Abstract
EXORIBONUCLEASE4 (XRN4), the Arabidopsis thaliana homolog of yeast XRN1, is involved in the degradation of several unstable mRNAs. Although a role for XRN4 in RNA silencing of certain transgenes has been reported, xrn4 mutant plants were found to lack any apparent visible phenotype. Here, we show that XRN4 is allelic to the unidentified components of the ethylene response pathway ETHYLENE-INSENSITIVE5/ACC-INSENSITIVE1 (EIN5/AIN1) and EIN7. xrn4 mutant seedlings are ethylene-insensitive as a consequence of the upregulation of EIN3 BINDING F-BOX PROTEIN1 (EBF1) and EBF2 mRNA levels, which encode related F-box proteins involved in the turnover of EIN3 protein, a crucial transcriptional regulator of the ethylene response pathway. Epistasis analysis placed XRN4/EIN5/AIN1 downstream of CTR1 and upstream of EBF1/2. XRN4 does not appear to regulate ethylene signaling via an RNA-INDUCED SILENCING COMPLEX–based RNA silencing mechanism but acts by independent means. The identification of XRN4 as an integral new component in ethylene signaling adds RNA degradation as another posttranscriptional process that modulates the perception of this plant hormone.
INTRODUCTION
The gaseous hormone ethylene regulates a wide range of developmental processes in plants and their response to stress and pathogens (Johnson and Ecker, 1998). Moreover, manipulation of the ethylene response pathway is of agronomic importance given its role in fruit ripening and floral abscission. Current knowledge of this signaling pathway is based on the extensive characterization of Arabidopsis thaliana mutants with altered ethylene responses (Alonso and Stepanova, 2004) (see Supplemental Figure 1 online). Ethylene signal transduction begins with ethylene binding to and inactivating a family of ethylene receptors. In the absence of ethylene, these receptors activate CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), a mitogen-activating protein kinase kinase kinase (MAPKKK) that negatively regulates the pathway (Kieber et al., 1993). After CTR1 inactivation, ETHYLENE-INSENSITIVE2 (EIN2), a positive regulator of the signaling cascade, which shares homology with the N-Ramp family of metal transporters (Alonso et al., 1999), promotes ethylene responses via the downstream transcription factor EIN3 and most likely also via EIN3-like1 (EIL1) and other EILs. EIN3 and EILs then upregulate primary target genes of the ethylene transcriptional cascade.
EIN3 (and possibly EILs) is regulated at the posttranslational level by the two closely related F-box proteins EIN3 BINDING F-BOX PROTEIN1 (EBF1) and EBF2 (for EIN3 binding F-BOX PROTEIN1 and -2), which are the substrate binding subunits of ubiquitin-ligating SCF complexes (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). In the absence of ethylene, the EIN3 protein is constitutively ubiquitylated and degraded in an EBF1/2-dependent manner. However, in the presence of ethylene, or in ebf1 ebf2 double mutant plants, EIN3 is stabilized and accumulates (Guo and Ecker, 2003; Potuschak et al., 2003; Yanagisawa et al., 2003; Gagne et al., 2004). Conversely, plants that ectopically overexpress EBF1 or EBF2 are less sensitive to ethylene because of increased degradation of EIN3 protein (Guo and Ecker, 2003; Potuschak et al., 2003). We previously showed that the accumulation of EIN3 regulates EBF1 and EBF2 mRNA levels, which presumably provides a feedback mechanism to limit the accumulation of EIN3 (Potuschak et al., 2003). Consistent with this mechanism, the transcript levels of EBF2 and, to a lesser extent, EBF1, show a pattern of ethylene-dependent accumulation that is disrupted in an ein3 mutant background (Potuschak et al., 2003).
At the protein level, ethylene-dependent accumulation of EIN3 protein is completely abolished in ein2 mutant plants and reduced in ein5 and ein6 mutant plants, suggesting that these genes act upstream of EIN3 in the ethylene response pathway (Guo and Ecker, 2003). EIN5, also known as AIN1 (for ACC-INSENSITIVE1), and EIN6 (Van Der Straeten et al., 1993; Roman et al., 1995; Smalle et al., 1997) are recessive ethylene-insensitive mutants whose corresponding genes still remain unknown. Because these loci have been tentatively placed upstream of EIN3 in the ethylene response pathway, we sought to investigate their role in the EBF1/2-dependent turnover of EIN3 protein. Here, we show that EIN5 is allelic to the cytoplasmic EXORIBONUCLEASE4 (XRN4), encoding a homolog of XRN1 of budding yeast. Epistasis analysis places EIN5 downstream of CTR1 and upstream of EBF1/2. Unlike other ethylene-insensitive mutants, which accumulate fewer EBF1/2 mRNAs, presumably because of less EIN3 protein, ein5-1 mutant plants accumulate higher levels of EBF1 and EBF2 mRNAs. ein5-1 seedlings have concomitantly reduced levels of EIN3 protein and are less ethylene-sensitive, which are both reminiscent of EBF1-overexpressing plants. XRN4 was recently implicated in the microRNA (miRNA)–mediated turnover of some Arabidopsis transcripts (Souret et al., 2004) and in the small interfering RNA (siRNA)–mediated turnover of transgene-derived transcripts (Gazzani et al., 2004). With respect to ethylene perception, XRN4 does not affect RNA-INDUCED SILENCING COMPLEX (RISC)–based silencing pathways but likely acts by independent means.
