Abstract
The roles of two cytosolic maize glutamine synthetase isoenzymes (GS1), products of the Gln1-3 and Gln1-4 genes, were investigated by examining the impact of knockout mutations on kernel yield. In the gln1-3 and gln1-4 single mutants and the gln1-3 gln1-4 double mutant, GS mRNA expression was impaired, resulting in reduced GS1 protein and activity. The gln1-4 phenotype displayed reduced kernel size and gln1-3 reduced kernel number, with both phenotypes displayed in gln1-3 gln1-4. However, at maturity, shoot biomass production was not modified in either the single mutants or double mutants, suggesting a specific impact on grain production in both mutants. Asn increased in the leaves of the mutants during grain filling, indicating that it probably accumulates to circumvent ammonium buildup resulting from lower GS1 activity. Phloem sap analysis revealed that unlike Gln, Asn is not efficiently transported to developing kernels, apparently causing reduced kernel production. When Gln1-3 was overexpressed constitutively in leaves, kernel number increased by 30%, providing further evidence that GS1-3 plays a major role in kernel yield. Cytoimmunochemistry and in situ hybridization revealed that GS1-3 is present in mesophyll cells, whereas GS1-4 is specifically localized in the bundle sheath cells. The two GS1 isoenzymes play nonredundant roles with respect to their tissue-specific localization.
INTRODUCTION
The cereals, including maize (Zea mays), wheat (Triticum aestivum), and rice (Oryza sativa), account for 70% of worldwide food production (http://apps.fao.org; Shewry and Jones, 2005). When such crops are grown for protein content, they require large quantities of nitrogenous fertilizers to attain maximal yields. In the past few years, there has been considerable interest in nitrogen use efficiency, which can be defined as the kernel yield per unit of nitrogen (N) in the soil and the N utilization efficiency, which is the yield per N taken up (Good et al., 2004; Hirel and Lemaire, 2005). A number of physiological and agronomic studies have been undertaken to identify which are the limiting steps in the control of N uptake, assimilation, and recycling during plant growth and development (Jeuffroy et al., 2002; Lawlor, 2002), including cereals such as maize (Hirel et al., 2001, 2005b; Gallais and Hirel, 2004), rice (Yamaya et al., 2002), and wheat (Kichey et al., 2006; Lopes et al., 2006). As maize carries out the NAD–malic enzyme type of C4 photosynthesis, it has a greater capacity to assimilate and metabolize C and N compounds than plants undergoing C3 photosynthesis and ultimately has a larger sink for starch and protein storage in the seeds (Oaks, 1994). Using maize as a model crop, Hirel et al. (2005a, 2005b) have investigated the changes in metabolite concentration and enzyme activities involved in N metabolism within a single leaf at different stages of leaf growth and at different periods of plant development during the kernel-filling period. It was concluded that total N, chlorophyll, soluble protein content, and Gln synthetase (GS) activity are strongly interrelated and are indicators that mainly reflect the metabolic activity of individual leaves with regard to N assimilation and recycling, whatever the level of N fertilization. In maize, the putative role of GS for kernel productivity has also been highlighted using quantitative genetic approaches, since quantitative trait loci (QTL) for the leaf enzyme activity have been shown to be coincident with QTL for yield. One QTL for a thousand kernel weight was coincident with the Gln3 locus (corresponding to the gene encoding cytosolic GS Gln1-4 studied in this work; Hirel et al., 2001), and two QTL for a thousand kernel weight and yield were coincident with the Gln4 locus (corresponding to the gene encoding cytosolic GS Gln1-3 studied in this work; Hirel et al., 2001). Such strong coincidences are consistent with the positive correlation observed between kernel yield and GS activity (Gallais and Hirel, 2004). Further QTL for GS gene loci have been identified in relation to remobilization of N from the leaf, stem, and whole plant, postanthesis N uptake (Gallais and Hirel, 2004), and germination efficiency (Limami et al., 2002).
Since all of the N in a plant, whether derived initially from nitrate, ammonium ions, N fixation, or generated by other reactions within the plant that release ammonium (decarboxylation of Gly during photorespiration, metabolism of N transport compounds, and the action of Phe ammonia lyase), is channeled through the reactions catalyzed by GS, it is somehow logical to find that in cereals, the enzyme is likely to be a major checkpoint controlling plant growth and productivity (Miflin and Habash, 2002; Hirel et al., 2005b; Tabuchi et al., 2005; Kichey et al., 2006). Thus, an individual N atom can pass many times through the GS reaction (Coque et al., 2006), following uptake from the soil, assimilation, and remobilization (Gallais et al., 2006) to final deposition in a seed storage protein.
GS in higher plants, including maize, can be readily separated by standard chromatographic, localization, and protein gel blotting techniques into cytosolic (GS1) and plastidic (GS2) forms (Hirel and Lea, 2001). However, this distinction is not as simple as was first thought because although only one gene has been shown to encode the plastidic form, a small family of up to five genes is now known to encode the cytosolic form (Cren and Hirel, 1999). Initial experiments indicated that the five cytosolic GS genes were differentially expressed in the roots, stems, and leaves of maize (Sakakibara et al., 1992, 1996; Li et al., 1993). In a more recent study of maize leaves performed by Hirel et al. (2005b), it was shown that two of the five genes encoding GS1 (Gln1-3 and Gln1-4) were highly expressed regardless of the leaf age and the level of N fertilization, although there was an increase in Gln1-4 transcripts in the older leaves. It has been suggested that Gln1-4 encodes a GS isoform that is involved in the reassimilation of ammonium released during the remobilization of leaf proteins, whereas Gln1-3 encodes a GS isoform, which plays a housekeeping role during plant growth (Hirel et al., 2005a). GS1-2, which is an important GS isoenzyme of the developing kernel, is abundant in the pedicel and pericarp but has also been shown to be present in immature tassels, dehiscing anthers, kernel glumes, ear husks, cobs, and stalks of maize plants (Muhitch et al., 2002; Muhitch, 2003). Compared with the four other genes encoding GS1, Gln1-5 is expressed at a very low level in leaves, roots, and stems (Sakakibara et al., 1992; Li et al., 1993). The plastidic GS (GS2) encoded by Gln2 was only expressed in the early stages of plant development, presumably to reassimilate ammonium released during photorespiration, which is at a much lower rate in a C4 plant compared with a C3 plant (Ueno et al., 2005).
Despite the information available, as outlined above, concerning the expression of the five cytosolic GS1 genes in maize and the evidence of their importance from the demonstration of QTL, the precise role of the individual GS1 isoenzymes in controlling the flux of ammonium into Gln is not known. Using the maize Mutator (Mu) system, individual maize mutants have been isolated that have insertions in three of the five GS1 genes (Hanley et al., 2000; S. Haines and K.J. Edwards, unpublished results). In all cases, the insertion position of the transposon within the GS1 gene is approximately midpoint and as such is expected to eliminate the production of a functional transcript. The insertion lines of Gln1-3∷Mu and Gln1-4∷Mu have undergone extensive backcrossing to the wild-type non-Mu line, and homozygous, heterozygous, and null mutant lines have been obtained. In addition, a double mutant (gln1-3 gln1-4) of the Gln1-3 and Gln1-4 genes has been produced. In this article, we have investigated the role of the cytosolic GS isoenzyme products of the Gln1-3 and Gln1-4 genes by studying the properties of the insertion mutants at the molecular, biochemical, and physiological levels and by examining the impact of the mutation on kernel yield and its components.
RESULTS
Isolation and Characterization of gln1-3– and gln1-4–Deficient Mutants and gln1-3 gln1-4 Double Mutant
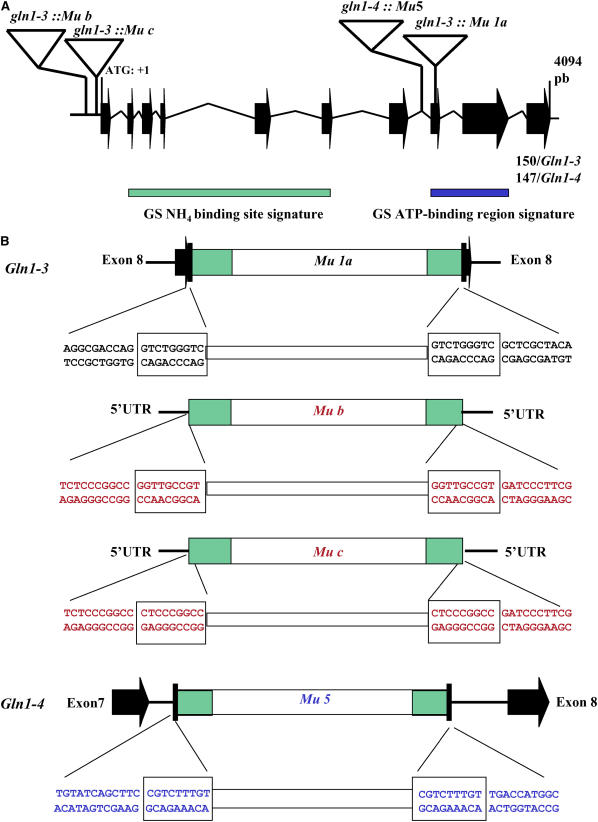
A Mu insertion event within Gln1-3 (Gln1-3∷Mu1a) was first identified using the services of the maize targeted mutagenesis program (http://mtm.cshl.edu/; May et al., 2003), and two further single Mu insertion events (Gln1-3∷Mub and Gln1-3∷Muc) were identified using the collection of mutants produced at Biogemma. The Mu insertion event within Gln1-4 was identified by the random sequencing of Mu-tagged fragments as described by Hanley et al. (2000). Sequence analysis of both events indicated that in the case of gln1-3, Mu1 (mutant a) had inserted within exon 8, and in the case of gln1-4, Mu5 had inserted within the intron separating exons 7 and 8 (Figure 1). In the cases of gln1-3 mutants b and c, Mu had inserted just upstream of the translation initiation codon in the 5′-untranslated region (Figure 1). In both cases, heterozygous plants had similar phenotypes to other randomly chosen Mu-active lines in the genetic background represented by line B73. During the various backcrosses, crosses, and selfings, no distorted segregation patterns were observed as monitored using a PCR assay designed to detect heterozygous and homozygous plants for the Gln1-3 and Gln1-4 insertion events (data not shown). In the case of gln1-3 mutants a and b after three rounds of crosses with the line FV2 and selfing, a segregating very small or aborted kernel phenotype was recorded, thus confirming a similar effect of the mutation on two other allelic mutants in a different genetic background represented by line FV2 (see Supplemental Figure 1 online). In all further experiments, only the homozygous gln1-3a single mutant, which exhibited the strongest phenotype, was studied.
