form (Figure 3).
We found that RhoGAP1 interaction with the CA RAC5 G15V mutant in the yeast system is much stronger than seen with the wild type. This is in contrast with results obtained by Wu et al. (2000), who showed better interaction of RopGAP1 with wild-type Rop1 and that Arabidopsis GAP3 interacts with the transitional state of Arabidopsis ROP1 using aluminum tetrafluoride. We can only explain this difference in results by the difference in GTPases and GAPs used in these experiments.
Unfortunately, in vitro interaction studies between RhoGAP1 and RAC5 are difficult because RhoGAP1 is very unstable when produced in bacteria as a full-length protein, no matter what fusion protein was used for purification. Clearly, more efforts will be needed to produce both prenylated GTPases and stable GAP protein for interaction studies.
We have shown GAP activity of RhoGAP1 toward both plant and mammalian RAC proteins (Figure 4B). This is in contrast with experiments in which Arabidopsis GAPs were tested on mammalian Cdc42, where GAP activity was only observed when the CRIB domain was removed (Wu et al., 2000). It is conceivable that plant RhoGAPs show different affinities and activities toward different kinds of mammalian Rho proteins. The CRIB domain may contribute to the affinity and have DN effects due to overly strong interactions between the two proteins.
We have also tested whether RhoGAP1 changes the localization of YFP-RAC5 in vivo. Although the interpretation of such experiments is somewhat complicated by the toxicity of both RhoGAP1 and RAC5, we could not detect any changes in RAC5 localization (data not shown), which is compatible with our model (see below).
Importance of the CRIB Domain
The exact definition of what constitutes a CRIB domain seems to vary between proteins (Burbelo et al., 1995). In RhoGAP1, the CRIB domain necessarily ends shortly before the GAP domain and must therefore be limited in its size, unlike in other proteins where C-terminal extensions have been demonstrated to contribute to binding (Burbelo et al., 1995). However, it may be difficult to delimit the CRIB domain and RhoGAP domain, as mouse RhoGAP1 has extended homologies N-terminally of the RhoGAP domain with the plant proteins, which contain a CRIB domain in this area (Figure 1). On the other hand, the CRIB domain and the RhoGAP domain may be acting together to ensure GTPase binding.
The function of CRIB domains in plant RhoGAPs is not entirely clear. They appear to bind active GTPase and thus help in assuring GAP activity (Figure 4), but they may also be inhibitory, as shown in heterologous assays and in animal and yeast systems (Wu et al., 2000; Pirone et al., 2001). This may be due to the fact that GAPs are catalytically active, and a rapid dissociation of the protein from the cognate GTPase determines the rate with which it can turn over to the next molecule. On the other hand, CRIB domains may represent a module to confer specificity to the GTPase interactor by modulating its affinity because only proteins that bind with the appropriate avidity will balance turnover and binding adequately. However, if binding is too strong, turnover may be hampered by slowed-down release and lead to reduced GAP activity. In this way, one protein may be inhibitory for one GTPase, while promotive for another, with subtle differences.
Our results also indicate that CRIB domains of proteins cannot be exchanged without losing either specificity or the correct binding affinity. This also means that the use of homologous systems, in which proteins from one species are used only in cell types where they are actually expressed, may be advisable.
GAP Localization and Membrane Association
RhoGAP1 localization is striking in that it is found at membranes not at the very apex, where Rho-type GTPases are thought to signal, but subapically, where it seems to restrict GTPase action. For this localization, the N terminus and the CRIB domain are essential, both of which are not strictly required for GAP activity. As RhoGAP1 contains no obvious sequence motif that would imply association with membranes, its localization must be through other proteins, of which prenylated GTPases are obvious candidates.