RESULTS
EBF1 and EBF2 Act Downstream of EIN5, and Their Transcript Levels Are Upregulated in an ein5 Mutant Background
Our previous analysis of the transcriptional accumulation of EBF1/2 in ethylene mutants, which led to the establishment of the EIN3-EBF1/2 feedback loop (Potuschak et al., 2003), did not include the ein5 mutant. Similar to other ethylene-insensitive mutants (Potuschak et al., 2003), we expected to find a downregulation of EBF1/2 transcripts in ein5-1 seedlings. Instead, we found that EBF1 and EBF2 transcript levels were moderately increased in the ein5-1 background compared with wild-type Arabidopsis (Figures 1A and 1B). Coupled with the reduced and delayed accumulation of EIN3 protein in the ein5-1 background, as described by Guo and Ecker (2003), this observation led us to speculate that EIN5 decreases EBF1/2 levels, which in turn promotes ethylene response by decreasing EIN3 protein degradation. In support of this notion, ectopic overexpression of EBF1 in wild-type Arabidopsis plants was shown previously to reduce ethylene sensitivity (Guo and Ecker, 2003; Potuschak et al., 2003). Under this scenario, EIN5 should act upstream of EBF1/2 in the ethylene response pathway. To test this possibility, we generated the triple mutant combination ein5-1 ebf1-1 ebf2-1 (Figures 1C and 1D). We used here the hypomorphic ebf2-1 allele (Potuschak et al., 2003), as complete loss-of-function mutants of both EBF1 and EBF2 exhibit a substantial growth arrest (Gagne et al., 2004). The ein5-1 ebf1-1 ebf2-1 plants displayed a constitutive ethylene response, also named a triple response, because it consists of three features: a short and thick hypocotyl, an exaggerated apical hook, and a short root. This phenotype (Figures 1C and 1D) is nearly identical to that of the ebf1-1 ebf2-1 double mutants when germinated in the dark, indicating that EIN5 acts upstream of EBF1 and EBF2. Adult ein5-1 ebf1-1 ebf2-1 plants also showed a severe constitutive ethylene response phenotype like that of the ebf1-1 ebf2-1 double mutant (Potuschak et al., 2003) when grown under normal light conditions (Figure 1E). In addition, ein5-1 ebf1-1 ebf2-1 plants show delayed bolting compared with the already late-flowering ebf1-1 ebf2-1. It is worth noting that ACC-insensitive1 (ain1)/ein5 plants were found to have a mild late-flowering phenotype (Van Der Straeten et al., 1993). The additive effect of ein5-1 and ebf1 ebf2 on this aspect of plant development suggests that some roles of EIN5 appear to be independent of EBF1/2.
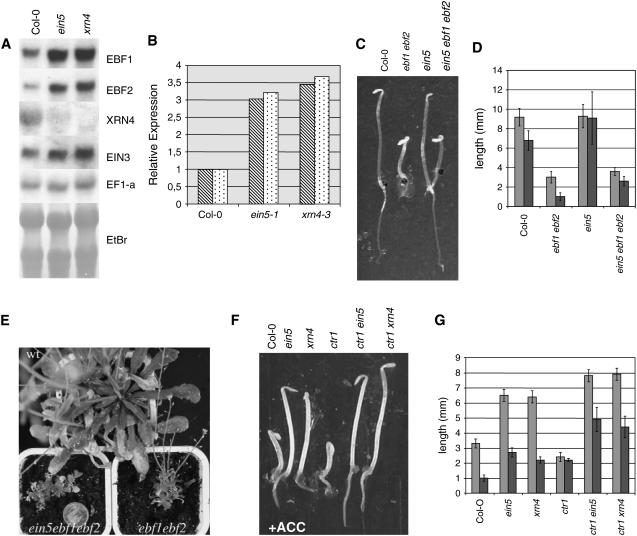
Figure 1.
EBF1 and EBF2 Transcript Levels Are Upregulated in ein5 Mutant Plants, and Epistasis Analysis of ein5.
(A) RNA gel analysis of EBF1 and EBF2 transcript accumulation in ecotype Columbia (Col-0), ein5-1, and xrn4-3. EtBr, ethidium bromide.
(B) Relative EBF1 (hatched bars) and EBF2 (dotted bars) transcript accumulation in Col-0, ein5-1, and xrn4-3 by quantification using a phosphor imager.
(C) Phenotypes of 3-d-old etiolated seedlings of the indicated genotypes grown without ACC.
(D) Hypocotyl (light gray) and root (dark gray) length measurements of 3-d-old dark-grown Col-0, ebf1-1 ebf2-1, ein5-1, and ein5-1 ebf1-1 ebf2-1 seedlings germinated in the absence of ACC. Values shown are average lengths (means ± se) of >10 hypocotyls or roots.
(E) Phenotypes of mature ebf1-1 ebf2-1, ein5-1 ebf1-1 ebf2-1, and wild-type (Col-0) plants grown on soil. ein5-1 plants are similar to wild-type plants on soil and therefore are not shown.
(F) Phenotypes of 3-d-old etiolated seedlings of the indicated genotypes grown on Murashige and Skoog (MS) medium supplemented with 10 μM ACC.
(G) Hypocotyl (light gray) and root (dark gray) length measurements of 3-d-old dark-grown Col-0, ein5-1, xrn4-3, ctr1-1, ctr1-1 ein5-1, and ctr1-1 xrn4-3 seedlings in the presence of 10 μM ACC. Values shown are average lengths (means ± se) of >10 hypocotyls or roots.
Roman et al. (1995) previously reported that EIN5 acts downstream of CTR1 in the ethylene signaling pathway. We confirmed this position by recreating ctr1-1 ein5-1 mutant plants (Figures 1F and 1G). Indeed, EIN5 is epistatic to CTR1, as the ein5-1 mutation suppresses the constitutive triple response of ctr1 seedlings. In conclusion, EIN5 acts downstream of CTR1 but upstream of EBF1/2.
EIN5 Is Allelic to the Exoribonuclease XRN4
A recent microarray analysis by Souret et al. (2004) identified the EBF1 mRNA (designated FBL6) among a small set of transcripts that are possible targets of the Arabidopsis XRN4 exoribonuclease. xrn4 knockout mutants were shown to accumulate slightly higher levels of EBF1/FBL6 mRNA as well as a shorter RNA fragment that might be a cleavage product. In striking contrast with comparable exoribonuclease mutants in other model organisms, no obvious phenotype was found for xrn4-deficient plants (Souret et al., 2004). Because ein5 mutant plants also upregulate EBF1 transcript levels and are phenotypically normal in the absence of ethylene, we compared EBF1 mRNA accumulation in ein5 and xrn4 plants (Figures 1A and 1B). Both lines showed a similar upregulation of the EBF1 transcript as well as an upregulation of the EBF2 transcript. To investigate whether this upregulation of EBF1/2 mRNAs in an xrn4-3 background might affect ethylene signaling, we germinated the xrn4-3 mutant in the dark on medium containing the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Figures 1F and 1G). Indeed, xrn4-3 plants were ethylene-insensitive to a similar level as ein5-1 plants. Moreover, like ein5-1 ctr1 plants, xrn4-3 ctr1-1 double mutant plants were ethylene-insensitive.