Figure 1.
Characterization of the Gln1-3∷Mu and Gln1-4∷Mu Insertion Events.
(A) Insertion position of the Mu elements within the Gln1-3 and Gln1-4 genes. For clarity and as the Gln1-3 and Gln1-4 genes show high sequence similarity, they are shown as a single structure. The Gln1-3 and Gln1-4 gene structure and exon sizes were determined by sequencing genomic DNA PCR products using primers designed from the corresponding cDNA sequences (Li et al., 1993): the Mu insertion sequences and the rice genomic clone AC105364. The rice and Gln1-3 and Gln1-4 genes consist of 10 exons (black arrows), ranging in size from 40 to 252 bp, and nine introns (black lines). The size of the last exon was 150 bp in Gln1-3 and 147 bp in Gln1-4. The intron and exon structure is drawn to scale. The gene numbering starts with the ATG codon (1 bp) and continues to the stop codon (position 3857 bp). The four Mu insertion events are indicated by triangles (not to scale). Also shown is the relative position of the GS-NH4+ binding site signature (green rectangle) and the GS-ATP binding region signature (blue rectangle).
(B) Sequence characterization of Mu insertions in Gln1-3 and Gln1-4. The nucleotide sequences flanking the 5′ and 3′ ends of the inserted transposons in Gln1-3∷Mu1 and Gln1-4∷Mu5, respectively, are presented and in each case show the Mu-specific 9-bp duplication of the target DNA (boxed). Sequence analysis of the flanking regions surrounding the Mu elements indicated that in the case of Gln1-3∷Mu1a, the insertion had occurred within exon 8, and in the case of Gln1-4∷Mu5, the insertion had occurred within the intron separating exon 7 and exon 8, while Gln1-3∷Mub and c are both located within the 5′-untranslated region. Black lettering indicates sequences derived from exons, and blue lettering indicates sequences derived from introns.
The analysis of genetic background homology is important for establishing the phenotypic compatibility of the wild-type and three mutant lines. Our results suggest that the genetic background of the wild type and the three mutants is similar since the genotyping of the gln1-3 and gln1-4 mutants and gln1-3 gln1-4 double mutant revealed that the rate of homology between the three mutants and the wild type, which should reach a maximum of 90%, was high. However, it was variable according to the mutant examined (67% for the gln1-3 mutant, 79% for the gln1-3 gln1-4 mutant, and 84% for the gln1-4 mutant). When this rate of homology was considered per chromosome, it was between 67 and 100% for the gln1-4 mutant, between 50 and 100% for the gln1-3 gln1-4 mutant, and between 33 and 100% for the gln1-3 mutant. The whole-genome rate of homology between the three mutants was also high but variable, 88% between gln1-4 and gln1-3 gln1-4 and ∼0.50% between gln1-3 and gln1-4 and between gln1-3 and gln1-3 gln1-4 (Table 1).
Table 1.
Whole-Genome Rate of Homology between the Wild Type and the Cytosolic GS-Deficient Mutants
| Wild Type | gln1-4 | gln1-3 | |
|---|---|---|---|
| gln1-4 | 84 | – | 50 |
| gln1-3 | 67 | 50 | – |
| gln1-3 gln1-4 | 79 | 88 | 47 |
The rate of homology is expressed as a percentage of the wild type. It was derived from the analysis of 34 SSR markers and was equal to the number of common SSR alleles divided by the total number of SSRs genotyped (34 without missing data).
GS Gene Transcription, Protein Expression, and Activity in gln1-3, gln1-4, and Double Mutants
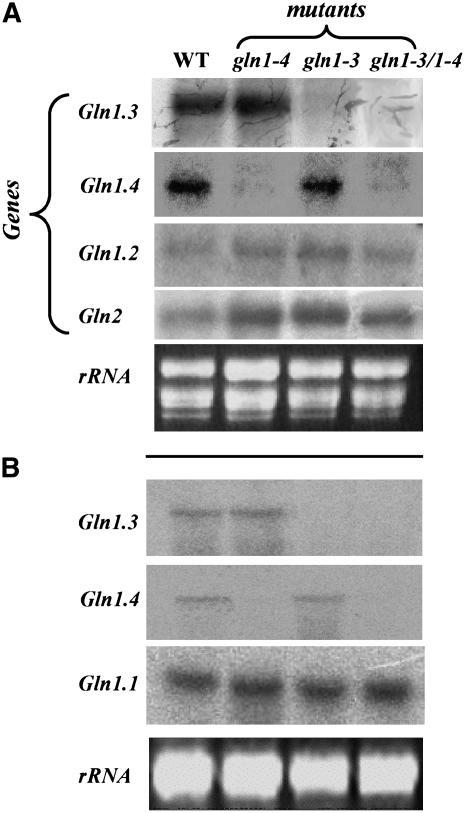
The steady state amounts of the five different mRNAs encoding GS1 were determined in both leaves and roots of the wild type, gln1-3, gln1-4, and gln1-3 gln1-4 mutants by RNA gel blot analysis using 32P-labeled specific 3′ ends of the corresponding cDNAs (Sakakibara et al., 1992; Hirel et al., 2005a). In leaves of wild-type plants at the vegetative stage (VS; 10 to 11 expanded leaves), among the five genes encoding GS1, the highest level of expression was observed for Gln1-3 and Gln1-4. Although present in a lower amount, Gln1-2 mRNAs were slightly more abundant in the three mutants compared with the wild type (Figure 2A). Transcripts for Gln1-1 and Gln1-5 were not detected (data not shown). Transcripts for plastidic GS (GS2, encoded by Gln2) were also present in leaves of young vegetative plants (Figure 2A), and again larger amounts were present in all three mutants when compared with the wild type. Similar results were obtained in leaves of plants harvested 15 or 55 d after silking (DAS), except that the level of expression of Gln2 mRNAs was much lower (data not shown). Compared with the wild type, Gln1-3 transcripts were present in equal amounts in the gln1-4 mutant but were not detectable in either the gln1-3 mutant or the gln1-3 gln1-4 mutant. Similarly, transcripts for Gln1-4 were not detected in either the gln1-4 mutant or the gln1-3 gln1-4 mutant (Figure 2A).
Figure 2.
Steady State Level of Transcripts for GS in GS1-Deficient Mutants.
(A) RNA gel blot showing the steady state level of the genes encoding cytosolic GS (Gln1-2, Gln1-3, and Gln1-4) and plastidic GS (Gln2) in leaves of the wild type and in gln1-3, gln1-4, and gln1-3 gln1-4 mutants.
(B) RNA gel blot showing the steady state level of the genes encoding cytosolic GS (Gln1-1, Gln1-3, and Gln1-4) in roots of the wild type and in gln1-3, gln1-4, and gln1-3 gln1-4 mutants.
The transcripts were detected by hybridization with cDNA 32P-labeled probes corresponding to the 3′ noncoding regions specific for each gene (Sakakibara et al., 1992). Each sample contains 10 μg of total RNA. Ethidium bromide–stained gels (rRNA) are presented at the bottom of each figure to show that similar amounts of total RNA were loaded in each track. Experiments were repeated three times using three different plants with similar results.
In the roots of the wild type, the transcript for Gln1-1 was the most abundant. Gln1-3 and Gln1-4 mRNAs were also detected, but their relative amounts were much lower (Figure 2B). The mRNAs for Gln1-2 and Gln1-5 were not detected (data not shown). As shown above in the leaves, the Gln1-3 transcripts were not present in the roots of gln1-3 and gln1-3 gln1-4 mutants, and Gln1-4 mRNAs were not identified in the roots of the gln1-4 and gln1-3 gln1-4 mutants. The high steady state level of expression of Gln1-1 mRNA was similar in the wild type and all three mutants (Figure 2B).
