Abstract
Secondary walls in fibers and tracheary elements constitute the most abundant biomass produced by plants. Although a number of genes involved in the biosynthesis of secondary wall components have been characterized, little is known about the molecular mechanisms underlying the coordinated expression of these genes. Here, we demonstrate that the Arabidopsis thaliana NAC (for NAM, ATAF1/2, and CUC2) domain transcription factor, SND1 (for secondary wall–associated NAC domain protein), is a key transcriptional switch regulating secondary wall synthesis in fibers. We show that SND1 is expressed specifically in interfascicular fibers and xylary fibers in stems and that dominant repression of SND1 causes a drastic reduction in the secondary wall thickening of fibers. Ectopic overexpression of SND1 results in activation of the expression of secondary wall biosynthetic genes, leading to massive deposition of secondary walls in cells that are normally nonsclerenchymatous. In addition, we have found that SND1 upregulates the expression of several transcription factors that are highly expressed in fibers during secondary wall synthesis. Together, our results reveal that SND1 is a key transcriptional activator involved in secondary wall biosynthesis in fibers.
INTRODUCTION
Secondary walls are the major constituent of wood, which is the most abundant biomass produced by plants. Secondary walls are composed mainly of cellulose, lignin, and hemicelluloses, including heteroxylans and glucomannans. During secondary wall formation, the biosynthesis of these cell wall components is highly coordinated. Three cellulose synthase (CesA) genes have been shown to be required for cellulose synthesis in secondary walls of Arabidopsis thaliana and rice (Oryza sativa) (Tanaka et al., 2003; Taylor et al., 2004). Most of the genes involved in the biosynthetic pathway of lignin have been isolated and functionally characterized (Boerjan et al., 2003). Recently, several genes were shown to participate in the biosynthesis of glucomannan and xylan (Dhugga et al., 2004; Liepman et al., 2005; Zhong et al., 2005). In addition, genomic analysis revealed a number of novel glycosyltransferases that might be involved in secondary wall synthesis (Aspeborg et al., 2005; Brown et al., 2005; Persson et al., 2005). Despite these advances in the study of secondary wall biosynthetic genes, little is known about the molecular mechanisms underlying the coordinated expression of these genes during wood formation.
Recent genomic studies in both poplar (Populus species) and Arabidopsis have identified a number of transcription factors that are potentially involved in the differentiation of xylem tracheary elements and fibers. It has been shown that several shoot apical meristem genes are also expressed in vascular cambium (Schrader et al., 2004), indicating a possible conservation of the molecular mechanisms underlying meristematic functions. Transcriptional profiling of genes differentially expressed during in vitro xylem differentiation in Zinnia (Demura et al., 2002) and Arabidopsis (Kubo et al., 2005) or during secondary growth (the formation of both secondary xylem and secondary phloem) in Arabidopsis stems and roots (Oh et al., 2003; Zhao et al., 2005) has led to the identification of diverse families of transcription factors, which are possible regulators of xylem differentiation or secondary growth. Similarly, microarray analysis showed that 182 transcription factors are differentially expressed during different developmental stages of Arabidopsis inflorescence stems (Ehlting et al., 2005). Although the exact functions of most of these xylem- or secondary growth–associated transcription factors are unknown, they provide useful tools to dissect the molecular mechanisms controlling the complex process of xylem development, including the initiation of differentiation, cell elongation, secondary wall thickening, and programmed cell death.
Noteworthy among these xylem- or secondary growth–associated transcription factors are a group of NAC (for NAM, ATAF1/2, and CUC2) domain transcription factors. NAC proteins are plant-specific transcription factors characterized by a conserved NAC domain located at the N-terminal region and a divergent C-terminal activation domain (Olson et al., 2005). In Arabidopsis, ∼105 NAC genes have been identified, and some of them have been shown to play diverse roles in plant growth and development and plant defense (Ooka et al., 2003; Olson et al., 2005). By microarray analysis of genes expressed during in vitro xylem differentiation in Arabidopsis, Kubo et al. (2005) found seven NAC genes, VND1 (for vascular-related NAC domain1) to VND7, whose expression is associated with xylem differentiation. Further study demonstrated that whereas dominant repression of VND6 or VND7 ;resulted in inhibition of the development of metaxylem or protoxylem in roots, respectively, overexpression of VND6 or VND7 led to ectopic differentiation of metaxylem or protoxylem, respectively. It was concluded that VND6 and VND7 are transcriptional switches for metaxylem and protoxylem vessel differentiation.
Recently, two other NAC genes, NST1 (for NAC secondary wall thickening promoting factor1) and NST2, have been shown to be essential for normal anther dehiscence in Arabidopsis (Mitsuda et al., 2005). Anthers develop a layer of secondary wall–containing endothecium during maturation, which is required for the rupture of the stomium during anther dehiscence. Dominant repression or double knockout of NST1 and NST2 causes a loss of secondary wall thickening in the endothecium and, concomitantly, an anther indehiscence phenotype. Overexpression of NST1 or NST2 results in ectopic formation of vessel-like cells, a phenotype similar to that caused by VND6 and VND7 overexpression. It was suggested that NST1 and NST2 are regulators of secondary wall thickening in anther endothecium.
Because secondary walls largely determine wood quality, and the most abundant secondary wall–containing cells in wood of dicot species are fibers, it is essential to reveal transcriptional regulators regulating secondary wall synthesis in fibers. Although a number of genes encoding NAC and other transcription factors have been shown to be upregulated during xylem differentiation and secondary growth, none of them has been demonstrated to regulate secondary wall synthesis in fibers. Arabidopsis inflorescence stems develop interfascicular fibers with massive secondary wall thickening, which can be used as an excellent model to study the transcriptional control of secondary wall synthesis. A previous study demonstrated that INTERFASCICULAR FIBERLESS1 (IFL), a homeodomain Leu-zipper transcription factor, is essential for the normal differentiation of interfascicular fibers in Arabidopsis (Zhong and Ye, 1999). It was shown that IFL1 is involved in the initiation of interfascicular fiber differentiation. In this study, we demonstrate an essential role of a NAC domain transcription factor, SND1, in secondary wall thickening of fibers. We show that expression of the SND1 gene is associated with secondary wall thickening in fibers and that dominant repression of SND1 leads to a severe decrease in the secondary wall thickness of fibers. Overexpression of SND1 activates the expression of secondary wall biosynthetic genes and results in ectopic secondary wall deposition. We further show that the expression of several fiber-associated transcription factors is upregulated by SND1 overexpression. Together, our results indicate that SND1 is a transcriptional switch for the developmental program of secondary wall thickening in fibers.