RhoGAP1 localization in many ways overlaps with the transition of cell wall components at the very apex of the pollen tube and those found in the flanks of the tubes. Pectin components of the cell wall, which are unusually abundant in pollen tubes, abruptly change from high concentrations of esterified pectins at the very apex to unesterified pectins at the flanks (Bosch et al., 2005). Lipid components of the membrane were also found to change in this area of the pollen tube apex (Kost et al., 1999). As RhoGAP1 is membrane localized near the area of highly active membrane deposition, it is also possible that RhoGAP1 directly or via lipid-sensitive proteins is restricted to areas of a particular lipid arrangement. The 14-3-3 protein implicated in RhoGAP1 localization presented here may be important for the association and dissociation of RhoGAP1 with such lipid-sensitive proteins. Furthermore, an actin fringe has been reported to localize immediately adjacent to the membrane in which RhoGAP1 is found (Lovy-Wheeler et al., 2005). An interaction with this scaffold may be direct via proteins interacting both with microfilaments and membranes but also indirect through local ion or pH gradients. This means that cell wall, membrane, cytoskeletal, and cytoplasmic components may interact to maintain a transition between growth zone and shank of the tube.
Implications of a Mutant with Reduced GAP Activity
Overexpression of the R183A mutant of RhoGAP1 shows strong ballooning of pollen tubes, which recalls the effect of RAC5 overexpression in tobacco pollen tubes (Klahre et al., 2006), although the balloons never quite acquired the round shape with a large central vacuole observed in the latter case. The phenotype probably is due to a DN effect of the interaction of the mutant with either the GTPase directly or with proteins that affect GTPase signaling activity. Given the fact that endogenous active RhoGAP1 is still present, the effect is probably due to the direct interaction with RAC5. The possibility of an interaction with endogenous RhoGAP1 in a complex formed by heterodimerization is unlikely, as we are not aware of any report describing such an association for similar proteins. However, in yeast two-hybrid assays, RhoGAP1 appears to weakly interact with itself (our unpublished data).
The DN effect of the R183A mutant of RhoGAP1 may be partly due to the fact that this protein cannot release RAC5 as quickly as the wild-type protein because it does not have a high GTPase-activating activity. It would be predicted that this would lead to an interruption of the quick recycling of RAC5, concomitant with the GTPase in a prolonged GTP bound, possibly signaling-active form. This may in turn lead to an enlargement of the zone in which active RAC5 is found, leading to depolarized growth.
Our data also suggest that the mutant interacts more strongly with RAC5, leading to a delayed dissociation of the two proteins. This explanation is, of course, not easy to distinguish from the latter explanation, as one leads to the other. However, if binding is stronger, blocking GTPase release, we would expect more active RAC5 at the flanks of pollen tubes. If RhoGAP1-RAC5 complexes are signaling-active, this would increase the growth area even further.
Interaction with a 14-3-3 Protein and Maintenance of the Subapical Localization
The interaction of RhoGAP1 with a 14-3-3 protein appears to fit quite well with the purported role of this class of proteins, namely, that they interact transiently with proteins that are relocalized between cellular compartments (Aitken, 2002). They have been implicated in cell cycle functions, signal transduction, cytoskeletal regulation, transcription, and the regulation of subcellular localization. In plants, their interactions are regulated through a C-terminal EF hand by charged molecules (Camoni et al., 2001; Athwal and Huber, 2002).
In animal cells, an interaction between a GAP and a 14-3-3 protein has also been observed (Benzing et al., 2000). The authors suggest that a portion of the available pool of this GAP for a heterotrimeric G-protein is kept in a dormant state by the interaction with the 14-3-3 protein, and they showed that GAP activity is reduced by this interaction. In our assays, GAP activity was not reduced by Nt 14-3-3 b-1 in vitro. This may mean that in pollen tubes, the effect of interaction may differ from the animal system in that the GAP protein is relocalized rather than sequestered by the 14-3-3 protein. However, if Nt 14-3-3 b-1 removes RhoGAP1 from the membrane for relocalization, this may similarly remove a portion of the protein from its site of action at the plasma membrane. Thereby 14-3-3 b-1 may counteract the overexpression phenotype of RhoGAP1 (Figure 9).