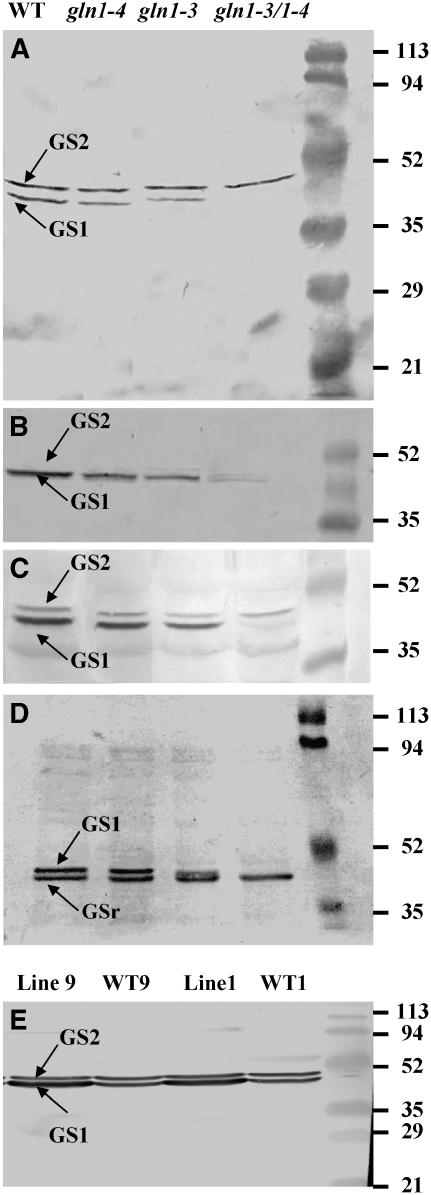
The GS isoenzyme protein content of roots and leaves of the wild type and three mutants was examined following one-dimensional polyacrylamide gel electrophoresis and protein gel blot analysis using antibodies raised against tobacco (Nicotiana tabacum) GS2. This technique provides a reliable method of estimating the relative amounts of both GS1 and GS2 in a crude protein extract of maize leaves (Becker et al., 2000). In the leaves of wild-type plants at VS, two polypeptides of similar relative abundance were detected. The 44-kD polypeptide corresponds to the plastidic form of GS, whereas the 40-kD polypeptide corresponds to the cytosolic form of GS. In the gln1-3 and gln1-4 mutants, a decrease in the amount of GS1 protein was observed, which was more pronounced in the former. In gln1-3 gln1-4, the GS1 protein was barely detectable. Similar amounts of GS2 protein were visible in the wild type and in all three mutants (Figure 3A). The decrease in GS1 protein contents of the gln1-3, gln1-4, and gln1-3 gln1-4 mutants at the VS was confirmed using antibodies raised against Phaseolus vulgaris root nodule GS (Cullimore and Miflin, 1984). However, since this antibody preparation is apparently more specific for cytosolic GS, GS2 protein was less readily visible on the protein gel blot (Figure 3B). When the GS proteins were analyzed in the leaves 55 DAS using the tobacco antibody, a similar pattern of decrease in GS1 protein content was obtained in the mutants. However, at this later stage of plant development, the amount of GS2 protein isolated from all four lines was much lower (Figure 3C).
Figure 3.
GS Subunit Composition of GS1-Deficient Mutants and Overexpressing Lines.
(A) Protein gel blot analysis of the GS subunit composition of leaves of the wild type, gln1-3, gln1-4, and gln1-3 gln1-4 harvested at VS using tobacco GS antibodies (Hirel et al., 1984). The top band (molecular mass of 44 kD) corresponds to the plastidic GS (GS2) subunit, and the bottom band (molecular mass of 39 kD) corresponds to the cytosolic GS (GS1) subunit. On the right side of the panel is the position of the protein molecular mass markers.
(B) Protein gel blot analysis of GS subunit composition of leaves of the wild type, gln1-3, gln1-4, and gln1-3 gln1-4 harvested at VS using P. vulgaris GS antibodies (Cullimore and Miflin, 1984).
(C) Protein gel blot analysis of GS subunit composition in leaves of the wild type, gln1-3, gln1-4, and gln1-3 gln1-4 harvested at 55 DAS using tobacco GS antibodies.
(D) Protein gel blot analysis of GS subunit composition in roots of the wild type, gln1-3, gln1-4, and gln1-3 gln1-4 harvested at VS using tobacco GS antibodies. The two bands (molecular masses of 40 and 38 kD) correspond to the GS1 and GSr forms of cytosolic GS.
(E) Protein gel blot analysis of GS subunit composition of leaves of the two null segregants (WT1 and WT9) and the two transgenic lines (Lines 1 and 9) with expression of Gln1-3 cDNA under the control of the CsVMV promoter. Plants were harvested at VS, and tobacco GS antibodies were used in the experiment. The top band (molecular mass of 44 kD) corresponds to the plastidic GS (GS2) subunit, and the bottom band (molecular mass of 39 kD) corresponds to the cytosolic GS (GS1) subunit.
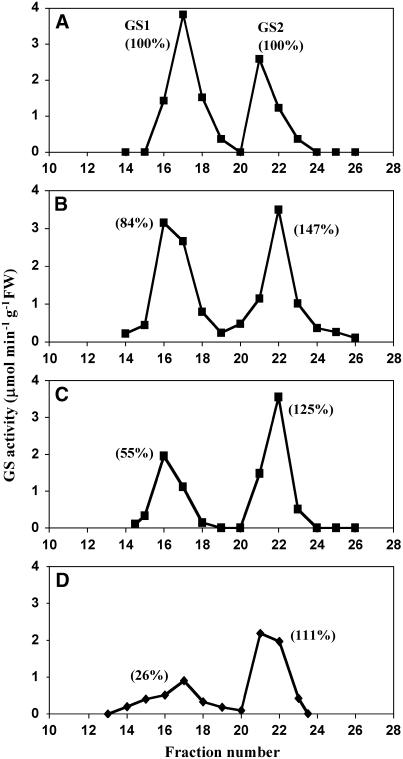
The GS isoenzyme activity content of the wild type and the three mutants was examined following ion exchange chromatography using plants at the VS (Figure 4). Compared with the wild type, a 16% decrease in GS1 activity was observed in the gln1-4 mutant (Figure 4B). The decrease in GS1 activity was higher in the gln1-3 mutant (45%) (Figure 4C). Only 26% of the wild-type GS1 activity remained in the gln1-3 gln1-4 mutant (Figure 4D). Interestingly, an increase in GS2 activity ranging from 11 to 47% was observed in all three mutants (Figure 4).
Figure 4.
GS Isoenzyme Composition of GS1-Deficient Mutants.
Ion exchange chromatography of GS activity in leaf extracts from the wild type (A), gln1-4 (B), gln1-3 (C), and gln1-3 gln1-4 (D). The first peak of GS activity corresponds to cytosolic GS (GS1) and the second peak to plastidic GS (GS2). The relative amounts of GS1 and GS2 activity indicated in parentheses were calculated using as a maximum the value measured for the amount of GS1 and GS2 activities in the wild type. GS activity was measured using the transferase reaction (Lea et al., 1999). Similar results were obtained with three independent extractions. FW, fresh weight.
In the root tissue of wild-type plants, two polypeptides of 38 and 40 kD corresponding to two cytosolic forms of GS were detected (Figure 3D). As already described by Sakakibara et al. (1992), the upper band corresponds to a GS1 protein of a similar size to that found in the leaves, whereas the lower band is a root-specific cytosolic GS (GSr). A small decrease in the amount of the GS1 (40 kD) polypeptide was just visible in the gln1-4 mutant, while a considerable decrease was clearly detectable in the gln1-3 mutant. In the gln1-3 gln1-4 double mutant, the 40-kD GS1 polypetide was not detected. Similar amounts of the GSr (38 kD) polypeptide were detected in the wild type and in all three mutants (Figure 3D). Compared with the wild type, root GS activities in the three mutants measured using the synthetase reaction at the VS were similar (3.2 ± 0.06 μmol·min−1·g−1 dry weight (DW) for the wild type, 3.4 ± 0.3 for the gln1-4 mutant, 2.8 ± 0.05 for the gln1-3 mutant, and 3.1 ± 0.05 for the gln1-3 gln1-4 mutant). This result indicates that root GS activity is mostly represented by the GSr protein, which is encoded by the Gln1-1 gene, the mRNA of which, is most abundantly expressed in roots (Figure 2B).
Identification of GS Protein Sequences
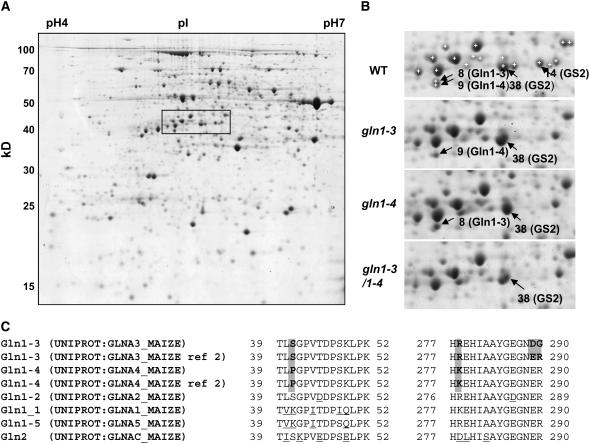
Using plants harvested at the VS, leaf proteins from the wild type, the gln1-3, the gln1-4, and the gln1-3 gln1-4 mutant were separated by two-dimensional gel electrophoresis (Figure 5A). To unambiguously demonstrate that the corresponding GS protein was lacking in the gln1-3 and gln1-4 mutants, 36 protein spots in the range of the pI and molecular mass of GS were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Protein identification revealed that out of these 36 spots, four of them (spots 8, 9, 14, and 38) contained a GS protein.
Figure 5.
Proteomic Analysis of GS1-Deficient Mutants.
(A) Two-dimensional gel of proteins extracted from the leaves of the wild type. The rectangle shows the region containing GS spots that is enlarged in (B).
(B) Identification of GS1-3, GS1-4, and GS2 proteins in the wild type and in gln1-3, gln1-4, and gln1-3 gln1-4. White crosses with no black arrowheads in the wild type show the spots that were analyzed by LC-MS/MS but did not contain a GS protein.