Because EIN5 was mapped like XRN4 to chromosome 1 in Arabidopsis, we speculated that EIN5 and XRN4 are allelic. This was indeed the case (Figures 2A to 2C), as F1 plants derived from a cross between the two homozygous recessive parents ein5-1 and xrn4-3 were also ethylene-insensitive. Sequencing of the XRN4 open reading frame (ORF) from the ein5-1 background revealed a 1-bp deletion at nucleotide position 1658 causing a frame-shift mutation (Figure 2D). This mutant should generate a truncated protein of 552 residues plus 13 additional residues before termination of the protein if translated. It is noteworthy that XRN4 transcripts are downregulated in ein5-1 and truncated and downregulated in xrn4-3 alleles (Figure 1A), with the latter suggesting that nonsense-mediated mRNA decay subsequently breaks down the XRN4 transcript.
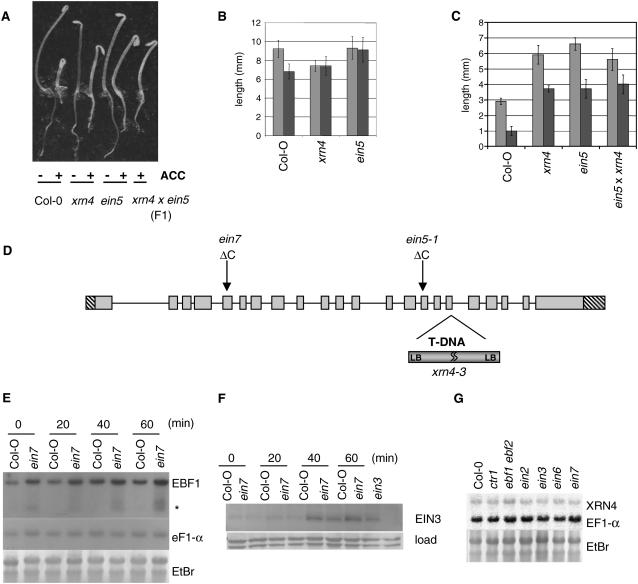
Figure 2.
ein5 and ein7 Are Mutants in the Exoribonuclease XRN4.
(A) Phenotypes of 4-d-old etiolated seedlings of the indicated genotypes grown on MS medium.
(B) Hypocotyl (light gray) and root (dark gray) length measurements of 4-d-old dark-grown Col-0, xrn4-3, and ein5-1 seedlings germinated in the absence of ACC. Values shown are average lengths (means ± se) of >10 hypocotyls or roots.
(C) Hypocotyl (light gray) and root (dark gray) length measurements of 4-d-old dark-grown Col-0, xrn4-3, ein5-1, and F1 seedlings of a xrn4-3 × ein5-1 cross in the presence of 10 μM ACC. Values shown are average lengths (means ± se) of >10 hypocotyls or roots.
(D) Gene structure of XRN4. Introns are indicated by lines. Shaded and hatched boxes indicate coding regions and 5′ and 3′ untranslated regions, respectively. The positions of ein5-1 and ein7 mutations are indicated and correspond to cytosine deletions creating truncated proteins in both cases. The position of the T-DNA of the xrn4-3 allele is also indicated.
(E) EBF1 transcript levels in Col-0 and ein7 at different time points during treatment with 50 μM ACC. RNA was extracted from 3-week-old Col-0 and ein7 seedlings at different time points of the ACC treatment, subjected to RNA gel blot analysis, and hybridized with the indicated probes. EtBr, ethidium bromide.
(F) EIN3 protein accumulation in Col-0 and ein7 at different time points during 50 μM ACC treatment. Total protein extracts from the same sample as indicated in (E) were subjected to immunoblot assays.
(G) RNA gel analysis of XRN4 transcript accumulation in different mutant backgrounds as indicated.
ein7 is a poorly characterized ethylene-insensitive mutant that has been proposed to be allelic to ein5 (Roman et al., 1995). However, to date, allelism between EIN5 and EIN7 has not been described, presumably because of the reported semidominant nature of the ein7 locus. As with ein5-1 and xrn4-3, ein7 seedlings have a moderately increased level of EBF1 transcript levels compared with the wild type that was maintained after ACC treatment (Figure 2E). Consistent with these results, the accumulation of EIN3 protein after ACC treatment was also attenuated in the ein7 background (Figure 2F). We obtained comparable data with ein5-1 and xrn4-3 (see Supplemental Figure 2 online). Thus, ein7 seedlings, like ein5-1 (Guo and Ecker, 2003), were able to accumulate EIN3 after ethylene stimulation, but to a lower level than wild-type seedlings. We sequenced the XRN4 ORF from the ein7 background and identified a 1-bp deletion causing a frame shift in the fifth exon at nucleotide 671. This change should lead to a truncation of the XRN4 peptide after residue 223 and create a short tail of 13 residues before termination of the protein (Figure 2D). As a result, we conclude that ein7, like ein5-1, disrupts production of the exoribonuclease XRN4. Because XRN4 acts downstream of CTR1, we tested whether XRN4 is transcriptionally misregulated in ctr1-1 or other mutants in the ethylene response pathway. However, XRN4 mRNA accumulated to similar levels in several ethylene signaling mutants, indicating that if XRN4 is regulated in an ethylene-dependent manner, this must occur at the posttranscriptional level (Figure 2G).