RESULTS
Developmental Expression of SND1 in Interfascicular Fibers and Xylem Cells
Arabidopsis inflorescence stems develop three to four layers of interfascicular fibers between vascular bundles. After cessation of stem elongation, the interfascicular fiber cells undergo massive secondary wall thickening (Ye et al., 2002). During this stage, genes encoding secondary wall biosynthetic enzymes are coordinately turned on. To study the transcriptional regulators responsible for the coordinated expression of these genes, we searched for transcription factors that are highly expressed in interfascicular fiber cells. Among a group of NAC genes that are expressed at a much higher level in stems than in other organs (Figure 1A), we have found that At1g32770 is specifically expressed in stems with no detectable expression in seedlings, leaves, flowers, and roots (Figure 1C). Further expression analysis in laser-microdissected cell types (Figures 1D and 1E) revealed that although At1g32770 was not expressed in pith parenchyma cells, it was highly expressed in developing interfascicular fibers and xylem cells, two cell types that undergo secondary wall thickening. Because of its close association with the development of secondary wall–containing cells, we named At1g32770 SND1 (for secondary wall–associated NAC domain protein). SND1 belongs to a subgroup of NAC transcription factors that includes VND6, VND7, NST1, and NST2 (Figure 1B; see Supplemental Figure 1 online).
Figure 1.
Gene Expression and Phylogenetic Analyses of NAC Genes.
(A) Preferential expression of a group of NAC genes in stems. The expression data are derived from the AtGenExpress project. Leaves and roots are from 7-day-old plants, and stems are from second internodes of 21-day-old plants.
(B) Phylogenetic analysis of stem-associated NAC genes together with other characterized NAC genes. The branch lengths of the tree are proportional to divergence. The 0.1 scale represents 10% change. Bootstrap values are shown in percentages at nodes.
(C) RT-PCR analysis showing the expression of SND1 in stems but not in other organs. The expression of the EF1α gene was used as an internal control.
(D) Cross section of a stem showing the areas (circled) microdissected by laser for RT-PCR analysis. if, interfascicular fiber; pi, pith cell; xy, xylem bundle.
(E) RT-PCR analysis of laser-microdissected cells showing the expression of SND1 in interfascicular fibers and xylem cells but not in pith cells.
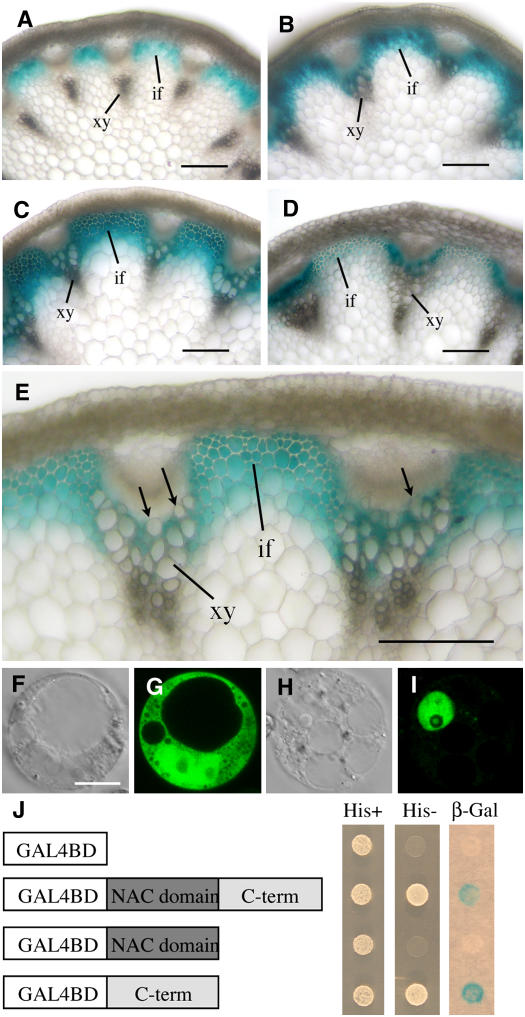
The developmental expression pattern of SND1 was examined in transgenic Arabidopsis plants expressing the SND1 gene fused with the β-glucuronidase (GUS) reporter gene. The SND1 gene used for expression analysis contains a 2.8-kb 5′ upstream sequence, the entire exon and intron region, and a 2-kb 3′ downstream sequence. Therefore, the observed GUS reporter gene expression should represent the endogenous SND1 gene expression pattern. Analysis of transgenic Arabidopsis plants expressing SND1:GUS revealed GUS staining exclusively in stems but not in other organs. Detailed examination in stems showed that in rapidly elongating internodes, the GUS staining was present only in elongating fiber cells in which no secondary wall was evident (Figure 2A). No GUS staining was seen in protoxylem. In internodes that are near the cessation of elongation, GUS staining was observed in both interfascicular fibers and developing metaxylem (Figure 2B). In nonelongating internodes, intensive GUS staining was seen in interfascicular fibers and metaxylem cells, both undergoing massive secondary wall thickening (Figure 2C). Close examination of metaxylem revealed that GUS staining was evident in xylary fiber cells but absent in developing vessels (Figure 2E). At the late stage of stem development, although strong GUS staining was evident in developing secondary xylem, GUS staining in interfascicular fibers became much less intense (Figure 2D). These results demonstrate that the expression of the SND1 gene is associated with the process of secondary wall thickening in interfascicular fibers and xylary fibers. The SND1 protein is targeted to the nucleus when expressed in carrot (Daucus carota) cells (Figures 2F to 2I) and able to activate transcription in yeast cells (Figure 2J), indicating that it is a transcription activator.
Figure 2.
Developmental Gene Expression Pattern and Transcriptional Activation Analysis of SND1.