(C) Sequence of the peptides that allowed the distinction between GS1-3 and GS1-4. Entry names of the sequence in the UNIPROT Database are given. Ref 2 of GS1-3 and GS1-4 refers to the allelic form sequenced by Sakakibara et al. (1992). Positions are relative to the first Met for all GS except for GS2, where it is relative to the first amino acid after the signal peptide. Shaded background shows the differences between GS1-3 and GS1-4 and between GS1-3 allelic forms. Differences between GS1-3 and GS1-4 and the rest of GS proteins are underlined.
By comparison with the protein profile of the wild type, it was observed that the gln1-3 mutant and the gln1-4 mutant were lacking spots 8 and 9, respectively, and that they were both absent in the gln1-3 gln1-4 mutant (Figure 5B). Spots 8 and 9 were very close to each other and exhibited a partial overlap. They had the same apparent pI but slightly differed by their apparent molecular mass. This finding is not surprising because according to the sequences of the genes isolated by Li et al. (1993), GS1-3 and GS1-4 proteins have the same pI and exhibit only a 188-D difference in their mass. Both products of the allelic forms isolated by Sakakibara et al. (1992) (GS112 and GS107 for GS1-3 and GS1-4, respectively) have one more basic amino acid and thus also exhibit similar pIs.
Two sites of sequence polymorphism were used to distinguish the GS1-3 and GS1-4 proteins from each other: the amino acid at position 41 is a Ser in GS1-3 and a Pro in GS1-4, and the amino acid at position 278 is an Arg in GS1-3 and a Lys in GS1-4. Peptides containing these sites of polymorphism were identified: TLSGPVTDPSK (and the miscleaved TLSGPVTDPSKLPK) allowed the identification of GS1-3, while TLPGPVTDPSK (and TLPGPVTDPSKLPK) allowed the identification of GS1-4 according to the S/P polymorphism at position 41, and HREHIAAYGEGNER and HKEHIAAYGEGNER allowed the identification of GS1-3 and GS1-4 proteins, respectively, according to the R/K polymorphism at position 278. It should be noted that the peptide HREHIAAYGEGNER is similar to the allelic form of GS1-3 sequenced by Sakakibara et al. (1992) but not to that sequenced by Li et al. (1993), in which the two last amino acids of the peptide, ER, are substituted by DG (Figure 5C).
Most of the GS spots were excised twice, and the numbers of peptides indicated below refer to the analyses that identified the largest number of peptides. According to the presence of the discriminating peptides, the excision of spot 8 from gels from the gln1-4 mutant allowed the identification of the GS1-3 protein (eight GS1 peptides from which three were specific to GS1-3), and the excision of spot 9 from the gln1-3 mutant allowed the identification of the GS1-4 protein (12 GS1 peptides from which three were specific to GS1-4). When excised from the wild type, no discrimination between the two proteins was possible because of a partial overlap of the two spots. Spots 14 and 38, which both exhibit a higher mass compared with GS1, were found to contain the plastidic GS2 protein (9 and 23 peptides allowed the identification of GS2 in spots 38 and 14, respectively). Interestingly, spot 14 was not visible in gln1-3, gln1-4, or the gln1-3 gln1-4 mutant (Figure 5B). It is worth noting that the intensity of spot 38 in the three mutants and the wild type was visibly proportional to GS2 activity (Figure 5).
Immunolocalization of GS
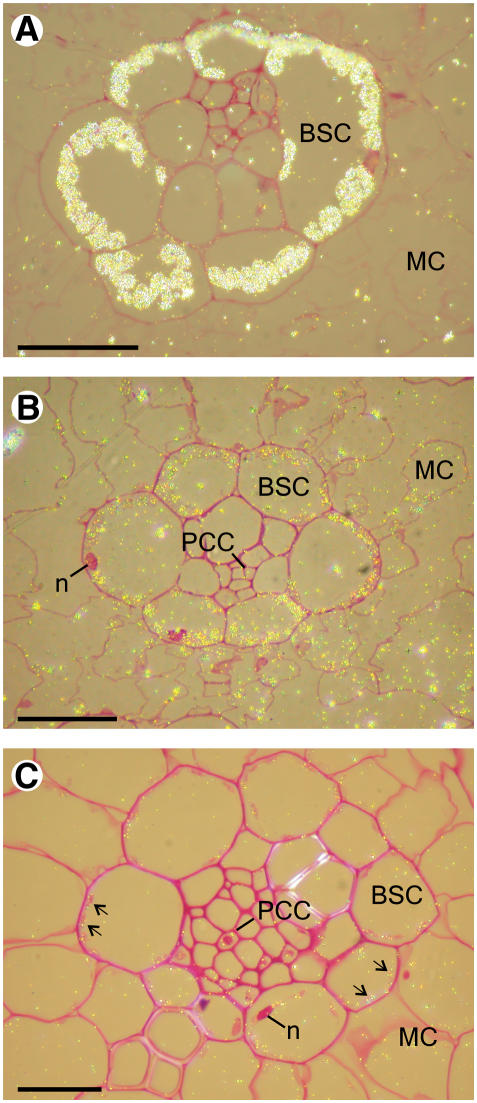
Immunocytochemical studies using both light (Figure 6) and electron microscopy (Figure 7) techniques were employed to determine in which leaf tissue GS was impaired in the three GS mutants. GS was first localized in leaf sections of the wild type and the gln1-3 gln1-4 mutant using immunogold labeling, followed by silver enhancement. A bright-field microscope emitting epipolarized light was used for these experiments, and the silver-enhanced gold particles appeared as a bright yellow color (Figure 6). In the wild-type leaf tissue sections treated with gold-labeled tobacco anti-GS antibodies, the presence of GS both in the bundle sheath cells (BSCs) and mesophyll cells (MCs) was clearly visible. The phloem companion cells (PCCs) located in the central part of the BSC were also faintly stained (Figure 6B). In leaf sections of the gln1-3 gln1-4 mutant, the fluorescent signal was much weaker and apparently restricted to the plastids (arrows) of both BSCs and MCs (Figure 6C). In addition, faint labeling was still detected in the PCC. Similar results were obtained when leaf sections of the gln1-3 and gln1-4 mutants were treated with the gold-labeled GS antiserum (data not shown). However, the resolution of the technique did not allow a clear differentiation of a specific signal in the various cell types and cellular compartments of the three mutants. In a control leaf section treated with ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) antibodies, a fluorescent signal was detected in plastids of the BSC, whereas the MCs were unstained, thus allowing a clear identification of the two cell types (Figure 6A).
Figure 6.
Histological Immunolocalization of Rubisco and GS in Leaf Transversal Sections of the Wild Type and the gln1-3 gln1-4 Mutant Stained by Fuchsin and Examined under Bright-Field Epipolarized Light.
(A) Immunolocalization of Rubisco in the leaf of the wild type.
(B) Immunolocalization of GS in the leaf of the wild type.
(C) Immunolocalization of GS in the leaf of the gln1-3 gln1-4 mutant.
n, nucleus. Bars = 100 μm.
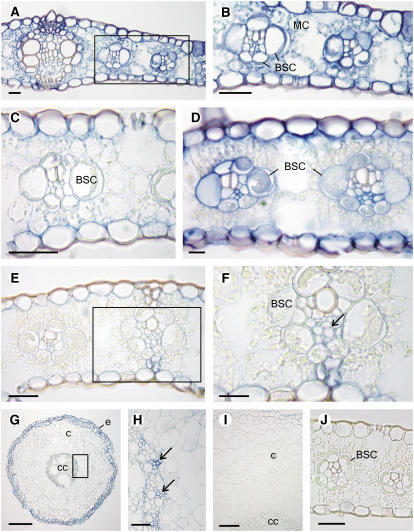
Figure 7.
Transmission Electron Microscopy Immunolocalization of GS in Maize Leaf Sections at VS.
(A) to (C) Localization of GS in parenchyma cells of the wild type (A). Magnification of the zone containing MC (B) and BSC (C).
(D) to (F) Localization of GS in parenchyma cells of the gln1-3 gln1-4 mutant (D). Magnification of the zone containing an MC (E) and a BSC (F).
(G) and (H) Localization of GS in MCs of the gln1-3 mutant (G) and in the BSC of the gln1-4 mutant (H).
(I) and (J) Localization of GS in PCC in the wild type (I) and in the gln1-3 gln1-4 mutant (J).
(K) Control section in the wild type treated with preimmune serum.
cy, cytosol; p, plastid; m, mitochondrion; s, starch granules. Arrows indicate the presence of GS protein.
Bars = 1 μm in (A) and (C) and 0.5 μm in (B) and (D) to (K).
To improve the resolution of the immunolocalization technique, immunogold transmission electron microscopy experiments were performed on leaf blade sections of the wild type and the three mutants to refine the cellular and subcellular localization of GS. In the wild type, GS was located both in the cytosol and the plastids of the MC and of the BSC (Figures 7A to 7C). However, while the amount of plastidic GS protein was similar in the MC and in the BSC, the amount of GS in the cytosol in the MC (Figure 7B) was less than half that present in the BSC (Figure 7C). Quantification of gold particles confirmed these observations of differences (Table 2). In the wild type and the three mutants, similar amounts of GS protein were detected in the plastids (Table 2). In the gln1-3 gln1-4 mutant (Figures 7D to 7F), a considerable decrease in the amount of cytosolic GS was observed both in the MC (Figure 7E) and the BSC (Figure 7F). In the gln1-3 mutant, the decrease in cytosolic GS protein was restricted to the mesophyll (Figure 7G, Table 2), whereas in the gln1-4 mutant, the amount of cytosolic GS was decreased only in the BSC (Figure 7H, Table 2).
Table 2.