xrn4/ein5 Plants Are Deficient in EIN3/EIL1-Dependent Growth Inhibition by Ethylene
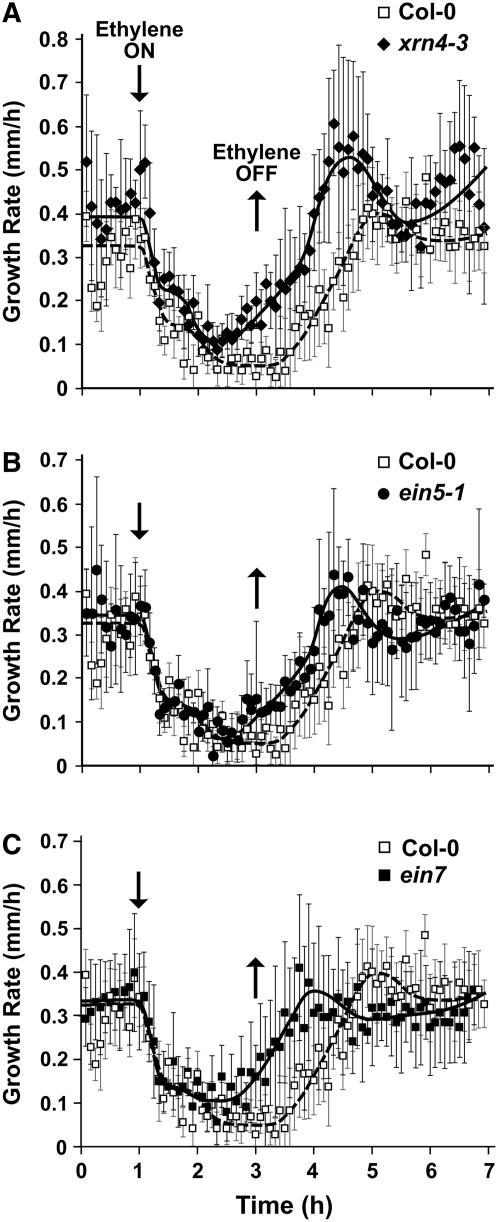
Earlier studies of etiolated Arabidopsis seedlings revealed two phases of ethylene-induced growth inhibition for wild-type Col-0 hypocotyls (Binder et al., 2004a, 2004b). The first phase of inhibition begins ∼10 min after ethylene is applied. Thirty minutes after the addition of ethylene, a new steady state growth rate is reached, which is maintained for ∼20 min before a second phase of growth inhibition begins, which reduces the growth rate to a lower steady state. At saturating concentrations of hormone, this lower growth rate is maintained for as long as the hormone is present. If ethylene is removed, hypocotyl elongation recovers by ∼90 min to pretreatment growth rates by a dampening oscillation. EIN2 is essential for both phases. Whereas the first phase of growth inhibition is independent of EIN3 and EIL1, the second phase response requires both of these transcription factors (Binder et al., 2004b).
Because both the xrn4-3 and ein5-1 mutants increase EBF1/2 mRNA levels and accumulate lower levels of EIN3 protein in response to ethylene or its precursor ACC, we examined the ethylene growth response kinetics of these mutants. As shown in Figure 3, wild-type Col-0 hypocotyls have a growth inhibition response similar to that described in previous reports (Binder et al., 2004a, 2004b). Recovery after the removal of ethylene was slightly slower in wild-type seedlings in this study, taking ∼105 min to recover to the initial, pretreatment growth rates. Both the xrn4-3 and ein5-1 mutants showed onset kinetics indistinguishable from those of the wild type after ethylene addition, with both phases of growth inhibition being evident (Figure 3). However, both mutants recovered markedly faster than the wild type after ethylene removal and appeared to begin growth recovery even before the ethylene was removed (Figures 3A and 3B). Experiments with ein7 generated similar kinetics (Figure 3C). These data were consistent with our observed upregulation of EBF1/2 RNA levels in xrn4/ein5/ein7-deficient plants and the subsequent reduced accumulation of EIN3 protein in these lines.
Figure 3.
Growth Kinetics of Etiolated Arabidopsis Hypocotyls in Response to Ethylene.
Growth rates were recorded for 1 h in air followed by a 2-h exposure to 10 μL/L ethylene. This was followed by 5 h in air. The responses of wild-type Col-0 hypocotyls are shown in both panels (open squares) for comparison with the homozygous mutants (closed symbols) xrn4-3 (A), ein5-1 (B), and ein7 (C). All data represent averages of at least five seedlings ± sd.
The xrn4 Mutation Does Not Affect EBF1/2 mRNA Turnover
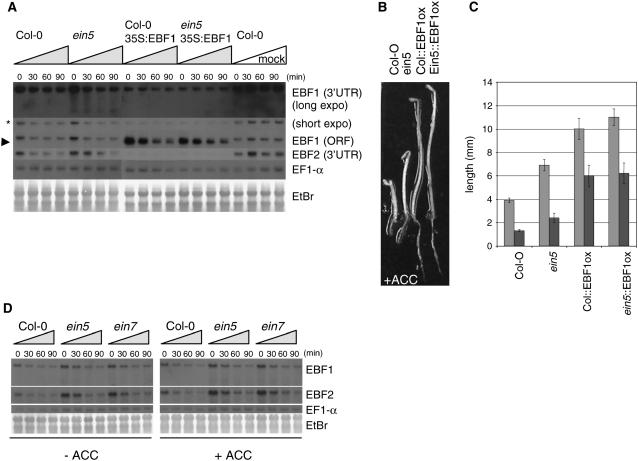
Because XRN4 functions in mRNA decay, one obvious scenario is that the upregulation of EBF1 in the xrn4-3 background is the consequence of increased EBF1 transcript stability. To test this possibility, we monitored the stability of the EBF1 and -2 mRNAs, using the adenosine analogue cordycepin (3′-deoxyadenosine) to block transcription. Under these conditions, both mRNAs were equally short-lived in wild-type, ein5-1, and ein7 plants, suggesting that the mode of action of XRN4 must be more complex (Figure 4A; data not shown). This finding is in agreement with that described by Souret et al. (2004), who also failed to find a stabilization of FBL6/EBF1 transcripts in xrn4 mutants. We further investigated the stability of ectopically expressed EBF1 mRNA under the control of the cauliflower mosaic virus 35S promoter and lacking the endogenous 5′ and 3′ untranslated regions of the transcript. The transgenic line selected showed a higher EBF1 mRNA level compared with the ein5 mutant (Figure 4A) and consistently exhibited a stronger ethylene-insensitive phenotype than the latter (Figures 4B and 4C). Moreover, when the transgene was expressed in the ein5 mutant background, the ethylene-insensitive phenotype of ein5 was much stronger, further demonstrating the correlation between the EBF1 expression levels and ethylene insensitivity. Surprisingly, the transcript produced from the synthetic construct was also short-lived in both wild-type and ein5 plants (Figure 4A). Furthermore, when EBF1 was overexpressed ectopically, wild-type transcript levels of EBF1 and EBF2 were greatly reduced, as shown previously (Potuschak et al., 2003), and were apparently less affected by the cordycepin treatment (Figure 4A). Nevertheless, this change in the turnover is not affected by the ein5-1 mutation.
Figure 4.
EBF1 and EBF2 mRNAs Remain Short-Lived in xrn4/ein5 Mutants, and Their Turnover Is Ethylene Independent.