Why the overexpression of 14-3-3 b-1 does not block RhoGAP1 action and leads to an only weak phenotype in pollen tubes is not entirely clear. It appears that even in wild-type tubes rather large amounts of 14-3-3 b-1 are expressed compared with RhoGAP1 (cf. Figures 2 and 8B). This may mean that the interaction between the two proteins is highly regulated, as implicated by the phosphorylation-dependent and calcium-sensitive binding of 14-3-3 proteins. Thus, apart from large amounts of 14-3-3 protein, signals derived from decreased GTPase signaling may be necessary for 14-3-3 activity. It appears particularly appealing to argue that RAC-associated kinases phosphorylate Nt GAP1 and thereby trigger its removal from the membrane.
In reports on Arabidopsis 14-3-3 proteins similar to Nt 14-3-3 b-1, some nuclear localization was observed (Paul et al., 2005). In pollen tubes, we could not detect any YFP fusion protein in the nucleus, which is in agreement with a function of Nt 14-3-3 b-1 in the cytoplasmic compartment. In Arabidopsis, 14-3-3 mutants generally lack a phenotype (Ferl, 2004), which may be explained by functional redundancy, even though interactions are generally considered to be specific (Aitken, 2002). This may also make it difficult to test whether 14-3-3 function is absolutely required for RhoGAP1 localization using Arabidopsis mutants. Furthermore, it complicates RNA knockdown approaches because targeting single proteins in tobacco is difficult without the necessary genomic information, and timing of the knockdown may be crucial and difficult in haploid pollen (Lee et al., 1996).
The 14-3-3 proteins are regulated by divalent charged polyamines and cations (Athwal and Huber, 2002). Furthermore, they regulate several ion channels (Bunney et al., 2002) and modulate protein kinase C–related pathways. The presence of a calcium gradient at the tip of pollen tubes makes it possible that the interaction between 14-3-3 proteins and their targets is modulated by pH or ion differences (see below).
In yeast two-hybrid interaction experiments, we have not found any difference between the two S158 mutants of RhoGAP1 (data not shown), and in vitro interaction studies were hampered by low stability of full-length RhoGAP1 protein. It is also possible that 14-3-3 relocalization of RhoGAP1 is not dependent only on the interaction between the two proteins but rather a protein complex. This would explain the modulation of S158 in protein interaction seen in vivo (Figure 9C).
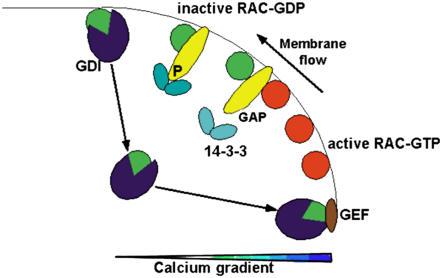
A Model for Spatial Confinement of GTPase Activity at the Pollen Tube Tip
Combining the above results and considerations into a model, we propose the following scenario depicted in Figure 10. Once the polarity of the pollen tube is established in the pollen grain, small GTPases are localized to the tip of pollen tubes. Due to the rapid integration of new membrane and cell wall material at the tip, small GTPases are swept toward the side, where they are inactivated by RhoGAP1. Inactive GTPase is then recycled from the membrane by a pump-like mechanism involving RhoGDI2 as previously described (Klahre et al., 2006), followed by reactivation of the GTPase by tip-localized GEFs (Gu et al., 2006). This three-step mechanism assures the maintenance of active GTPase at the tip of pollen tubes.
Figure 10.
Model for the Recycling of GTPase to the Pollen Tube Tip through GAPs, GDIs, and GEFs.
This model postulates that tip-localized GTPase flows with the newly inserted membrane toward the flanks of the pollen tube, where it gets inactivated by RhoGAP1. Localization of RhoGAP1 is dependent on helper proteins, such as 14-3-3, which are sensitive to lipid composition, pH, or ion gradients. At the flank of the tube, inactive, GDP-bound GTPase is then detached from the membrane by a GDI, which then transports it back to the apex, where it gets reactivated by a GEF. Interaction between components of the cell wall, the actin cytoskeleton, the membrane, and the cytoplasm may synergistically maintain this cycle on several planes of action.