Quantification of GS Protein in Different Tissue Sections of a Leaf
| No. of Gold Particles/μm−2
|
|||||
|---|---|---|---|---|---|
| Cytosol
|
Plastid
|
||||
| BSC | MC | PCC | BSC | MC | |
| Wild type (line B73) | 24.9 ± 6.1 | 10.4 ± 3.9 | 10.8 ± 3.1 | 8.9 ± 3.1 | 9.1 ± 3.3 |
| gln1-4 | 6.5 ± 3.7a | 8.2 ± 1.6b | 10.9 ± 3.6b | 9.6 ± 5.4b | 10.1 ± 3.8b |
| gln1-3 | 20.1 ± 4.2b | 5.4 ± 3.0a | 9.6 ± 1.8b | 11.3 ± 3.6b | 9.6 ± 4.3b |
| gln1-3 gln1-4 | 3.6 ± 2.3a | 1.6 ± 0.8a | 11.4 ± 3.3b | 9.9 ± 2.9b | 8.3 ± 0.7b |
| Wild type (line FV2) | 19.5 ± 6 | 9.5 ± 2.5 | 11.0 ± 3.0 | 7.0 ± 2.0 | 8.5 ± 2.5 |
| CsVMV-Gln1-3 | 28.0 ± 8.2b | 19.5 ± 6.8a | 25.5 ± 4.0a | 7.2 ± 2.5b | 8.5 ± 3.0b |
Immunolocalization of the enzyme was performed using transmission electron microscopy on leaves at the VS. Values are the mean ± sd of gold particles counted on 10 to 15 different sections.
Significantly different from the wild type.
Not significantly different from the wild type at 0.05 probability level.
Similar amounts of cytosolic GS were detected in the PCC of the wild type (Figure 7I) and in the gln1-3 gln1-4 mutant (Figure 7J). Quantification of gold particles in the three mutants confirmed this observation (Table 2). A control section treated with preimmune serum shows that both the cytosol and the plastids of a MC were free of label (Figure 7K).
In Situ Hybridization of Cytosolic GS Transcripts
In situ hybridization was performed in leaves of the wild type and the three GS mutants to confirm at the transcript level that Gln1-4 and Gln1-3 were expressed in two different cell types. Tissue sections of the wild type were hybridized with digoxygenin-labeled sense and antisense RNA probes from the coding region of Gln1-3, which shares >85% homology with the four other genes encoding GS1 (Sakakibara et al., 1992). In all tissue sections, hybridization of the probe was observed by the presence of a blue signal corresponding to the reaction catalyzed by the alkaline phosphatase. In transverse leaf sections of the wild type, the antisense probe from the coding region of Gln1-3 hybridized with all the different leaf tissues, including the MC, the BSC, and the two epidermal cell layers (Figures 8A and 8B). In the gln1-4 mutant, there was no significant hybridization signal in the BSC (Figure 8C). In the gln1-3 mutant, the signal was much stronger in the BSC (Figure 8D). In the gln1-3 gln1-4 double mutant, the GS1 probe hybridized with some of the vascular cells present in the BSC, whereas both the MC and BSC remained unstained (Figures 8E and 8F). In the three mutants, a weak signal was always visible in both the lower and upper epidermis (Figures 8C to 8E).
Figure 8.
In Situ Localization of Cytosolic GS Transcripts in Leaf and Root Sections of the Wild Type and the gln1-4, gln1-3, and gln1-3 1-4 Mutants.
(A) and (B) Localization of GS1 transcripts in the leaves of the wild type (A). Magnification of the zone containing the MC and BSC cells (B).
(C) Localization of GS1 transcrips in the gln1-4 mutant showing the absence of staining in the BSC.
(D) Localization of GS1 transcripts in the gnl1-3 mutant showing staining in the BSC.
(E) and (F) Localization of GS1 transcripts in the gln1-3 gln1-4 mutant.
(F) Magnification of the zone containing the BSC showing the presence of staining in the vascular tissue (arrow).
(G) and (H) Localization of GS1 transcripts in the roots of the wild type showing the presence of a strong signal in the epidermis and in the two outermost cortical cell layers.
(H) A magnification of the root central cylinder showing the presence of signal in the vascular tissue (arrows).
(I) Transverse section of a root tissue hybridized with the digoxygenin-labeled sense probe showing the absence of signal.
(J) Transverse section of a leaf tissue hybridized with the digoxygenin-labeled sense probe showing the absence of signal.
c, cortex; cc, central cylinder; e, epidermis. Bars = 100 μm.
In transverse root sections of the wild type, a strong hybridization signal was detected in the epidermis and the two outermost cell layers of the cortex (Figure 8G). The GS probe from the coding region of Gln1-3 also weakly hybridized with some of the cells located in the central cylinder (Figure 8H). Similar results were obtained with the three mutants (data not shown). No hybridization was observed when similar root (Figure 8I) or leaf sections (Figure 8J) were incubated with the sense probe.
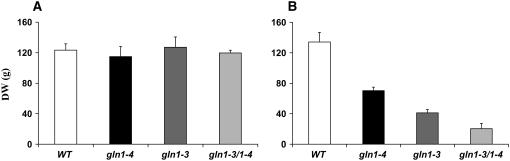
Phenotype and Kernel Production
To determine the impact of the mutations on plant phenotype and kernel production, plants were grown in a glasshouse until maturity under suboptimal N feeding conditions. No significant differences were seen in the dry matter production of the vegetative parts of the shoot (Figure 9A), which exhibited a similar phenotype (Figure 10). By contrast, a major decrease in the dry matter content of the ear was observed, which when compared with the wild type was 68% for gln1-3, 48% for the gln1-4 mutant, and 84% for the gln1-3 gln1-4 mutant. A reduction of ear size was observed in gln1-3 and gln1-4 mutants and was even more severe in the gln1-3 gln1-4 mutant (Figure 9B). The main agronomic characteristics of the three GS mutants grown under N suboptimal conditions are presented in Table 3. In both gln1-3 and gln1-4 mutants, a strong reduction in kernel yield was observed, which was more important in the former, thus confirming the phenotype shown in Figure 11A. In the gln1-3 gln1-4 mutant, the kernel yield was reduced to only 12% of the wild type (Table 3). In gln1-4, there was a greater reduction in kernel weight but a larger kernel number when compared with gln1-3. In the gln1-3 gln1-4 mutant, both yield components were strongly reduced compared with the wild type and the two single mutants (Table 3). Compared with the wild type, an ∼20% increase in N kernel content was observed in the three mutants (Table 3).
Figure 9.
Shoot and Ear DW Accumulation in GS1-Deficient Mutants.
(A) Total DW of shoot vegetative parts of wild type, gln1-3, gln1-4, and gln1-3 gln1-4. Plants were harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
(B) Total ear DW of wild type, gln1-3, gln1-4, and gln1-3 gln1-4. Plants were harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
The plants were grown in the glasshouse on soil and watered daily with the medium described by Coïc and Lesaint (1971). Values are the mean ± se of three individual plants.
Figure 10.
Phenotype of the Shoot in GS1-Deficient Mutants.
The wild type, gln1-3, gln1-4, and gln1-3 gln1-4 maize plants (line B73) at a developmental stage corresponding to 55 d after flowering. Plants were grown in the glasshouse on soil and watered daily with the medium described by Coïc and Lesaint (1971). Note that both the number of remaining leaves and the position of the ear were similar in the wild type and the three mutants.
Table 3.
Kernel Yield and Its Components in Maize Mutants Deficient in Cytosolic GS and Transgenic Plants Overexpressing the Enzyme
| Grain Yield (g) | Grain Number | Thousand Kernels Weight (g) | Grain N Content (% DW) | |
|---|---|---|---|---|
| Wild type (line B73) | 117 ± 15 (100) | 464 ± 63 (100) | 252 ± 12 (100) | 2.10 ± 0.12 (100) |
| gln1-4 | 59 ± 4.5 (50)a | 331 ± 25 (71)a | 178 ± 11 (70)a | 2.49 ± 0.04 (118)a |
| gln1-3 | 27 ± 9.5 (23)a | 120 ± 43 (26)a | 220 ± 5 (87)a | 2.66 ± 0.07 (126)a |
| gln1-3 gln1-4 | 14 ± 8 (12)a | 89 ± 51 (19)a | 158 ± 4 (63)a | 2.48 ± 0.08 (118)a |
| Wild type (line FV2) | 26.05 ± 3.5 (100) | 102.5 ± 17.4 (100) | 259.9 ± 8.2 (100) | 2.16 ± 0.02 (100) |
| CsVMV-Gln1-3 | 36.9 ± 3.8 (142)a | 127.9 ± 13.6 (125)a | 276.9 ± 6.7 (106) | 2.19 ± 0.06 (101) |
Each value is the mean ± sd obtained from three individual plants for the mutants and six individual plants for the transgenics grown on soil in a glasshouse and watered daily with the medium described by Coïc and Lesaint (1971). The values indicated in parentheses are expressed as percentages of the value in the wild type. For the plants overexpressing Gln1-3, the values are the mean of the two transgenic lines and of the corresponding null segregants.
Significantly different from the wild type at 0.05 probability level.
Figure 11.
Phenotype of the Ear in GS1-Deficient Mutants and Overexpressing Lines.
(A) Ears of wild type, gln1-3, gln1-4, and gln1-3 gln1-4 of maize plants (line B73) harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
(B) Ears of wild-type null segregants and T4 transgenic lines (Lines 1 and 9) overexpressing the Gln1-3 cDNA. Maize plants (line FV2) were harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
The ear in the control null segregants WT1 and WT9 was smaller compared with that of the wild type shown in (A). This is due to the fact that the European line FV2 used for generating the transgenic maize plants produces fewer kernels than the North American line B73.