The expression of the SND1 gene during different stages of stem development was studied using the GUS reporter gene. Transgenic Arabidopsis plants expressing SND1:GUS were examined for GUS activity. For subcellular localization, green fluorescent protein (GFP)–tagged SND1 was expressed in carrot protoplasts and its subcellular location was examined with a laser confocal microscope. For transcriptional activation analysis, full-length or partial sequence of SND1 fused with the GAL4 DNA binding domain was expressed in yeast and tested for the activation of expression of the His3 and LacZ reporter genes. if, interfascicular fiber; xy, xylem. Bars = 150 μm in (A) to (D) and 14 μm in (E) for (E) to (H).
(A) Cross section of a rapidly elongating internode showing GUS staining in developing interfascicular fiber cells.
(B) Cross section of an internode near the cessation of elongation showing GUS staining in both interfascicular fibers and metaxylem.
(C) Cross section of a nonelongating internode showing intensive GUS staining in both interfascicular fibers and metaxylem, both undergoing massive secondary wall thickening.
(D) Cross section of a mature internode showing high GUS staining in secondary xylem but weaker staining in interfascicular fibers.
(E) High magnification of a stem section showing GUS staining in xylary fiber cells but absent in developing vessels (arrows) in metaxylem.
(F) and (G) Differential interference contrast (DIC) image (F) and the corresponding fluorescent signals (G) of a carrot cell expressing GFP alone.
(H) and (I) DIC image (H) and the corresponding fluorescent signals (I) of a carrot cell expressing SND1-GFP. Note that SND1-GFP is targeted to the nucleus.
(J) Transactivation analysis of different regions of SND1 fused with the GAL4 DNA binding domain in yeast. Note that the full-length and C-terminal region of SND1 were able to activate the expression of His3 and LacZ reporter genes.
Dominant Repression of SND1 Causes a Dramatic Reduction in Secondary Wall Thickening of Fiber Cells
To investigate the potential role of SND1 in secondary wall thickening, the wall thickness of interfascicular fibers, xylem vessels, and xylary fibers in the stems of T-DNA knockout lines of SND1 (SALK_015495 and SALK_037737) was examined, and no alterations were observed (data not shown). This finding suggests that other NAC genes might compensate for the loss of SND1 expression. A similar scenario was observed for the VND genes that are expressed during vessel development (i.e., knockout of individual VND genes did not cause any phenotypes) (Kubo et al., 2005). It has been demonstrated that transcriptional factors fused with the EAR repression domain act as dominant repressors to inhibit the expression of their target genes (Hiratsu et al., 2003, 2004), and dominant repression of several redundant NACs, such as RD26, VND6, VND7, NST1, and NST2, results in loss-of-function phenotypes (Fujita et al., 2004; Kubo et al., 2005; Mitsuda et al., 2005). To overcome the problem of genetic redundancy, we used the same dominant repression strategy to study the SND1 functions. SND1 fused with the EAR repression domain was expressed under the control of a cauliflower mosaic virus (CaMV) 35S promoter in transgenic Arabidopsis plants. Among 64 transgenic plants in the first generation, the inflorescence stems of 15 of them did not stand as straight as wild-type stems (Figure 3B). RT-PCR analysis confirmed the presence of the SND1 repressor transcripts in these plants (Figure 3A). Cross sections of the basal part of stems revealed that the wall thickness of interfascicular fibers in the SND1 repressors was reduced drastically compared with that of the wild type (Figures 3E to 3H, Table 1). The wall thickness of xylary fibers was also reduced dramatically, whereas the vessel walls in metaxylem were only slightly affected (Figures 3I to 3L, Table 1). Longitudinal sections of stems showed that the length of fiber cells in the SND1 repressors was not altered compared with that of the wild type (Figures 3C and 3D). These results demonstrate that dominant repression of SND1 causes a specific inhibition in the secondary wall thickening of fiber cells.
Figure 3.
Effects of Dominant Repression of SND1 on Secondary Wall Thickening in Fibers.
The full-length SND1 cDNA was fused in-frame with the dominant EAR repression sequence and transformed into Arabidopsis plants. The phenotypes of the transgenic plants were examined. co, cortex; if, interfascicular fiber; ve, vessel; xf, xylary fiber. Bars = 75 μm in (C) and (D), 35 μm in (E), (F), (I), and (J), and 2.4 μm in (G) for (G), (H), (K), and (L).
(A) RT-PCR analysis showing expression of the SND1 repressor (SND1-SRDX) in the stems of three representative transgenic Arabidopsis lines. The expression of the endogenous SND1 gene (SND1) is shown for comparison.
(B) Wild type plant (left) and a transgenic Arabidopsis plant expressing the SND1 repressor (right).
(C) and (D) Longitudinal sections of the interfascicular region of stems showing the similar length of fiber cells in the wild type (C) and the SND1 repressors (D).
(E) and (F) Cross sections of the interfascicular region showing that the interfascicular fibers of SND1 repressors (F) had thin walls compared with those of the wild type (E).
(G) and (H) Transmission electron micrographs of interfascicular fiber walls of the wild type (G) and the SND1 repressors (H).
(I) and (J) Cross sections of the vascular bundle region of the wild type (I) and the SND1 repressors (J).
(K) and (L) Transmission electron micrographs of xylem cells showing that although the wall thickness of vessels was not changed, that of the xylary fibers was reduced severely in the SND1 repressors (L) compared with the wild type (K).
Table 1.
Wall Thickness of Vessels and Fibers in Stems of the Wild Type, SND1 Repressors, and SND1 Overexpressors
| Plant | Interfascicular Fibers | Vessels | Xylary Fibers |
|---|---|---|---|
| Wild type | 2.14 ± 0.30 | 1.26 ± 0.17 | 0.60 ± 0.13 |
| SND1 repressors | 0.63 ± 0.11 | 1.10 ± 0.28 | 0.33 ± 0.09 |
| SND1 overexpressors | 0.24 ± 0.06 | 1.48 ± 0.36 | 0.12 ± 0.04 |
Wall thickness was measured from transmission electron micrographs of fibers and vessels. Data are means (μm) ± se from 25 cells.