(A) RNA gel analysis of a time-course experiment for EBF1 and EBF2 transcript half-life determination in Col-0 and the ein5-1 mutant. The blot was hybridized with either the 3′ untranslated region (UTR; detecting endogenous transcripts) or ORF probes (also detecting ectopically expressed EBF1) as indicated, and EBF1/2 transcript half-lives were determined relative to EF1-α, which is a stable transcript. The asterisk indicates the possible EBF1 mRNA cleavage product as indicated by Souret et al. (2004), and the arrowhead indicates the shorter EBF1 transcript originating from the 35S:EBF1 transgene. EtBr, ethidium bromide.
(B) Strong EBF1 overexpression exacerbates the ethylene-insensitive phenotype of ein5. An Arabidopsis T-DNA–transformed line that expresses a high level of EBF1 mRNA (see [A]) exhibits strong ethylene insensitivity when germinated in the dark in the presence of 10 μM ACC. This phenotype is maintained when the transgene is introduced into the ein5-1 mutant.
(C) Hypocotyl (light gray) and root (dark gray) length measurements of 3-d-old dark grown Col-0, ein5-1, Col-0∷EBF1ox, and ein5-1∷EBF1ox seedlings in the presence of 10 μM ACC. Values shown are average lengths (means ± se) of >10 hypocotyls or roots.
(D) RNA gel analysis of a time-course experiment for EBF1 and EBF2 endogenous transcript half-life determination in Col-0 and in ein5-1 and ein7 mutants pretreated with or without 50 μM ACC. EBF1/2 transcript half-lives were determined relative to EF1-α.
We next asked whether ethylene might change the half-life of EBF1 and/or EBF2 mRNAs in ein5/ein7 mutant backgrounds. However, EBF1 and EBF2 mRNAs remained short-lived in both ein5 and ein7 with and without preincubation with the ethylene precursor ACC (Figure 4D). From these experiments, we conclude that the higher accumulation of EBF1 and EBF2 mRNAs in the exoribonuclease mutants most likely is not the consequence of an altered turnover of these transcripts.
XRN4 Function in Ethylene Signaling Is Independent of Known Mutants in miRNA and siRNA Pathways
Although we cannot totally exclude a direct role of XRN4 in the turnover of EBF1/2 mRNAs, our results suggest that XRN4 modulates EBF1/2 indirectly, either by positively regulating a repressor or by negatively regulating an activator of EBF1/2 transcription. Such a regulation of EBF1/2 mRNA abundance by XRN4 could occur via a miRNA- or siRNA-dependent mechanism, as several known miRNA substrates of XRN4 are miRNA targets (Souret et al., 2004) and because the suppression of SHOOT MERISTEMLESS and WUSCHEL overexpression in an xrn4 mutant background requires RDR6/SDE1 (for RNA-DEPENDENT RNA POLYMERASE6/SILENCING DEFECTIVE1), the RNA-dependent RNA polymerase involved in siRNA-dependent transgene silencing (Gazzani et al., 2004). Therefore, we tested mutants that are impaired in miRNA and siRNA pathways, such as hen1, rdr2, dcl2, dcl3, sde1, and the recently described dcl4-1 (for hua enhancer1, RNA-dependent RNA polymerase2, and dicer-like 2/3/4) (Park et al., 2002; Boutet et al., 2003; Xie et al., 2004; Gasciolli et al., 2005) for their response toward ethylene (see Supplemental Figure 3 online). None showed a significant degree of ethylene insensitivity (data not shown) or higher EBF1/2 transcript levels under the conditions tested (see Supplemental Figure 3A online). We generated ein5 sde1 and ein5 rdr2 double mutant plants and found that they were indistinguishable from ein5 when germinated in the presence of ACC (see Supplemental Figures 3B and 3C online). Finally, we created an xrn4-3 hen1 double mutant that was also ethylene-insensitive (see Supplemental Figures 3D and 3E online). The slight increase in hypocotyl length observed for this double mutant compared with the xrn4 single mutant might be attributed to the mix of two different ecotypes (Col and Landsberg erecta). We conclude from these experiments that the ethylene-insensitive phenotype of xrn4 does not require functional HEN1, RDR6/SDE1, or RDR2.
DISCUSSION
The control of RNA turnover is a crucial aspect of gene expression in all organisms (Parker and Song, 2004; Wilusz and Wilusz, 2004). In animal cells, XRN1, a 5′→3′ processive exoribonuclease, plays an important role in the ARE (for AU-rich element in the 3′ untranslated region) mRNA degradation decay pathway (Stoecklin et al., 2006) and in nonsense-mediated mRNA decay, a pathway required to destroy aberrant RNAs harboring premature translation termination codons (Lejeune et al., 2003; Gatfield and Izaurralde, 2004). It appears to function by processively degrading RNAs from the 5′ end after removal of the 5′ cap, thus requiring a first cleavage event by another ribonuclease before substrate recognition. In Arabidopsis, the cytoplasmic XRN4 appears to represent the plant ortholog of XRN1 (Kastenmayer and Green, 2000). Arabidopsis also possesses two homologs of XRN4, called XRN2 and XRN3, that localize to the nucleus and therefore act on a different population of RNA substrates than XRN4 (Kastenmayer and Green, 2000). Although the function(s) of XRN4 is not completely resolved, it was recently implicated in the degradation of several, but not all, miRNA-directed cleavage products (Souret et al., 2004). A role for XRN4 in transgene silencing via RNA interference in Arabidopsis has also been proposed (Gazzani et al., 2004). However, in contrast with other model organisms (Tishkoff et al., 1995; Newbury and Woollard, 2004), Arabidopsis xrn4 null mutants are viable, with no apparent deleterious phenotype (Souret et al., 2004).