Physiology of the GS1-Deficient Mutants
The effect of decreased levels of GS1 activity on representative markers of leaf N and C metabolism (Hirel et al., 2005b) was examined in the three maize mutants. At the VS, the effect of the reduction in GS1 activity on the level of free ammonium ions (NH4+) was investigated. Table 4 shows that the reduction in GS1 activity in gln1-3 and gln1-4 mutants resulted in an almost twofold increase in the quantity of free NH4+ in the leaves, while in the gln1-3 gln1-4 mutant, this amount was approximately four times higher compared with the wild type. At the VS, no significant differences in the NO3−, soluble sugars, free amino acids (both qualitatively and quantitatively), and total N contents of the leaves were observed between the wild type and the three mutants (Table 4). Although less marked, an increase in the quantity of free NH4+ was still observed 15 DAS in the three GS mutants. At this stage of plant development, the relative concentrations of NO3−, amino acids, and soluble sugars were also reduced, but no significant changes between the wild type and the mutants were detected (data not shown). As reported earlier (Hirel et al., 2005b), the amounts of NO3−, NH4+, and soluble sugars in the leaves 55 DAS were at least four times lower in both the wild type and the three GS mutants. However, no marked differences were observed between the wild type and the three mutants. By contrast, at the 55-DAS stage of plant development, there was a marked increase in the leaf total amino acid content of the leaves of the three mutants (Table 5). This increase was due in part to an increase in Asn and Glu contents, which were up to 20- and 10-fold higher, respectively, in the gln1-3 gln1-4 mutant. An increase in the amount of total N was also observed in the leaves of the three mutants (Table 5).
Table 4.
Concentration of Nitrogen and Carbon Metabolites in a Mature Leaf of Maize Mutants Deficient in Cytosolic GS
| Metabolite Concentration (μmol g DW−1) and Total N (%)
|
||||
|---|---|---|---|---|
| Metabolites | Wild Type | gln1-4 | gln1-3 | gln1-3 gln1-4 |
| NH4+ | 24.22 ± 1.9 | 41.5 ± 3.1a | 43.6 ± 2.3a | 99.15 ± 11.4a |
| NO3− | 64 ± 2.5 | 57.3 ± 11.4 | 61.5 ± 6.7 | 64 ± 2.1 |
| Soluble sugars | 2350 ± 110 | 1930 ± 220 | 1980 ± 410 | 2030 ± 306 |
| Amino acids | 73.20 ± 17 | 93.03 ± 6.5 | 83.1 ± 3 | 98.9 ± 30 |
| Total N | 4.36 ± 0.08 | 4.31 ± 0.1 | 4.28 ± 0.1 | 4.42 ± 0.3 |
Metabolites were quantified in maize plants at VS. The plants were grown hydroponically on the medium described by Coïc and Lesaint (1971). Values are the mean of three plants ± sd. For soluble sugars, sucrose represented 98% of the total.
Significant changes at the 0.05 probability level compared with the wild type.
Table 5.
Concentration and Proportion of Amino Acids and Nitrogen Content in the Leaf below the Ear of the Wild Type and Cytosolic GS-Deficient Mutants
| Amino Acid Concentration (μmol g DW−1) and Proportions (%)a
|
||||
|---|---|---|---|---|
| Amino Acid | Wild Type | gln1-4 | gln1-3 | gln1-3 gln1-4 |
| Asp | 1.85 ± 0.52 (11) | 4.19 ± 1.24 (10) | 1.61 ± 0.31 (6) | 1.07 ± 0.24 (3) |
| Asn | 0.58 ± 0.07 (3) | 6.18 ± 1.61 (15)b | 1.58 ± 0.10 (6)b | 11.91 ± 1.60 (29)b |
| Glu | 1.03 ± 0.15 (6) | 2.27 ± 0.22 (5)b | 3.56 ± 0.55 (14)b | 9.57 ± 3.88 (23)b |
| Gln | 3.56 ± 1.01 (21) | 6.85 ± 1.63 (16)c | 3.57 ± 0.10 (14)c | 1.96 ± 0.44 (5)c |
| Ala | 4.80 ± 1.41 (29) | 10.40 ± 1.58 (25) | 4.02 ± 0.12 (16) | 4.61 ± 1.59 (11) |
| GABAd | 0.11 ± 0.02 (1) | 0.15 ± 0.04 (1) | 0.21 ± 0.02 (1) | 0.30 ± 0.08 (1) |
| Pro | 0.86 ± 0.32 (5) | 1.25 ± 0.46 (3) | 1.17 ± 0.22 (5) | 0.64 ± 0.09 (2) |
| Others | 3.88 ± 0.67 (23) | 10.63 ± 1.61 (25) | 9.50 ± 0.51 (37) | 10.90 ± (27) |
| Total | 16.67 ± 3.66 (100) | 41.92 ± 7.20 (100)b | 25.22 ± 0.6 (100)b | 40.96 ± 4.73 (100)b |
| Total N (%) | 2.43 ± 0.16 | 2.84 ± 0.18b | 2.80 ± 0.16b | 2.77 ± 0.14b |
Amino acids were separated and quantified in maize plants 55 d after flowering. The plants were grown in the glasshouse on soil and watered daily with the medium described by Coïc and Lesaint (1971). Values are the mean of three plants ± sd. Relative amino acid proportions are given in parentheses.
Significant changes in the amino acid content and N content compared with the wild type at the 0.05 probability level.
No significant changes compared with the wild type.
GABA, γ-aminobutyrate.
Analysis of the phloem sap composition at the VS revealed that in the three mutants there was a general decrease in the concentration of all the main amino acids except for Glu. This decrease was approximately twofold for Asp, Gln, and Pro and at least fourfold for Asn and Ala. However, the relative proportions of the amino acids were not markedly modified except for Glu, for which a slight increase was observed (Table 6). No marked changes were observed for the free amino acid content of the kernel, except a twofold decrease in Asn in the mutants, which represented ∼14% of the total in the wild type (data not shown).
Table 6.
Concentration and Proportion of Amino Acids in the Phloem Sap of the Wild Type and Cytosolic GS-Deficient Mutants
| Amino Acid Concentration (μmol μL−1) and Proportions (%)a
|
||||
|---|---|---|---|---|
| Amino Acid | Wild Type | gln1-4 | gln1-3 | gln1-3 gln1-4 |
| Asp | 81.1 ± 10.1 (10) | 43.9 ± 8.4 (13)b | 60.5 ± 6.8 (16)b | 46.9 ± 4.6 (14)b |
| Asn | 21.2 ± 10.5 (3) | 5.2 ± 1.3 (2)b | 2.6 ± 0.4 (1)b | 4.3 ± 0.4 (1)b |
| Glu | 79.5 ± 15.1 (10)c | 46.8 ± 9.5 (14)c | 79.5 ± 13 (21)c | 56.3 ± 6.2 (17)c |
| Gln | 159.8 ± 42.5 (19) | 70.5 ± 19.4 (21)b | 66 ± 32 (18)b | 67.1 ± 6.6 (20)b |
| Ala | 85.6 ± 1.4 (4.6) | 22.1 ± 4.5 (7)b | 10.9 ± 12 (3)b | 18.3 ± 1.2 (5)b |
| GABA | 48.0 ± 29.1 (6) | 9.2 ± 5.3 (3) | 4.9 ± 2.4 (1) | 4.5 ± 1.1 (1) |
| Pro | 9.9 ± 2.1 (1) | 5.2 ± 0.9 (2)b | 2.8 ± 0.6 (2)b | 2.8 ± 2.8 (1)b |
| Others | 289.7 ± 24.7 (35) | 116.3 ± 9.3 (35) | 136.4 ± 10.8 (38) | 126.8 ± 26 (37) |
| Total | 825 ± 28.2 (100) | 329 ± 64b | 367 ± 54 (100)b | 335 ± 24 (100)b |
Amino acids were separated and quantified in the phloem sap of maize plants at VS. The plants were grown hydroponically on the medium described by Coïc and Lesaint (1971). Values are the mean of three plants ± sd. Relative amino acid proportions are given in parentheses.
Significant changes in the amino acid content compared with the wild type at the 0.05 probability level.
No significant changes compared with the wild type.
To determine if the roots were able to provide enough N assimilates to the shoots in the three mutants, the amino acid composition of the xylem sap was analyzed. A slight decrease in the total amino acid concentration of the xylem sap at the VS was observed, particularly in gln1-3, due mainly to a decrease in Asn, the most abundant amino acid in the xylem sap. However, this decrease was not significant (Table 7). Interestingly, the amount of Gln, which is almost as abundant as Asn, was not significantly modified in the mutants compared with the wild type.
Table 7.
Concentration and Proportion of Amino Acids in the Xylem Sap of the Wild Type and Cytosolic GS-Deficient Mutants
| Amino Acid Concentration (μmol μL−1) and Proportions (%)a
|
||||
|---|---|---|---|---|
| Amino Acid | Wild Type | gln1-4 | gln1-3 | gln1-3 gln1-4 |
| Asp | 13.8 ± 7.8 (7.8) | NAb | 19.8 ± 2.8 (17.1) | 13.6 ± 7.0 (9.1) |
| Asn | 57.0 ± 18.5 (32.1) | 18.0 ± 4.2 (15.5)c | 38 ± 13.6 (25.5)c | |
| Glu | 5.6 ± 2.5 (3.1) | 2.2 ± 0.4 (1.8)c | 4.6 ± 1.8 (3.0)c | |
| Gln | 42.4 ± 8.5 (23.9) | 32.6 ± 3.2 (28.1)c | 35.6 ± 1.6 (23.8)c | |
| Ala | 8.2 ± 1.4 (4.6) | 1.0 ± 2 (0.8) | 9.2 ± 2.8 (6.1) | |
| GABA | 1.0 ± 0.6 (0.5) | 0.2 ± 0.4 (0.2) | 1.4 ± 1.0 (0.9) | |
| Pro | 0 ± 0 (0) | 0 ± 0 (0) | 0 ± 0 (0) | |
| Others | 49.4 ± 11.4 (28) | 42.4 ± 4.4 (36.5) | 46.6 ± 5.8 (31.6) | |
| Total | 177.4 ± 8.9 (100) | 116.2 ± 2.0 (100) | 149.0 ± 7.3 (100) | |
Amino acids were separated and quantified in the xylem sap of detopted maize plants at VS. The plants were grown hydroponically on the medium described by Coïc and Lesaint (1971). Values are the mean of three plants ± sd. Relative amino acid proportions are given in parentheses.