Overexpression of SND1 Induces Ectopic Deposition of Secondary Walls in Normally Nonsclerenchymatous Cells
The dominant repression study suggests that SND1 might be a transcriptional activator involved in secondary wall biosynthesis in fibers. To test whether overexpression of SND1 induces secondary wall synthesis, we generated transgenic Arabidopsis plants with overexpression of SND1. Among 57 transgenic lines, 28 of them exhibited a prominent visual phenotype that shows small rosette size and stunted growth of leaves with severely upward-curling blades (Figures 4B and 4C). RT-PCR analysis demonstrated that although the SND1 mRNA was not detected in wild-type seedlings, it was highly accumulated in the seedlings of these transgenic plants (Figure 4A). Examination of the leaf epidermal cells and mesophyll cells of SND1 overexpressors revealed that a large number of these cells had massive deposition of lignified secondary walls (Figure 5), which was likely the cause of the stunted growth of leaves. The ectopically deposited secondary walls formed thick bands in the epidermis (Figures 5C, 5D, 5G, and 5H) or a reticulated pattern in mesophyll cells (Figures 5E, 5F, 5I, and 5J), which are different from the helical pattern of vessels in wild-type leaves (Figures 5A and 5B).
Figure 4.
Overexpression of SND1 Affects Overall Plant Development.
The full-length SND1 cDNA driven by the CaMV 35S promoter was expressed in transgenic Arabidopsis plants.
(A) RT-PCR analysis showing SND1 overexpression in the seedlings (top panel) and stems (bottom panel) of several representative lines of transgenic plants.
(B) Seedlings of the wild type (left) and an SND1 overexpressor (right).
(C) Leaves of the wild type (left) and SND1 overexpressors (middle and right).
(D) Adult plants of the wild type (left) and an SND1 overexpressor (right).
(E) Flowers of the wild type (left) and an SND1 overexpressor (right).
Figure 5.
Overexpression of SND1 Induces Ectopic Deposition of Lignified Secondary Walls in Epidermal and Mesophyll Cells of Leaves.
Leaves of 3-week-old transgenic plants were observed with a confocal microscope for secondary walls and lignin autofluorescence, and differential interference contrast (DIC) and confocal images were collected. Bars = 42 μm in (A) to (F) and 21 μm in (G) to (J).
(A) and (B) DIC image (A) and lignin autofluorescent signals (B) of a wild-type leaf showing the helical secondary wall thickening in veins.
(C) and (D) DIC image (C) and lignin autofluorescent signals (D) of leaf epidermis of SND1 overexpressors showing the massive ectopic secondary wall thickening.
(E) and (F) DIC image (E) and lignin autofluorescent signals (F) of leaf mesophyll cells of SND1 overexpressors showing the ectopic deposition of secondary walls.
(G) and (H) High magnification of (C) and (D) showing the band-like secondary wall thickening in epidermis.
(I) and (J) High magnification of (E) and (F) showing the reticulated secondary wall thickening in mesophyll cells.
The ectopic secondary wall thickening was also observed in the floral organs and stems of SND1 overexpressors. The flowers of SND1 overexpressors were often sterile and had severely shortened sepals, petals, stamens, and carpels (Figure 4E). In these organs, many cells that are parenchymatous in the wild type had lignified secondary wall thickening in SND1 overexpressors (Figures 6B and 6C; data not shown). In the stems, ectopic lignified secondary wall thickening was evident in many epidermal cells (Figures 6E and 6G) but seldom seen in cortical and pith cells. Similar to those in the epidermal cells of leaves, the ectopic secondary walls in the epidermal cells of stems are deposited as thick bands (Figure 6G). Examination of cotyledons, hypocotyls, and roots of SND1 overexpressors revealed that the ectopic secondary wall thickening was only occasionally observed in the epidermal cells of hypocotyls and cortical cells of roots but was not seen in other cell types of these organs.
Figure 6.
SND1 Overexpression Causes Ectopic Deposition of Lignified Secondary Walls in Normally Nonsclerenchymatous Cells of Flowers and Stems.
For visualization of lignified secondary walls, tissues were stained with phloroglucinol-HCl before observation with a microscope. co, cortex; ep, epidermis; if, interfascicular fiber; ve, vessel; vn, vascular strand; xf, xylary fiber; xy, xylem. Bars = 92 μm in (A) and (B), 32 μm in (C), 160 μm in (D) and (E), 53 μm in (F) and (G), and 4.4 μm in (H) to (K).
(A) Wild-type carpel showing lignin staining in vascular strands.
(B) Carpel of SND1 overexpressors showing ectopic deposition of lignified secondary walls in parenchyma cells.
(C) High magnification of a carpel of SND1 overexpressors showing the helical secondary wall thickening.
(D) and (E) Cross sections of stems showing ectopic lignin staining in the epidermis of SND1 overexpressors (E) compared with the wild type (D).
(F) and (G) Lignin staining of the epidermal peel of stems showing ectopic deposition of thick secondary walls (inset) in SND1 overexpressors (G) compared with the wild type (F).
(H) and (I) Transmission electron micrographs of interfascicular fiber walls of the wild type (H) and SND1 overexpressors (I).
(J) and (K) Transmission electron micrographs of xylem cells of the wild type (J) and SND1 overexpressors (K).
The Secondary Wall Thickening of Fibers Is Drastically Inhibited in SND1 Overexpressors
Despite the fact that many epidermal cells in the stems of SND1 overexpressors had ectopic secondary wall thickening, it was surprising to find that the inflorescence stems were often pendent after they bore siliques (Figure 4D). Cross sections of stems revealed that interfascicular fiber cells, which normally deposit a massive amount of secondary walls in the wild type, had extremely thin walls in the SND1 overexpressors (Figures 6H and 6I, Table 1). Examination of xylem cells showed that xylary fibers, which also have a thick layer of secondary walls in the wild type, nearly lacked secondary walls, whereas the thickness of vessel walls appeared to be increased slightly in the SND1 overexpressors (Figures 6J and 6K, Table 1). Longitudinal sections showed that the length of fiber cells in the SND1 overexpressors was indistinguishable from that of the wild type (data not shown). It was confirmed that SND1 was indeed overexpressed in the stems of the SND1 overexpressors (Figure 4A). These results demonstrate that although SND1 overexpression induces ectopic secondary wall deposition in cells that are normally parenchymatous, excess SND1 apparently inhibits normal secondary wall thickening in fibers.