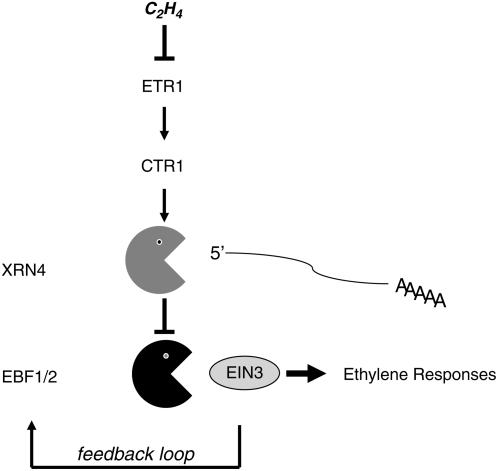
The work presented here confers a physiological function to a member of the XRN1/XRN4 exoribonuclease family in hormone signaling. Based on our genetic analysis, XRN4 is an integral component of the ethylene response pathway in Arabidopsis. It likely acts downstream of the MAPKKK CTR1 and upstream of the F-box proteins EBF1/2, given the ability of xrn4 mutants to suppress the constitutive ethylene response seen in ctr1 but not the constitutive ethylene response phenotype of the ebf1-1 ebf2-1 double mutants (Figure 5). We also present evidence that XRN4 loss of function results in increased levels of EBF1 and EBF2 transcripts, which encode two F-box proteins that target EIN3 for ethylene-dependent degradation. This provides a rationale for the reduced accumulation of EIN3 protein observed in ein5 mutant plants (Guo and Ecker, 2003). This reduced accumulation of EIN3 also explains the faster recovery in growth after the removal of ethylene that we observed. It is worth noting that ain1/ein5 mutant plants have no obvious signaling defects for other hormones (Van Der Straeten et al., 1993), suggesting that XRN4/EIN5 has a specific role in ethylene signaling.
Figure 5.
Representation of the Ethylene Signaling Cascade Including XRN4/EIN5.
Ethylene is perceived by the ETHYLENE RESPONSE (ETR/ERS) receptors located in the endoplasmic reticulum membrane. Binding of ethylene to the receptors results in the inactivation of both the receptors and CTR1, thereby causing the derepression of positive regulatory factors, such as EIN2 (data not shown) and XRN4/EIN5. How CTR1 regulates EIN2 and XRN4/EIN5 and the relationship of EIN2 with XRN4/EIN5 in the ethylene signaling cascade are unknown. Strikingly, our model proposes the existence of two degradation pathways in ethylene signaling: one RNA decay pathway into the cytosol that indirectly controls the steady state levels of EBF1/2 mRNAs, which are components of a protein degradation pathway that controls EIN3 stability in the nucleus.
The molecular functions of several components in the ethylene signaling pathway remain rather mysterious (Alonso and Stepanova, 2004). Thus, after the ethylene signal has been transmitted from the receptors to the Raf-like protein kinase CTR1, there are few insights into the molecular mechanism that leads to the nuclear accumulation of the EIN3 transcription factor. In particular, the MAPK module predicted to be downstream of CTR1 still remains to be characterized. Moreover, neither the function nor the subcellular location of EIN2, which is known to act downstream of CTR1 but upstream of EBF1/2, has been solved. This work adds another level of complexity to this pathway with the identification of a component, XRN4, that is involved in RNA decay. XRN4 is known to act in the cytosol to regulate the transcript levels of EBF1 and EBF2. These two F-box proteins operate in a proteolytic degradation pathway in the nucleus (Figure 5). Although both EIN2 and XRN4/EIN5 are epistatic to CTR1, it is unknown whether they act in the same or independent pathways. Interestingly, it was recently found that EIN2, but not EIN5, is required for insect resistance; by contrast, EIN5, but not EIN2, was required for the growth enhancement induced by bacterial harpin proteins (Dong et al., 2004). Thus, EIN2 and XRN4/EIN5 might act independently in at least some pathways, although they are both necessary for an efficient EIN3 accumulation after ethylene perception (Guo and Ecker, 2003).
One attractive hypothesis was that EBF1/2 mRNAs are directly targeted by a miRNA or a siRNA and that this mechanism requires XRN4 for the initial cleavage and the turnover of the cleavage products. However, several lines of evidence do not support such a model. First, EBF1/2 remain short-lived mRNA species in xrn4/ein5 mutants (Souret et al., 2004; this work). Second, exhaustive sequencing of small RNAs in Arabidopsis failed to identify sequences matching EBF1 or EBF2 mRNAs (Lu et al., 2005); in particular, none was found to fit the predicted EBF1 cleavage site (Souret et al., 2004). Third, previous extensive screens failed to detect ethylene-insensitive mutants that fall into the miRNA, siRNA, and transacting small interfering RNA classes of mutants. Indeed, the mutants that we tested (hen1, rdr2, sde1, dcl2, and dcl3) and that affect these different RNA silencing pathways (Brodersen and Voinnet, 2006) show a normal accumulation of EBF1/2 transcripts. Nevertheless, even if these mutants do not result in ethylene insensitivity, it is still possible that one of them is required for the ethylene-insensitive phenotype seen in xrn4/ein5 mutant plants. Such a scenario would be reminiscent of the suppressor phenotype of xrn4 mutations, which acts through siRNA-dependent silencing of a reporter gene and requires the RNA-dependent RNA polymerase SDE1 (Gazzani et al., 2004). A similar pathway could act on an upstream regulator of EBF1/2, but not directly on EBF1/2 (or cauliflower mosaic virus 35S promoter–driven EBF1), as they are not silenced in an xrn4/ein5 background. Nevertheless, this also seems unlikely, as the hen1 xrn4-3, ein5-1 sde1, and ein5-1 rdr2 mutants generated were found to have an ein5-like ethylene response. Thus, we conclude that XRN4 indirectly regulates EBF1/2 mRNA levels by a mechanism that does not seem to require RISC-based RNA silencing pathways.
Interestingly, proteins involved in RNA metabolism are increasingly being shown to be important in plant hormone regulation (Fedoroff, 2002). The Arabidopsis HYPONASTIC LEAVES1 gene encodes a nuclear double-stranded RNA binding protein, and its loss of function increases plant sensitivity to abscisic acid but reduces the sensitivity to auxin and cytokinin (Lu and Fedoroff, 2000). Other RNA binding proteins that affect abscisic acid signaling are the Sm-like protein SUPERSENSITIVE TO ABA AND DRAUGHT (Xiong et al., 2001) and the mRNA cap binding protein ABA HYPERSENSITIVE1 (Hugouvieux et al., 2001). The cloning of the abscisic acid–hypersensitive mutant ABA-hypersensitive germination2 revealed a gene encoding a poly(A)-specific ribonuclease supposed to function in mRNA decay (Nishimura et al., 2005). Future analyses will certainly reveal the mechanisms for how these RNA binding and processing enzymes control plant hormonal pathways and also identify their target RNAs.