Correct RhoGAP1 localization itself might be ascertained by the calcium and phosphorylation-dependent interaction of RhoGAP1 with 14-3-3 b-1. Upon transport of RhoGAP1 by the membrane flow toward the flanks, regulated interaction with 14-3-3 b-1 may lead to RhoGAP1 relocalization to the cytoplasm, release by calcium-regulated 14-3-3 interaction, and binding to a new, active GTPase molecule nearer to the tip.
Exactly how relocalization occurs will be the topic of further investigations. Certainly, other components may be contributing to the relocalization of RhoGAP1. The curvature of the membrane could be sensed directly by RhoGAP1 or by adapter proteins, as has been described for an animal GAP (Bigey et al., 2003). A direct involvement of lipids on association and activities of GAPs, as seen in animals systems (Ligeti et al., 2004), will also have to be tested for plant proteins.
METHODS
Plant Maintenance
Tobacco plants (Nicotiana tabacum Petite Havana SR1) were grown in a greenhouse under standard conditions and pollen collected from fully grown plants.
Pollen Transient Expression and Growth
Pollen were harvested from mature anthers and grown on medium at room temperature as described (Kost et al., 1998). For RNA isolation from pollen or from pollen tubes harvested 3 h after imbibition, material was ground in liquid nitrogen and RNA isolated as described below. For transient expression of proteins in pollen tubes, pollen grains were bombarded with gold particles as described (Kost et al., 1998) except that water was used to suspend pollen grains. Two micrograms of plasmid DNA was bombarded per sample unless described otherwise. To ascertain equal amounts of plasmid bombarded and comparable amounts of protein expressed in transfections with combinations of plasmids, total amounts of plasmid were kept constant by the addition of LAT52:GUS encoding plasmids.
RNA Isolation, Gel Blotting, and Hybridization
RNA was isolated from separated plant tissues ground in liquid nitrogen using Trizol (Invitrogen) according to the manufacturer's recommendation. RNA gel blots, DIG-labeled probes for hybridization, and blot washing were prepared as described (Klahre et al., 2006). Radioactive probes were prepared using a Megaprime labeling kit (GE Healthcare-Amersham) following the manufacturer's recommendations.
Yeast Two-Hybrid Screens and Assays
Yeast two-hybrid screens and interaction analysis were performed using the Matchmaker GAL4 system (Clontech; manual PT3061-1) as described by the supplier. Saccharomyces cerevisiae HF7c cells were transformed using a lithium/polyethylene glycol/dimethylsulfoxide-mediated transformation as described in the manual. Chemicals for yeast media were bought from MP Biomedicals/Q Biogen (Eschwege) and agar and amino acids from Serva.
To verify two-hybrid interactions ∼2 μg of each plasmid were cotransformed into yeast and plated on agar plates with or without His and containing the indicated concentrations of 3-amino-1,2,5-triazole (Sigma-Aldrich). For photography, several single colonies were suspended individually in SD medium without His, Leu, and Trp and replated as indicated. Photographs were taken 3 to 6 d after replating.
DNA Techniques and Plasmid Construction
DNA cloning, analysis, and electrophoresis were performed as described (Sambrook et al., 1989). Enzymes for recombinant technologies were purchased from NEB. Large preparations of plasmid DNA for yeast transformation and plant cell bombardment were prepared using JetStar 2.0 maxipreps from Genomed.
For yeast two-hybrid analysis, protein encoding fragments were cloned into a derivative of pGAD-GH and pGBK-T7 (Clontech) with a modified polylinker. For YFP fusion protein expression, fragments were cloned into a pUC derivative containing a LAT52 promoter and NOS polyadenylation signal as described (Kost et al., 1998; Klahre et al., 2006). GST fusion proteins were prepared using pGEX-4T (GE Healthcare/Amersham/Pharmacia).
Truncations of RhoGAP1 were produced using PCR and DNA oligomers to produce fusion proteins containing the following parts of RhoGAP1: the N terminus (amino acids 1 to 95), the CRIB domain (amino acids 96 to 126), the C terminus (amino acids 127 to 391), or combinations thereof.