SND1 Overexpression Induces the Expression of Secondary Wall Biosynthetic Genes and Several Fiber-Associated Transcriptional Factors
To make lignified secondary walls in Arabidopsis, genes involved in the biosynthesis of cellulose, heteroxylan, and lignin need to be switched on. Quantitative PCR analysis showed that the seedlings of SND1 overexpressors had significantly induced expression of the cellulose synthase genes CesA7 and CesA8 (Taylor et al., 2004), the xylan biosynthesis–related gene FRAGILE FIBER8 (FRA8) (Zhong et al., 2005), and the lignin biosynthetic genes 4CL1 (for hydroxycinnamate CoA ligase) and CCoAOMT (for caffeoyl CoA O-methyltransferase) (Boerjan et al., 2003) (Figure 7A), suggesting that the expression of SND1 is capable of turning on a whole set of genes involved in secondary wall synthesis. This is consistent with the data showing that, in addition to lignin (Figures 5 and 6), both cellulose and xylan were abundant in the ectopically deposited secondary walls of epidermis in leaves and stems (Figure 8).
Figure 7.
Real-Time Quantitative PCR Analysis of Genes Induced by SND1 Overexpression.
(A) Relative expression level of genes involved in the biosynthesis of cellulose (CesA7 and CesA8), xylan (FRA8), and lignin (CCoAOMT and 4CL1) and genes involved in programmed cell death (XCP1, XCP2, and BFN1) in the seedlings of SND1 overexpressors compared with the wild type (control). The expression level of each gene in the wild type is set to 1. Error bars represent se of three replicates.
(B) Relative expression levels of fiber-associated transcription factors in the seedlings of SND1 overexpressors compared with the wild type (control).
Figure 8.
Detection of Cellulose and Xylan in Ectopically Deposited Secondary Walls in Leaves and Stems of SND1 Overexpressors.
Thin sections were stained with Calcoflour White for the detection of cellulose or probed with the LM10 xylan monoclonal antibody for the detection of xylan. co, cortex; ep, epidermis; if, interfascicular fiber; xy, xylem. Bars = 186 μm in (A) to (F) and 102 μm in (G) to (L).
(A) and (B) Toluidine blue staining of leaf sections showing thick walls of epidermal cells in SND1 overexpressors (B) compared with the wild type (A).
(C) and (D) Calcoflour White staining of cellulose in leaf sections of the wild type (C) and SND1 overexpressors (D).
(E) and (F) Detection of xylan in leaf sections of the wild type (E) and SND1 overexpressors (F).
(G) and (H) Toluidine blue staining of stem sections of the wild type (G) and SND1 overexpressors (H).
(I) and (J) Calcoflour White staining of cellulose in stem sections of the wild type (I) and SND1 overexpressors (J). The inset shows a high magnification of epidermis with intensive cellulose staining in walls.
(K) and (L) Detection of xylan in stem sections of the wild type (K) and SND1 overexpressors (L). The inset shows a high magnification of epidermis with intensive xylan staining in walls.
It is known that tracheary elements undergo secondary wall thickening followed by programmed cell death, whereas fibers are believed to produce secondary walls without immediate cell death. Because the ectopically deposited secondary walls in SND1 overexpressors shared similar patterns with those of tracheary elements, we were prompted to investigate whether SND1 overexpression also induces programmed cell death. Three hydrolase genes, XYLEM CYSTEINE PEPTIDASE1 (XCP1), XCP2, and BIFUNCTIONAL NUCLEASE1 (BFN1), have been shown to be associated with programmed cell death during xylem differentiation (Funk et al., 2002; Ito and Fukuda, 2002). Quantitative PCR analysis demonstrated that none of these genes was induced by SND1 overexpression (Figure 7A), indicating that SND1 specifically activates the developmental program of secondary wall thickening without induction of programmed cell death. This is consistent with the observation that although the strongly curled leaves of SND1 overexpressors had a large number of cells with massive deposition of secondary walls, they exhibited no chlorosis, indicating that these cells were still alive.
Although SND1 overexpression activates the expression of genes involved in secondary wall synthesis, it seems unlikely that all secondary wall biosynthetic genes are regulated directly by SND1. It has been shown that genes in the lignin biosynthetic pathway are regulated by MYB transcription factors (Tamagnone et al., 1998; Patzlaff et al., 2003; Goicoechea et al., 2005). We propose that different transcriptional regulators mediate the expression of genes participating in the biosynthesis of different secondary wall components, and SND1 is a key regulator coordinating the expression of these transcriptional factors. To test this hypothesis, we examined the expression of several transcription factors that are highly expressed in fibers during secondary wall synthesis (Z.-H. Ye, unpublished data). Quantitative PCR analysis showed that the expression of six transcription factors, including two NAC genes (At4g28500 and At1g28470), three MYB genes (At1g66230, At4g22680, and At1g63910), and a homeobox gene (KNAT7), was induced dramatically in the seedlings of SND1 overexpressors. By contrast, the expression of another fiber-associated homeobox gene, IFL1, was not induced by SND1 overexpression, consistent with the role of IFL1 in the initiation of fiber differentiation rather than in the regulation of secondary wall thickening (Zhong and Ye, 1999). Similarly, expression of the homeobox gene ATHB8, which was proposed to be involved in the early stage of xylem differentiation (Baima et al., 2001), was not induced in the SND1 overexpressors. In addition, it was found that SND1 overexpression did not alter the expression level of VND6 and VND7 genes (data not shown), indicating that the VND6 and VND7 genes are not downstream targets of SND1.
DISCUSSION
Secondary wall deposition is the most prominent hallmark of wood formation. It not only helps vascular plants to build powerful conduits for water transport but also provides strong mechanical force to support the heavy plant body, thus enabling trees to grow up to hundreds of feet tall. Understanding the molecular switches controlling secondary wall deposition in wood is of importance in basic plant biology as well as for potential genetic engineering of wood quality and quantity in tree species. Our finding that SND1 regulates secondary wall synthesis in fibers marks an important step toward the understanding of how secondary wall synthesis is regulated during wood formation.