METHODS
Arabidopsis Lines and Plant Crosses
All Arabidopsis thaliana lines used have been described elsewhere: ago4-1 (Zilberman et al., 2003); ctr1-1 (Kieber et al., 1993); ein5-1 and ein7 (Roman et al., 1995); hen1 (Chen et al., 2002); rdr2, dcl2-1, and dcl3-1 (Xie et al., 2004); dcl4-1 (Gasciolli et al., 2005); ebf1-1 and ebf2-1 (Potuschak et al., 2003); sde1 (Dalmay et al., 2000); xrn4-3 (SALK_014209) (Gazzani et al., 2004). Note that EIN5 has been shown to be allelic to AIN1 and that ein5-1 has been renamed ain1-10 (Smalle et al., 1997). However, the bulk of the current literature still uses the name EIN5; therefore, we continue to use this name to avoid confusion.
For the generation of ein5-1 ebf1-1 ebf2-1 triple mutant plants, ebf1-1 ebf2-1 plants were pollinated with ein5-1–derived pollen. In the F2 generation, ein5-1 homozygote lines were selected based on their ethylene-insensitive phenotype and tested by PCR for the ebf1-1 and ebf2-1 T-DNA insertions. No plants showing an ein5-1–like ethylene-insensitive phenotype were found to be also double homozygous for ebf1-1 and ebf2-1. Plants that were ein5-1 homozygous and double heterozygous for the ebf1-1 and ebf2-1 insertion were allowed to self, and F3 seeds were tested for their seedling phenotype. ein5-1 ebf1-1 ebf2-1 triple mutant plants were segregating at the expected ratio and confirmed by PCR testing. For the generation of ein5-1 ctr1-1 and xrn4-3 ctr1 double mutant plants, ctr1 plants were pollinated with pollen from ein5-1 or xrn4-3 donors; ctr1 homozygotes were then selected in the F2 generation based on their phenotype and genotyped for the ein5-1 mutation and xrn4-3 T-DNA insertions. No ctr1 ein5 or ctr1 xrn4-3 homozygotes were identified that way, but several ctr1 plants were found to be heterozygote for the ein5-1 mutation or the xrn4-3 T-DNA insertion. The plants were allowed to set seeds, and they segregated ethylene-insensitive plants in the next generation that were found to be double homozygotes. For the generation of ein5-1 sde1, ein5-1 rdr2-1, and xrn4-3 hen1 plants, F2 seedlings were genotyped directly and double homozygotes were allowed to set seeds. Ethylene responses of double homozygotes were measured with F3 and F4 seeds.
Sequencing of the XRN4 ORF from ein5-1 and ein7
Sequencing of RT-PCR amplification products of the XRN4 ORF from ein5-1 and ein7 material identified a 1-bp deletion at position 1658 and 671 for ein5-1 and ein7, respectively. In the case of ein5-1, the deletion causes the truncation of the XRN4 protein sequence after amino acid 552 and leads to the addition of 13 amino acids (RYLNFTLLILSLT) before a premature stop codon. In the case of ein7, the deletion leads to the termination of the XRN4 protein sequence after amino acid 223 and the addition of the short peptide QIHGIVYMVWMQI before a premature stop codon. To confirm the sequencing results, PCR markers for ein5-1 and ein7 were designed and tested with genomic DNA. For detection of the ein5-1 mutation, a derived cleaved amplified polymorphic sequence marker was designed using the primers ein5F (5′-GTTGATGACTGATCCCTCATCCT-3′) and ein5R (5′-GAGTGTCAACTATCCAGCATGAA-3′). ein5-1–specific PCR products were cleaved by Taq1, whereas wild-type specific PCR products were not cleaved. For detection of the ein7 mutation, a cleaved-amplified polymorphic sequence marker was designed with the primers ein7F (5′-TTCAAATGTTCCGGGAGAAG-3′) and ein7R (5′-GACGAAGCACCAACACCTTA-3′). ein7-derived PCR products were cleavable with the restriction enzyme BclI. PCR products derived from wild-type DNA were not cleaved.
Seedling Responses to Ethylene, and Growth Rate Measurements on Hypocotyls
Seedling responses to ethylene were measured exactly as described previously (Potuschak et al., 2003).
The effect of ethylene on the growth rate of hypocotyls was measured using etiolated Arabidopsis seedlings as described previously (Binder et al., 2004a, 2004b). Mutant and wild-type seeds were surface-sterilized by treatment with 70% ethanol for ∼30 s, placed on sterile filter paper to dry, and plated on half-strength MS medium, pH 5.7, containing 0.8% agar and B5 vitamins, consisting of inositol (100 mg/mL), nicotinic acid (1 mg/mL), pyridoxin HCl (1 mg/mL), and thiamine HCl (10 mg/mL) with no added sugar. Five micromolar l-α-(2-aminoethoxyvinyl)-glycine was included to inhibit the biosynthesis of ethylene.
Seeds were cold-treated for 2 to 4 d at 4°C and exposed to light for 2 to 8 h before being grown vertically in the dark for 2 d at 22°C. Growth rate measurements were performed as described (Binder et al., 2004b). After 1 h of treatment with air to establish a basal growth rate, ethylene was introduced at a flow rate of 10 mL/min, giving a final concentration of 10 μL/L. Ethylene was removed 2 h later. Gas flow was maintained at 100 mL/min using Side-Trak mass flow meters and controller (Sierra Instruments). Hypocotyl growth was measured from digital images that were captured every 5 min for 7 h with either an EDC-1000N CCD (Electrim) or an Infinity 2-1M camera (Luminera) and light provided by an infrared light-emitting diode. Growth rates were calculated using custom software generated by Edgar Spalding in LabVIEW 5.0 (National Instruments) as described previously (Parks and Spalding, 1999; Folta and Spalding, 2001). All data presented represent averages of at least five seedlings from a minimum of three separate experiments.
Plasmid Construction and Transformation
For the chimeric EBF1 construct, the ORF of EBF1 was modified by PCR and inserted via NcoI into the multiple cloning site of a modified pPily (Farras et al., 2001), which carried an additional six hemagglutinin epitope tags to facilitate protein detection. The expression cassette containing the 2× cauliflower mosaic virus 35S promoter, the epitope-tagged EBF1 ORF, and the nopaline synthase terminator was excised via NotI cleavage and inserted in a XmaI-digested pCAMBIA1380 binary vector (www.cambia.org). NotI and XmaI restriction sites were partially filled in before ligation. After transformation of Col-0 by flower dip (Clough and Bent, 1998), homozygote lines containing single inserts were selected. One such line was crossed into ein5-1. Again, plants homozygous for ein5-1 and the T-DNA insertion were selected.