Point mutations were introduced into existing cDNA fragments by PCR amplifications using DNA oligomers containing the indicated mutations and subcloned in the indicated vectors. The DNA sequence of all PCR-amplified parts and the introduction of the point mutations were verified by DNA sequencing (SeqLab).
Recombinant Protein Purification
GST fusion proteins were isolated from 500-mL cultures of BL21 Escherichia coli cells containing the appropriate plasmid grown at 30°C to an OD600 of 0.8 to 1, induced with 0.2 mM isopropylthio-β-galactoside (Roth), and grown for another 2 h. Cells were harvested by centrifugation, chilled, homogenized in a French press, and the debris sedimented at 6000g. GST fusion proteins were purified by incubation with glutathione-sepharose (Amersham Biosciences) for 1 h and harvested by centrifugation at 300g for 1 min. Sepharose beads were resuspended in storage buffer (50 mM Tris, 5 mM MgCl2, 250 mM NaCl, and 50 mM glycerol, pH 7.5), snap frozen in liquid nitrogen, and stored in aliquots at −80°C. Proteins were eluted from beads using 10 mM reduced glutathione (Sigma-Aldrich) and 50 mM Tris, pH 8, and immediately used.
GAP Assays
GAP activity assays were performed as described (Self and Hall, 1995). Five to ten micrograms of GTPase were preincubated in 50 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 10 mM EDTA, 5 mM DTT, 0.3 μM GTP, and 10 nM [γ-32P]GTP (Hartmann Analytic) in a total volume of 300 μL. After a 10-min incubation at room temperature, 5 μL of 1 M MgCl2 was added to stop nucleotide exchange. Samples (40 μL) were taken at this point and 5, 15, and 30 min after addition of the activating protein, and the samples were mixed with 1 mL of ice-cold assay buffer (50 mM Tris, pH 7.5, 50 mM NaCl, and 5 mM MgCl2). The samples were filtered through nitrocellulose filters (Pall Corporation) and the bound radioactivity counted in a LS6000SC scintillation counter (Coulter Beckmann).
Microscopic Analysis and Data Analysis
Transiently transformed pollen tubes were transferred upside down with the solid medium onto cover slips for analysis on inverted microscopes. Epifluorescence and light microscopy was performed on a DM IRB microscope (Leica) equipped with differential interference contrast optics, a 100-W mercury lamp, an fluorescein isothiocyanate filter block (excitation, 450 to 490 nm; dichroic 510-nm emission: 515 long-pass 13S; Leica), ×5 and ×40 lenses (N plan ×5/0.12 and HCX PL FL L ×40/0.6; Leica), and a digital cooled camera (DFC350FX R2; Leica). For tube measurements, ImageJ software (http://rsb.info.nih.gov/ij) was used.
Confocal laser scanning images were taken with an LSM510Meta (Zeiss) inverted microscope. YFP fluorescence was excited with the 514-nm argon laser line and imaged through a 405/514-nm dichroic mirror and a 530- to 600-nm band-pass emission filter.
Digital images were corrected for size, brightness, and contrast using Adobe Photoshop (Adobe Systems).
Computer programs and image analysis were done with the help of GCG analysis software (Accelrys). For similarity and identity determination, we used the GAP program (gap creation penalty 8; gap extension penalty 2).
Accession Numbers
Sequence data for RhoGAP1 can be found in the GenBank data library under accession number DQ813657.
Acknowledgments
We thank Katja Piiper for technical support and Stephen Greiner and Stephanie Cottier for discussions and critical reading of the manuscript. The plasmid for GST human RAC1 was a kind gift of Emmanuel Lemichez. Work in our lab was supported by the German Research Council (DFG; KO2278) and the state of Baden Wuerttemberg.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ulrich Klahre (uklahre@hip.uni-heidelberg.de).