We have demonstrated that SND1 is a transcriptional activator regulating secondary wall synthesis in fibers based on the following lines of evidence. First, the SND1 gene is specifically expressed in interfascicular fibers and xylary fibers in stems. Second, SND1 is targeted to the nucleus and able to activate transcription. Third, dominant repression of SND1 inhibits secondary wall deposition but not the elongation of fiber cells. Fourth, overexpression of SND1 induces the expression of secondary wall biosynthetic genes, leading to massive ectopic deposition of secondary walls in cells that are normally nonsclerenchymatous. Considering the facts that SND1 is not expressed in vessels and that dominant repression of SND1 causes a dramatic reduction in the secondary wall thickness of fibers but has only a slight effect on the wall thickness of vessels, we suggest that the secondary wall synthesis in vessels of stems is regulated by transcription factors other than SND1. This is consistent with the transcriptional profiling study of genes upregulated during tracheary element differentiation in cultured Arabidopsis cells, in which SND1 is not induced (Kubo et al., 2005). Although SND1 overexpression induces the ectopic deposition of secondary walls in parenchyma cells and causes a slight increase in the wall thickness of vessels, it inhibits secondary wall deposition in fibers. This indicates that a defined level of SND1 is critical for the normal deposition of secondary walls in fibers. There exist precedents for this type of phenomenon. For example, both downregulation and overexpression of the NAP gene, which also encodes a NAC transcription factor, inhibit cell elongation (Sablowski and Meyerowitz, 1998). Another possible explanation is that SND1 overexpression causes cosuppression of the endogenous SND1 gene in fiber cells, thus leading to defective secondary wall synthesis. However, this is unlikely, because the SND1 T-DNA knockout lines do not show any defects in secondary wall synthesis in fibers.
It is interesting that although SND1 overexpression induced many parenchyma cells in leaves and floral organs and epidermal cells in stems to produce secondary walls, ectopic secondary wall deposition was seldom seen in the parenchyma cells of other organs. This finding indicates that parenchyma cells in different organs exhibit differential competence to induction by SND1. The incompetence of some cells to induction by SND1 could be attributable to the lack of another factor that is required for SND1 functions, the presence of an SND1 inhibitor or the rapid degradation of the SND1 protein in these cells. Nevertheless, the finding that SND1 overexpression results in the massive ectopic deposition of secondary walls in many nonsclerenchymatous cells of leaves, stems, and floral organs indicates that the expression of SND1 is sufficient to induce the developmental program for secondary wall synthesis in certain cell types. Overexpression of NST1 or NST2, both of which are required for secondary wall thickening in endothecium of anthers, was also shown to induce the ectopic deposition of secondary walls in certain organs (Mitsuda et al., 2005). Because SND1, NST1, and NST2 are phylogenetically grouped closely to each other, it is likely that they share similar functions, albeit in different cell types. In addition, overexpression of VND6 and VND7, which are also in the same phylogenetic subgroup as SND1, has been demonstrated to induce the ectopic deposition of secondary walls. It was suggested that VND6 and VND7 regulate the differentiation of metaxylem and protoxylem, respectively (Kubo et al., 2005).
It was found that the deposition patterns of the ectopic secondary walls in SND1 overexpressors were different from the uniformly thickened walls of fibers. Instead, they resemble the patterns of tracheary elements. Walls of tracheary elements can have annular, helical, scalariform, reticulate, and pitted patterns. In SND1 overexpressors, the secondary walls in leaf epidermal cells are deposited in thick bands similar to the scalariform pattern, whereas those in leaf mesophyll cells form a reticulated pattern. However, the ectopic secondary wall thickening in the parenchyma cells of carpels tends to have a helical pattern. This indicates that the ectopic secondary walls induced by SND1 overexpression can be deposited in different patterns depending on the cell type; therefore, SND1 may not control the pattern of secondary wall deposition. Because secondary wall deposition is thought to be regulated by cortical microtubules (Baskin, 2001), it is possible that the patterns of ectopic secondary walls induced by SND1 overexpression are dictated by the intrinsic microtubule organization in different cell types.
In conclusion, we have demonstrated that the secondary wall–specific transcription factor, SND1, is essential for normal secondary wall synthesis in fibers and that overexpression of SND1 is able to activate secondary wall biosynthetic pathways, leading to the ectopic deposition of secondary walls. Identification of SND1 as a key switch for the developmental program of secondary wall thickening in fibers opens a new avenue to dissect the transcriptional networks coordinating the complex process of wood formation. Our finding that SND1 induces the expression of secondary wall–associated transcription factors suggests that SND1 is a key regulator in coordinating the expression of other transcription factors, which in turn might activate different sets of genes involved in secondary wall biosynthesis. A GenBank BLAST search revealed that SND1 homologs are present in several other plants species, including cotton (Gossypium hirsutum), poplar, and spruce (Picea abies) (data not shown), indicating that the regulation of secondary wall synthesis by SND1-like genes might be a common mechanism in plants. Because the most abundant secondary wall–containing cells in wood of dicot species are fibers, and wood is the most important raw material for traditional forest products and potentially for biofuel production, the identification of SND1 as a transcriptional switch for secondary wall formation in fibers has significant biotechnological implications.
METHODS
Gene Expression Analysis
The expression data of Arabidopsis thaliana NAC genes in different organs are derived from the AtGenExpress project (Schmid et al., 2005; http://www.weigelworld.org/resources/microarray/AtGenExpress/).
For RT-PCR analysis, total RNA was isolated using a Qiagen RNA isolation kit. The seedlings used were 2 weeks old. Mature leaves were from 6-week-old plants. Inflorescence apices and mature roots were from 8-week-old plants. Stems 1 and 2 were from 4- and 8-week-old plants, respectively. The purified RNA was treated with DNase I to remove any potential genomic DNA contamination before use for first-strand cDNA synthesis. The first-strand cDNA was used as a template for PCR amplification of the SND1 transcripts. PCR was performed for variable cycles to determine the logarithmic phase of amplifications for the samples. RT-PCR was repeated three times, and identical results were obtained. The expression level of the EF1α gene was used as an internal control to determine the RT-PCR amplification efficiency among different samples.