RNA Gel Analyses and Transcript Half-Life Measurements
Unless stated otherwise, RNA was extracted from 3-week-old light-grown seedlings. RNA preparation, RNA gel blotting, and hybridization were performed using standard protocols. Quantification of RNA gel blots were performed with a Fuji BAS 1000 Imager and MacBAS software version 2.1. For the estimation of half-life measurements with cordycepin, the protocol of Seeley et al. (1992) with modifications for Arabidopsis (Gutierrez et al., 2002) was followed. To test the influence of ACC on the stability of mRNAs, 50 μM ACC was added to the plant material submerged in the cordycepin incubation buffer (Seeley et al., 1992) 30 min before the addition of cordycepin. To test the influence of ACC on EIN3 protein accumulation or EBF1/2 transcript levels, 3-week-old light-grown plants were submerged into half-strength MS buffer (1% sucrose) and treated with 50 μM ACC for the indicated period of time.
Protein Gel Blots
Samples of 15 μg of proteins were separated on SDS gels and blotted onto Immobilon-P membranes (Millipore). The probing procedure and antibodies against EIN3 have been described (Guo and Ecker, 2003; Yanagisawa et al., 2003).
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure 1. Flow Chart of the Ethylene Response Pathway.
Supplemental Figure 2. Reduced EIN3 Protein Accumulation in ein5-1 and xrn4-3 Plants in Response to ACC Treatment.
Supplemental Figure 3. Mutants Deficient in miRNA, siRNA, and Transacting Small Interfering RNA Pathways Do Not Exhibit Higher EBF1 and EBF2 mRNA Levels and Do Not Significantly Alter the Ethylene-Insensitive Phenotype of ein5.
Supplementary Material
Acknowledgments
We thank Olivier Voinnet and members of his laboratory for insightful discussions and the gift of Arabidopsis strains and Patrick Achard for discussions and critically reading the manuscript. We thank Hongwei Guo and Joseph R. Ecker for an antibody against EIN3. This work was supported by the Centre National de la Recherche Scientifique and a grant from the French Ministry of Research (Grant ACI 2004 No. BCMS 167) to P.G. and by a grant from the U.S. National Science Foundation Arabidopsis 2010 Program (Grant MCB-0115870) to R.D.V.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pascal Genschik (pascal.genschik@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., and Stepanova, A.N. (2004). The ethylene signaling pathway. Science 306 1513–1515. [DOI] [PubMed] [Google Scholar]
- Binder, B.M., Mortimore, L.A., Stepanova, A.N., Ecker, J.R., and Bleecker, A.B. (2004. a). Short term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol. 136 2921–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, B.M., O'Malley, R.C., Wang, W., Moore, J.M., Parks, B.M., Spalding, E.P., and Bleecker, A.B. (2004. b). Arabidopsis seedling growth response and recovery to ethylene: A kinetic analysis. Plant Physiol. 136 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet, S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Chen, X., Liu, J., Cheng, Y., and Jia, D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Dong, H.P., Peng, J., Bao, Z., Meng, X., Bonasera, J.M., Chen, G., Beer, S.V., and Dong, H. (2004). Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol. 136 3628–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). RNA-binding proteins in plants: The tip of an iceberg? Curr. Opin. Plant Biol. 5 452–459. [DOI] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26 471–478. [DOI] [PubMed] [Google Scholar]
- Gagne, J.M., Smalle, J., Gingerich, D.J., Walker, J.M., Yoo, S.D., Yanagisawa, S., and Vierstra, R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., and Izaurralde, E. (2004). Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429 575–578. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677. [DOI] [PubMed] [Google Scholar]
- Gutierrez, R.A., Ewing, R.M., Cherry, J.M., and Green, P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. [DOI] [PubMed] [Google Scholar]
- Johnson, P.R., and Ecker, J.R. (1998). The ethylene gas signal transduction pathway: a molecular perspective. Annu. Rev. Genet. 32 227–254. [DOI] [PubMed] [Google Scholar]
- Kastenmayer, J.P., and Green, P.J. (2000). Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 97 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L.E. (2003). Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12 675–687. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Newbury, S., and Woollard, A. (2004). The 5′-3′ exoribonuclease xrn-1 is essential for ventral epithelial enclosure during C. elegans embryogenesis. RNA 10 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N., Kitahata, N., Seki, M., Narusaka, Y., Narusaka, M., Kuromori, T., Asami, T., Shinozaki, K., and Hirayama, T. (2005). Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 44 972–984. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and Song, H. (2004). The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11 121–127. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Spalding, E.P. (1999). Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc. Natl. Acad. Sci. USA 96 14142–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. [DOI] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley, K.A., Byrne, D.H., and Colbert, J.T. (1992). Red light-independent cinstability of oat phytochrome mRNA in vivo. Plant Cell 4 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Haegman, M., Kurepa, J., Van Montagu, M., and Straeten, D.V. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret, F.F., Kastenmayer, J.P., and Green, P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15 173–183. [DOI] [PubMed] [Google Scholar]
- Stoecklin, G., Mayo, T., and Anderson, P. (2006). ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff, D.X., Rockmill, B., Roeder, G.S., and Kolodner, R.D. (1995). The sep1 mutant of Saccharomyces cerevisiae arrests in pachytene and is deficient in meiotic recombination. Genetics 139 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Straeten, D., Djudzman, A., Van Caeneghem, W., Smalle, J., and Van Montagu, M. (1993). Genetic and physiological analysis of a new locus in Arabidopsis that confers resistance to 1-aminocyclopropane-1-carboxylic acid and ethylene and specifically affects the ethylene signal transduction pathway. Plant Physiol. 102 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., and Wilusz, J. (2004). Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20 491–497. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Gong, Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D., and Zhu, J.K. (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1 771–781. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S.D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425 521–525. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- While this manuscript was under review, a report by Olmeda et al. (2006) identified EIN5 as XRN4.
- Olmedo, G., Guo, H., Gregory, B.D., Nourizadeh, S.D., Aguilar-Henonin, L., Li, H., An, F., Guzman, P., and Ecker, J.R. (2006). ETHYLENE INSENSITIVE5 encodes a 5′ →3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103 13286–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.