References
- Aitken, A. (2002). Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol. Biol. 50 993–1010. [DOI] [PubMed] [Google Scholar]
- Athwal, G.S., and Huber, S.C. (2002). Divalent cations and polyamines bind to loop 8 of 14-3-3 proteins, modulating their interaction with phosphorylated nitrate reductase. Plant J. 29 119–129. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell, A., Yang, Z., Springer, P.S., and Bailey-Serres, J. (2002). Rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296 2026–2029. [DOI] [PubMed] [Google Scholar]
- Benzing, T., Yaffe, M.B., Arnould, T., Sellin, L., Schermer, B., Schilling, B., Schreiber, R., Kunzelmann, K., Leparc, G.G., Kim, E., and Walz, G. (2000). 14-3-3 interacts with regulator of G protein signaling proteins and modulates their activity. J. Biol. Chem. 275 28167–28172. [DOI] [PubMed] [Google Scholar]
- Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436 1176–1180. [DOI] [PubMed] [Google Scholar]
- Bernards, A., and Settleman, J. (2004). GAP control: Regulating the regulators of small GTPases. Trends Cell Biol. 14 377–385. [DOI] [PubMed] [Google Scholar]
- Bigey, J., Gounon, P., Robineau, S., and Antonny, B. (2003). Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature 426 563–566. [DOI] [PubMed] [Google Scholar]
- Borg, S., Podenphant, L., Jensen, T.J., and Poulsen, C. (1999). Plant cell growth and differentiation may involve GAP regulation of Rac activity. FEBS Lett. 453 341–345. [DOI] [PubMed] [Google Scholar]
- Bosch, M., Cheung, A.Y., and Hepler, P.K. (2005). Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 138 1334–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney, T.D., van den Wijngaard, P.W.J., and de Boer, A.H. (2002). 14-3-3 protein regulation of proton pump and ion channels. Plant Mol. Biol. 50 1041–1051. [DOI] [PubMed] [Google Scholar]
- Burbelo, P.D., Drechsel, D., and Hall, A. (1995). A conserved binding motif defines numerous candidate target proteins for both cdc42 and rac GTPases. J. Biol. Chem. 270 29071–29074. [DOI] [PubMed] [Google Scholar]
- Camoni, L., Visconti, S., Marra, M., and Aducci, P. (2001). Adenosine 5′-monophosphate inhibits the association of 14-3-3 proteins with the plant plasma membrane H(+)-ATPase. J. Biol. Chem. 276 1709–1712. [DOI] [PubMed] [Google Scholar]
- Carol, R.J., Takeda, S., Linstead, P., Durrant, M.C., Kakesova, H., Derbyshire, P., Drea, S., Zarsky, V., and Dolan, L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Cheung, A., and Wu, H.-M. (2003). Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.Y., Hasson, T., Kelley, P.M., Schwender, B.J., Schwartz, M.F., Ramakrishnan, M., Kimberling, W.J., Mooseker, M.S., and Corey, D.P. (1996). Molecular cloning and domain structure of human mysosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics 36 440–448. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., Chen, C.Y.-h., Tao, L.-z., Andreyeva, T., Twell, D., and Wu, H.-M. (2003). Regulation of pollen tube growth by Rac-like GTPases. J. Exp. Bot. 54 73–81. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420 629–635. [DOI] [PubMed] [Google Scholar]
- Feig, L.A. (1999). Tools of the trade: Use of dominant-inhibitory mutants of the Ras-family GTPases. Nat. Cell Biol. 1 E25–E27. [DOI] [PubMed] [Google Scholar]
- Ferl, R.J. (2004). 14-3-3 proteins: Regulation of signal-induced events. Physiol. Plant. 120 173–178. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, D.L., Eccleston, J.F., and Lowe, P.N. (1999). The conserved arginine in rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with Rho.GDP and aluminum fluoride. Biochemistry 38 985–991. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Friml, J., Swarup, R., Ljung, K., Sandberg, G., Terlou, M., Palme, K., Bennet, M.J., and Scheres, B. (2002). Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr. Biol. 12 329–334. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Fu, Y., Dowd, P., Li, S., Vernoud, V., Gilroy, S., and Yang, Z. (2005). A rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Li, S., Lord, E.M., and Yang, Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control rho GTPase-dependent polar growth. Plant Cell 18 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Vernoud, V., Fu, Y., and Yang, Z. (2003). ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54 93–101. [DOI] [PubMed] [Google Scholar]