The interfascicular fiber cells, xylem cells, and pith cells of Arabidopsis stems were captured using the PALM microlaser system (PALM Microlaser Technologies) as described (Nakazono et al., 2003). Internodes near the cessation of elongation from the inflorescence stems of 6-week-old plants were fixed in acetone and sectioned for laser microdissection. Total RNA was isolated from the captured cells using the PicoPure RNA isolation kit (Arcturus) and amplified using the RiboAmp HS RNA amplification kit (Arcturus) according to the manufacturer's instructions. The amplified RNA was used for RT-PCR analysis as described above.
The developmental expression pattern of the SND1 gene was studied using the GUS reporter gene. A 6-kb genomic DNA fragment containing a 2.8-kb 5′ upstream sequence, the entire SND1 exon and intron region, and a 2-kb 3′ downstream sequence was used. The GUS reporter gene was inserted in-frame just before the stop codon of the SND1 gene and then cloned into the binary vector pBI101 (Clontech) to create the SND1:GUS construct. The construct was transformed into Arabidopsis plants by the Agrobacterium tumefaciens–mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and used for expression analysis of the GUS reporter gene as described previously (Zhong et al., 2005). The plants used for GUS staining were ∼12 cm tall, and the sections were from internodes located ∼2, 5, 8, and 11 cm from the apex.
For quantitative PCR analysis, total RNA isolated from 2-week-old seedlings of wild-type plants and SND1 overexpressors was treated with DNase I and used for first-strand cDNA synthesis. The first-strand cDNA was used as a template for real-time quantitative PCR analysis with the QuantiTect SYBR Green PCR kit (Qiagen). PCR was performed in the Smart Cycler (Cepheid). Relative mRNA levels were determined by normalizing the PCR threshold cycle number of each gene with that of the EF1α reference gene. The expression level of each gene in the wild-type control was set to 1, and the data were the average of triplicate samples. The primers used for PCR analysis are as follows: CesA7 (5′-TTGTTGCAGGCATCTCAGATG-3′ and GCAGTTGATGCCACACTTGGA-3′), CesA8 (5′-TGAGCTTTACATTGTCAAATG-3′ and 5′-GCAATCGATCAAAAGACAGTT-3′), FRA8 (5′-GACTTGTTGAATCGGTGGCTC-3′ and 5′-GAAAGAGTTTGACCTTCTAAC-3′), CCoAOMT (5′-TCGTTGATGCTGACAAAGACA-3′ and 5′-ACTGATCCGACGGCAGATAG-3′), 4CL1 (5′-GGTTACCTCAACAATCCGGCA-3′ and 5′-CAAATGCAACAGGAACTTCAC-3′), XCP1 (5′-CAGAGGAACAATGAGATCAAC-3′ and 5′-CCCAACAGCTACCACATTGAC-3′), XCP2 (5′-GATGAGACTAACAAGAAAGGG-3′ and 5′-GCCCAACAACTTCCACAAGAG-3′), BFN1 (5′-CGTGGACAGAATGCAACGATC-3′ and 5′-ACCAGCAATAGCATGATCGTC-3′), IFL1 (5′-CCAAGCTGTGAATCTGTGGTC-3′ and 5′-CGATCTTTGAGGATCTCTGCA-3′), ATHB8 (5′-CTCAACATCAGCCTCGTGATG-3′ and 5′-TCCAGAGATCTGCAATCACGC-3′), At4g28500 (5′-CAGACTCAACCACGTCAATGC-3′ and 5′-AGGGATAAAAGGTTGAGAGTC-3′), At1g28470 (5′-CTACCAAACACAGCCTAGGCA-3′ and 5′-ATTCTTCATGAAGCTTTC-3′), At1g66230 (5′-AACAATGTCTTCATCCACGTC-3′ and 5′-CTAAGCTATGATCAATGGGAC-3′), At4g22680 (5′-TGTCTCAGTGGAACCAAAGAC-3′ and 5′-CCCAAAATCATGAACACCAAA-3′), At1g63910 (5′-CTAAGCTATGATCAATGGGAC-3′ and 5′-CGAAGAAGGAAGAGAAGAAGA-3′), VND6 (5′-CCCAACTACAATAATGCAAC-3′ and 5′-GCTCATGATTAGCTGAGAA-3′), VND7 (5′-GGACGAATAAAGATCAGAACGA-3′ and 5′-ATGCGGATGTATGACTTGTGTC-3′), and KNAT7 (5′-CGAGAATCGAAGATGTAAGAG-3′ and 5′-GTGTTTGCGCTTGGACTTCAA-3′).
Phylogenetic Analysis
The phylogenetic relationship of NAC proteins was analyzed with ClustalW version 1.75 (Thompson et al., 1994) and parsimony analysis. The phylogenetic tree was displayed using the TREEVIEW program (Page, 1996) in PHYLIP format and bootstrapped using 1000 bootstrap trials.
Dominant Repression of SND1
The full-length coding region of the SND1 cDNA was fused in-frame with the dominant EAR repression sequence (Hiratsu et al., 2004) and placed under the control of the CaMV 35S promoter in the binary vector pBI121 (Clontech). The construct was transformed into wild-type Arabidopsis plants. Transgenic plants were selected on kanamycin and examined for their phenotypes.
Overexpression of SND1
The full-length SND1 cDNA was ligated downstream of the CaMV 35S promoter in pBI121 and transformed into wild-type Arabidopsis plants. Transgenic plants were selected on kanamycin and used for further analysis.
Protein Targeting and Transcriptional Activation Analysis
The subcellular localization of SND1 was done by expression of GFP-tagged SND1 in carrot (Daucus carota) protoplasts. The full-length SND1 cDNA was fused in-frame with the GFP cDNA (ABRC; developed by S.J. Davis and R.D. Vierstra) and ligated between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 (Clontech). The SND1-GFP construct was transfected into carrot protoplasts according to Liu et al. (1994). The transfected protoplasts were incubated in darkness for 20 h before examination using a Leica TCs SP2 spectral confocal microscope (Leica Microsystems). Images were saved and processed with Adobe Photoshop version 7.0 (Adobe Systems).