Not available.
No significant changes in the amino acid content compared with the WT at the 0.05 probability level.
Overexpression of GS1
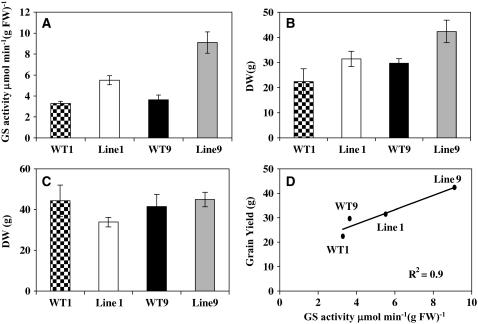
The cDNA encoding Gln1-3 was made constitutive by fusing it with the cassava vein mosaic virus promoter (pCsVMV) (see Supplemental Figure 2 online). This promoter is highly active in plant cells and directs constitutive gene expression in most tissue of transgenic plants (Verdaguer et al., 1998). After selection and regeneration, followed by three backcrosses with the line FV2, two T4 transgenic plants (Lines 1 and 9) overexpressing Gln1-3 and the corresponding null segregants (WT1 and WT9) were selected for further analysis. Compared with the two null segregants, a twofold and threefold increase in total leaf GS activity was observed in Lines 1 and 9, respectively (Figure 12A). The level of GS protein in the two transgenic lines was examined by protein gel blot analysis using the tobacco antibodies recognizing GS1 and GS2. Figure 3E shows a protein gel blot of crude protein extracts from leaves at the VS of the two null segregants (WT1 and WT9) and the two independent transformants (Lines 1 and 9). An increase in the amount of GS1 protein was clearly visible in the protein extracts of the two transformed plants, with that of Line 9 being noticeably higher. When the GS proteins were analyzed in the leaves 55 DAS, a similar pattern of increase in GS1 protein content was obtained (data not shown).
Figure 12.
Shoot and Grain Yield in Relation to GS Activity of Plants Overexpressing Gln1-3.
(A) Total leaf GS activity in the two null segregants (WT1 and WT2) and the corresponding two transgenic lines (Lines 1 and 9) overexpressing GS1.
(B) Kernel yield (DW) of two null segregants (WT1 and WT2) and two transgenic lines (Lines 1 and 9) overexpressing GS1. Plants were harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
(C) Total DW of shoot vegetative parts of two null segregants (WT1 and WT2) and two transgenic lines (Lines 1 and 9) overexpressing GS1. Plants were harvested at maturity and grown under N suboptimal conditions on a nutrient solution containing 10 mM NO3−.
(D) Scatterplots between leaf GS activity and kernel yield in the two null segregants (WT1 and WT2) and the corresponding two transgenic lines (Lines 1 and 9) overexpressing GS1. Each symbol corresponds to a leaf sample harvested at the VS.
Values are the mean ± se of six individual plants.
Immunogold transmission electron microscopy experiments were performed on leaf blade sections of the wild type and on the transgenic lines overexpressing Gln1-3. An increase in GS protein was only observed in the cytosol of the MC and of the PCC, while the amount of plastidic GS protein was similar in both cell types (see Supplemental Figure 3 online). Quantification of gold particles confirmed these observations of differences (Table 2).
To determine the impact of Gln1-3 overexpression on plant phenotype and kernel production, plants were grown in a glasshouse until maturity under suboptimal N feeding conditions. A significant increase in grain yield was observed, which when compared with two corresponding wild-type control plants was ∼30% for both Lines 1 and 9 (Figure 12C), thus confirming the phenotype shown in Figure 11B. Kernel number was the yield component that was mostly responsible for the increase in kernel production (Table 3). Linear regression and the resulting correlation coefficient were calculated for the level of leaf GS activity versus grain yield in the two null segregants and the two transgenic lines. Figure 12D shows that there is a strong relationship between leaf GS activity and grain yield (r2 0.91), thus indicating that the increase in yield is proportional to the increase in enzyme activity. By contrast, no significant differences were seen in shoot dry matter production between the two wild-type control plants and the two transgenic lines (Figure 12C).
DISCUSSION
GS and Yield
Using a reverse genetic approach, we have demonstrated the functional importance of cytosolic GS in the control of yield and its components in maize. Under suboptimal N feeding conditions, a reduction in kernel size and kernel number was observed in the gln1-3 and gln1-4 single mutants and was even more severe in the gln1-3 gln1-4 double mutant. In addition, we have shown that in the gln1-3 mutant, kernel number is the yield component that is preferentially affected, whereas in the gln1-4 mutant, it is kernel size. In transgenic plants, in which Gln1-3 was overexpressed in the MC, an increase in kernel number was observed. This finding confirms that the GS1-3 isoenzyme plays a major role in controlling yield through the production of kernels.
This finding is in agreement with the coincidence previously demonstrated with the QTL for the two yield components and the two GS1 structural genes (Hirel et al., 2001). Therefore, these observations suggest that the reduced N channeled through the reaction catalyzed by GS1-3 is more specifically invested in kernel set, whereas the reduced N arising from the reaction catalyzed by GS1-4 is rather used for kernel filling. The expression of Gln1-4 is enhanced during leaf aging (Hirel et al., 2005a). Thus, the isoform GS1-4 is likely to be involved in the reassimilation of NH4+ released during protein breakdown to provide Gln for kernel growth during the filling period. The housekeeping role of the Gln1-3 gene product is in agreement with its role in providing enough N assimilates to the developing ear to avoid kernel abortion (Below, 1987). The increase in kernel number observed in transgenic plants overexpressing Gln1-3 further supports this hypothesis. In the gln1-3 gln1-4 mutant, both kernel size and kernel number were reduced, thus demonstrating the cumulative effect of the two mutations.
Previous studies have shown that the supply of N assimilates facilitates the utilization of carbohydrates by the kernels (Below et al., 2000), thus influencing both kernel weight and yield (Below et al., 1981). The qualitative importance of N assimilate supply to the kernels has also been emphasized by Seebauer et al. (2004), who proposed that Gln and Asn, two molecules used for N transport to the kernel, play an important role during the kernel filling process.
The finding that vegetative biomass production is not affected in the two single GS mutants and the gln1-3 gln1-4 double mutant reinforces the conclusion that the leaf isoforms GS1-3 and GS1-4 are specifically involved in the synthesis of Gln that is further used for kernel set and kernel development. This therefore implies that the root GS (represented by the GSr isoform encoded by Gln1-1), whose activity is not notably affected in the mutants, is probably able to provide the major part of the reduced N that is used for vegetative growth. The contribution of GS2 and GS1-2 in providing Gln to the leaf, at least in the early phase of plant development before the silking period, can also be considered, since both the expression of corresponding genes and the presence of an active protein were found in all three mutants. Although the exact physiological function of GS1-5 remains to be determined, its role may be confined to photosynthetic epidermal cells under certain conditions of illumination (Sakakibara et al., 1992).
The importance of GS in controlling cereal productivity has recently been strengthened by a study performed in rice, in which a strong reduction in both growth rate and kernel yield was observed in a mutant deficient in cytosolic GS (Tabuchi et al., 2005).
Physiological Impact of Decreased GS Activity
The main impact of the reduction in GS activity in the mutants was an increase in the leaf amino acid content at a rather late stage during the kernel-filling period (55 DAS), which was primarily due to an increase in Asn and to a lesser extent Glu, notably in the gln1-3 gln1-4 mutant. The increase in Glu was proportional to the reduction of GS1 activity in the mutants, indicating that there was an accumulation of substrate as a result of lower enzyme activity. No major changes in the profile of N and C metabolites were observed before that period, except an accumulation of NH4+ in vegetative plants, which is likely to be the result of lower GS1 activity during full leaf growth. In legumes, an accumulation of Asn was observed when GS activity was impaired, suggesting that other enzymes (e.g., Asn synthetase) may be important in bypassing the flux of reduced nitrogen to avoid a toxic accumulation of NH4+, thus ensuring plant survival (Carvalho et al., 2003; Harrison et al., 2003; Wong et al., 2004). The question as to whether these alternative metabolic pathways are important in the control of plant productivity has been extensively studied in legumes and nonlegumes (Knight and Langston-Unkefer, 1988; Brears et al., 1993; Lam et al., 2003) but has still not been fully elucidated, particularly in cereals.