- Hepler, P.L., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Hwang, J.-U., Gu, Y., Lee, Y.-J., and Yang, Z. (2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16 5385–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A.B., and Hall, A. (2005). RhoGTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21 247–269. [DOI] [PubMed] [Google Scholar]
- Klahre, U., Becker, C., Schmitt, A.C., and Kost, B. (2006). Interaction with NtRhoGDI2 is essential for normal NtRAC5 localization and activity in tobacco pollen tubes. Plant J. 46 1018–1031. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 19 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16 393–401. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., Karunanandaa, B., McCubbin, A., Gilroy, S., and Kao, T.-h. (1996). PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 9 613–624. [Google Scholar]
- Lemmon, M.A., Ferguson, K.M., O'Brien, R., Sigler, P.B., and Schlessinger, J. (1995). Specific and high-affinity binding of inositol phophates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 92 10472–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, D.A., Lin, R., Cerione, R.A., and Manor, D. (1998). Biochemical studies of the mechnism of action of the Cdc42-GTPase-activating protein. J. Biol. Chem. 273 16210–16215. [DOI] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heat, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeti, E., Dagher, M.-C., Hernandez, S.E., Koleske, A.J., and Settleman, J. (2004). Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 279 5055–5058. [DOI] [PubMed] [Google Scholar]
- Lord, E.M. (2003). Adhesion and guidance in compatible pollination. J. Exp. Bot. 54 47–54. [DOI] [PubMed] [Google Scholar]
- Lord, E.M., and Russell, S.D. (2002). The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18 81–105. [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler, A., Wilsen, K.L., Baskin, T.I., and Hepler, P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221 95–104. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16 (suppl.), S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendjik, A.J., Ruperti, B., and Palme, K. (2004). Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 7 694–700. [DOI] [PubMed] [Google Scholar]
- Moon, S.Y., and Zheng, Y. (2003). Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13 13–22. [DOI] [PubMed] [Google Scholar]
- Parton, R.M., Fischer-Parton, S., Trewavas, A.J., and Watahiki, M.K. (2003). Pollen tubes exhibit regular periodic membrane trafficking events in the absence of apical extension. J. Cell Sci. 116 2707–2719. [DOI] [PubMed] [Google Scholar]
- Paul, A.-L., Sehnke, P.C., and Ferl, R. (2005). Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol. Biol. Cell 16 1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirone, D.M., Carter, D.E., and Burbelo, P.D. (2001). Evolutionary expansion of CRIB-containing Cdc42 effector proteins. Trends Genet. 11 370–373. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scheffzek, K., and Ahmadian, M.R. (2005). GTPase activating proteins: Structural and functional insights 18 years after discovery. Cell. Mol. Life Sci. 62 3014–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self, A.J., and Hall, A. (1995). Measurement of intrinsic nucleotide exchange and GTP hydrolysis rate. Methods Enzymol. 256 67–76. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., and Oppnenheimer, D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21 271–295. [DOI] [PubMed] [Google Scholar]
- Sorrell, D.Z., Marchbank, A.M., Chrimes, D.A., Dickinson, J.R., Rogers, H.J., Francis, D., Grierson, C.S., and Halford, N.G. (2003). The Arabidopsis 14-3-3 protein, GF14ω, binds to the Schizosaccharomyces pombe Cdc25 phsophatase and rescues checkpoint defects in the rad24- mutant. Planta 218 50–57. [DOI] [PubMed] [Google Scholar]
- Thompson, G., Owen, D., Chalk, P.A., and Lowe, P.N. (1998). Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry 37 7885–7891. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., Christensen, R., and Bones, A.M. (2000). Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Gu, Y., Li, S., and Yang, Z. (2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13 2841–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 124 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.-L., and Yang, Z. (2000). The rop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 5 298–303. [DOI] [PubMed] [Google Scholar]