For transcriptional activation analysis, the full-length or partial sequence of SND1 cDNA was fused in-frame with the GAL4 DNA binding domain in the pAS2-1 vector (Clontech). The constructs were transformed into yeast strain CG-1945 containing the His3 and LacZ reporter genes. The transformed yeast cells were grown on SD plates with or without His and subjected for β-galactosidase assay.
Microscopy
The basal part of inflorescence stems from 10-week-old plants was fixed in 2% formaldehyde and embedded in low-viscosity (Spurr's) resin (Electron Microscopy Sciences) (Taylor and Mims, 1991). One-micrometer-thick sections from embedded tissues were cut with a microtome, stained with toluidine blue, and observed with a bright-field light microscope. For transmission electron microscopy, 85-nm-thick sections were cut, poststained with uranyl acetate and lead citrate, and observed using a Zeiss EM 902A transmission electron microscope (Carl Zeiss).
Examination of Secondary Walls
For visualization of ectopically deposited secondary walls in SND1 overexpressors, methanol-cleared leaves were observed with a confocal microscope. Both differential interference contrast and lignin autofluorescence images were collected. The flower and stem samples were stained with phloroglucinol-HCl and observed using a microscope.
For detection of cellulose in the ectopically deposited secondary walls, one-micrometer-thick sections of leaves and stems were stained with 0.01% Calcoflour White and observed with a UV fluorescence microscope as described (Hughes and McCully, 1975). Under the conditions used, only secondary walls exhibited brilliant fluorescence.
For detection of xylan in the ectopically deposited secondary walls, 1-μm-thick sections of leaves and stems were incubated with the LM10 monoclonal antibody (McCartney et al., 2005) and fluorescein isothiocyanate–conjugated secondary antibodies. The fluorescence-labeled xylan signals were visualized with a confocal microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SND1 (EF101892), CesA7 (NM_121748), CesA8 (NM_117994), FRA8 (DQ182567), IFL1 (AF188994), ATHB8 (NM_119441), KNAT7 (AF308451), CCoAOMT (NM_119566), 4CL1 (NM_179462), XCP1 (NM_202958), XCP2 (NM_101938), BFN1 (NM_100991), AtNAC1 (NM_179486), CUC1 (NM_112380), CUC2 (NM_124774), CUC3 (NM_106292), TIP (NM_122367), NST1 (NM_130243), NST2 (NM_116056), VND1 (NM_127362), VND2 (NM_119783), VND3 (NM_126028), VND4 (NM_101098), VND5 (NM_104947), VND6 (NM_125632), VND7 (NM_105851), NAM (NM_104166), ATAF1 (NM_100054), and ATAF2 (NM_147856).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Alignment of the Deduced Protein Sequences of Arabidopsis NAC Genes Used for the Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank E.A. Richardson and G. Zhou for their help with microscopy, J.H. Leebens-Mack for his help with phylogenetic analysis, and the editors and reviewers for their constructive comments and suggestions. This work was supported by grants from the U.S. Department of Energy, Bioscience Division (Z.-H.Y.) and the Japan Society for the Promotion of Science (T.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zheng-Hua Ye (zhye@plantbio.uga.edu).
Online version contains Web-only data.
References
- Aspeborg, H., et al. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215 150–171. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A.H., Ellis, J., Goodacreb, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura, T., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. USA 99 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G.S., Dolan, M., Kinney, A.J., Tomes, D., Nichols, S., and Anderson, P. (2004). Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303 363–366. [DOI] [PubMed] [Google Scholar]
- Ehlting, J., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42 618–640. [DOI] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., Tran, L.-S.P., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 863–876. [DOI] [PubMed] [Google Scholar]
- Funk, V., Kositsup, B., Zhao, C., and Beers, E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128 84–94. [PMC free article] [PubMed] [Google Scholar]
- Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B., Verhaegen, D., Chaubet-Gigot, N., and Grima-Pettenati, J. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43 553–567. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321 172–178. [DOI] [PubMed] [Google Scholar]
- Hughes, J., and McCully, M.E. (1975). The use of an optical brightener in the study of plant structure. Stain Technol. 50 319–329. [DOI] [PubMed] [Google Scholar]
- Ito, J., and Fukuda, H. (2002). ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A.H., Wilkerson, C.G., and Keegstra, K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 102 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, L., Marcus, S.E., and Knox, J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53 543–546. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickening and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono, M., Qiu, F., Borsuk, L.A., and Schnable, P.S. (2003). Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: Identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S., Park, S., and Han, K.-H. (2003). Transcriptional regulation of secondary growth in Arabidopsis thaliana. J. Exp. Bot. 54 2709–2722. [DOI] [PubMed] [Google Scholar]
- Olson, A.N., Ernst, H.A., Leggio, L.L., and Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10 79–87. [DOI] [PubMed] [Google Scholar]
- Ooka, H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10 239–247. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L.J., Smith, C., Bevan, M.W., Mansfield, S., Whetten, R.W., Sederoff, R.R., and Campbell, M.M. (2003). Characterization of a pine MYB that regulates lignification. Plant J. 36 743–754. [DOI] [PubMed] [Google Scholar]
- Persson, S., Wei, H., Miline, J., Page, G.P., and Somerville, C.R. (2005). Identification of genes required for cellulose synthesis by repression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92 93–103. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schrader, J., Nilsson, J., Mellerowicz, E., Berglund, A., Nilsson, P., Hertzberg, M., and Sandberg, G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A., and Hirochika, H. (2003). Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J., and Mims, C.W. (1991). Fungal development and host cell responses to the rust fungus Puccinia substriata var. indica in seedlings and mature leaves of susceptible and resistant pearl millet. Can. J. Bot. 69 1207–1219. [Google Scholar]
- Taylor, N.G., Gardiner, J.C., Whiteman, R., and Turner, S.R. (2004). Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11 329–338. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220 225–256. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Pena, M.J., Zhou, G.K., Nairn, C.J., Wood-Jones, A., Richardson, E.A., Morrison, W.H., III, Darvill, A.G., York, W.S., and Ye, Z.-H. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.