According to Seebauer et al. (2004), Gln is the major amino acid entering the cob just after silking and is then further metabolized into Asn, which is used for optimal kernel growth and protein accumulation. Phloem sap analysis confirmed that Gln is a major N transport form in maize (Oaks, 1992) and that the concentration is considerably reduced in the three mutants. This reduction was also observed for most of the main amino acids of the phloem sap, which is a typical symptom of N deficiency in plants (Tercé-Laforgue et al., 2004). Consequently, less Gln and other amino acids were apparently transported to the developing kernels, thus causing a shortage in the amount of N translocated and therefore a reduction in both kernel number and kernel size. The higher leaf N content of the mutants at maturity confirms that N assimilates are not efficiently translocated to the ear. Since Asn accumulated in the leaves of the mutants during the kernel-filling period, it seems likely that Asn is synthesized at the expense of Gln but is not efficiently transported to the developing ear. This hypothesis was further confirmed by the finding that although there was an accumulation of Asn in the leaves of the mutants, the amino acid remained a relatively minor N compound transported in the phloem sap, the concentration in all three mutants being lower than that of the wild type. The lower relative content of soluble Asn in the kernels in the mutants compared with the wild type is also an argument in favor of this hypothesis.
The higher kernel N content of the mutants may be explained by the fact that there are less kernels to fill. Thus, the amount of N available to an individual kernel may be increased, even if the amino acids (mainly represented by Gln) translocated in the phloem are reduced.
The results so far show that at the VS, the impact of altered GS1 expression was minimal or absorbed by the system. This could due to support from the root in producing Gln and/or compensation by other isozymes (GS1-2, GS1-1, and GS2). However, there was a drastic phenotype at the reproductive stage, which implicates the deficient isoenzymes play major roles after VS and/or that the flux of ammonia through those enzymes is of magnitudes higher during the grain-filling period.
Now, the function of GS1-2 and GS1-5 remains to be determined, which may be involved in the control of Gln synthesis in reproductive tissues, such as the cobs (Seebauer et al., 2004), the kernels and other vascular-rich organs (Muhitch, 2003), and the leaf or stem epidermis (Li et al., 1993).
Cell-Specific Expression and Function of GS Isoenzymes in Leaves
Originally, Sakakibara et al. (1992) and Li et al. (1993) showed that there is an organ-specific expression of the different genes encoding GS1 and GS2. Later on, Becker et al. (2000) found that the relative proportions of GS1 and GS2 were different in the MC and BSC. This finding led these authors to propose that the source of Gln in both the MC and in the BSC originated from protein catabolism through the reaction catalyzed by GS1. GS2 in the MC was responsible for ammonia assimilation as the result of NO3− reduction occurring specifically in this cell type. In the BSC, GS2 in conjunction with GOGAT was involved in the reassimilation of ammonium arising from photorespiration.
In this work, the analysis of the cellular localization of GS in the wild type and in the three mutants revealed that in leaves, the tissue distribution of the individual cytosolic GS isoenzymes is different. The GS1-3 isoenzyme is localized in the MC, whereas the GS1-4 isoenzyme is restricted to the BSC. The in situ hybridization experiments confirmed the cell-specific expression of the corresponding transcripts.
Since the gene encoding GS1-3 (Gln1-3) is constitutively expressed until a very late stage of leaf development (Hirel et al., 2005a), we propose that GS1-3 in the MC is more specifically involved in the synthesis of Gln following NO3− reduction until plant maturity. The finding that GS1 protein is more abundantly expressed in the MC of transgenic plants overexpressing Gln1-3 further supports this hypothesis. This function is in agreement with the finding that postflowering NO3− uptake still occurs during the kernel-filling period and represents ∼50% of the N allocated to the grain (Martin et al., 2005). Therefore the major impact of the lack of GS1-3 activity would be a reduction in grain number due to the lack of N available to induce the setting of more kernels. This hypothesis is in agreement with the well-known phenomenon of kernel abortion as the result of N deficiency during ear development (Below, 1987).
The BS-specific GS1-4 isoform is encoded by a gene induced during leaf aging (Hirel et al., 2005a; Martin et al., 2005). Since Rubisco is located in this cell type and is a major source of N during the transition from sink to source (Esquivel et al., 2000), we hypothesize that GS1-4 has a catabolic function in the reassimilation of ammonium released during protein degradation in senescing leaves. In the gln1-4 mutant, kernel size was reduced, which suggests that the N originating from this catabolic process is more specifically used to fill the kernels.
The finding that only Gln1-2 transcripts were detected in the leaves of the three mutants and that GS is still detected in the PCCs suggests that this gene encodes a phloem-specific GS isoenzyme. We have also shown that in the double mutant, GS1 transcripts were detected in the leaf and root vascular tissue, which strengthens our hypothesis. A phloem-specific GS isoenzyme has only been identified previously in C3 plant species. Its physiological role appears to be related either to N storage under stress conditions (Brugière et al., 1999) or to N export during plant growth and development (Suarez et al., 2002; Tabuchi et al., 2005). In maize, it is likely that the GS1-2 isoenzyme contributes to N translocation but is not directly involved in supplying N to the kernels.
We have also demonstrated that a high level of GS1 transcripts were expressed in the root cortex and the two outermost cell layers of the cortex, thus confirming that the GS1-1 isoenzyme is likely to be involved in Gln synthesis following NO3− uptake and reduction in these two cell types, which have been shown to be directly involved in the process of NO3− uptake (Nazoa et al., 2003).
Although the signal was low, transcripts for GS1 were detected in the upper and lower leaf epidermis of the three mutants, thus suggesting that the fifth gene encoding GS1 (Gln1-5) may be expressed in these two cell layers. However, further experimentation is necessary to confirm this finding and identify the physiological function of GS1-5 isoenzyme.
The disappearance of a GS2 polypeptide exhibiting a different pI in the three mutants is an interesting finding that suggests that there is a regulatory control mechanism that regulates the balance between GS1 and GS2 synthesis and/or activity. The increase in GS2 activity observed in the three mutants indicates that the GS2 holoenzyme composed of only one polypeptide is more active. Since in maize, GS2 is encoded by a single gene (Gln5 locus located on chromosome 10 corresponding to Gln2 gene; Hirel et al., 2001), it is likely that the GS2 isoenzyme is subjected to posttranslational modifications. Such posttranslational modifications have previously been reported in C3 species, such as Medicago truncatula and tobacco, where the presence of isoelectric variants of GS2 is the result of protein phosphorylation (Riedel et al., 2001; Lima et al., 2005). The finding that a significant proportion of GS2 appears to be localized in the mitochondria (Taira et al., 2004) should also be taken into consideration in future studies with respect to its possible role during ammonia assimilation and or reassimilation. However, in the immunogold transmission electron microscopy experiments performed in this study, there was no evidence that any form of GS was present in the mitochondria.
Thus, the GS1-deficient mutants characterized in this work are not only of considerable value in identifying the key separate roles performed by the individual GS1 isoenzymes in a C4 plant, they also appear to be a good model for defining the roles of different GS isoenzymes in maize (Figure 13). This will hopefully lead to increases in nitrogen use efficiency and a better understanding of the relationship between N assimilation and crop productivity, which could lead to improved yield and fertilizer use in this economically important crop.
Figure 13.
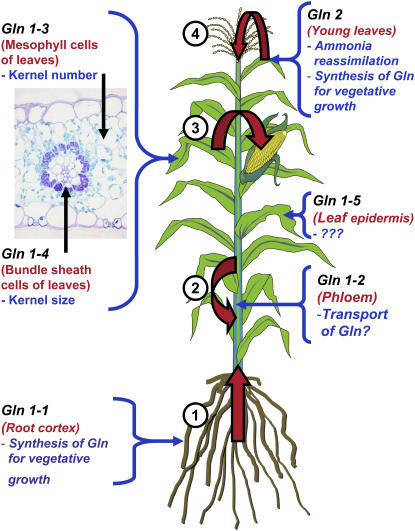
Schematic Representation Depicting the Expression and Possible Function of the GS Isoenzymes within the Maize Plant as Determined in This Work.
Gln1-1 to Gln 1-5 are the five genes encoding cytosolic GS and Gln2 the gene encoding plastidic GS. Their tissue or cellular localization is indicated in red, and their putative function is indicated in blue, deduced from the analysis of the gln1-3– and gln1-4–deficient mutants. The large red arrows indicate the flux of Gln occurring within the plant: 1, from the roots to the shoots (reaction catalyzed by GS1-1); 2, in the phloem (reaction catalyzed by GS1-2); 3, from the source leaves to the ear (reaction catalyzed by GS1-3 and GS1-4); and 4, from the young leaves to the other shoot parts (reaction catalyzed by GS2). The function of GS1-5 is unknown, as indicated by question marks.
METHODS
Isolation and Characterization of the Gln1-3∷Mu and Gln1-4∷Mu Insertion Events
The Gln1-3 Mu1-insertion event was isolated with the help of the maize targeted mutagenesis program (http://mtm.cshl.edu/; May et al., 2003) using the following primers: 5′-CCTTCCGTGGGTATTTCTTCAG-3′ and 5′-AGTGGCTTTGCAATCCGCTTC-3′.
The GS1-4∷Mu insertion event was isolated from the Bristol University Mu population using a random sequencing strategy as described by Hanley et al. (2000). B73 and the various Mu maize (Zea mays) lines of interest were grown in a glasshouse under a light/dark regime of 16/8 h and a temperature cycle of 25°C during the light period (210 μmol m−2 s−1) and 18°C at night. In all cases, maize lines carrying the Mu insertion event were twice backcrossed to a Mu-off line (May et al., 2003) before backcrossing to B73 (derived from a line originally obtained from Garst Seeds [Slater]) but maintained by selfing for seven generations. Single or double homozygous mutant lines were then generated by standard means. The number of nonspecific Mu insertion events was monitored at various stages by random sequencing of Mu-tagged fragments. For both the Gln1-3∷Mu and Gln1-4∷Mu insertion events, the number of nonspecific Mu sequences was reduced from ∼50 in the original lines to less than five (data not shown). In no case did the nonspecific Mu sequences appear to cosegregate with the GS insertion events. The exact insertion sites within both Gln1-3∷Mu and Gln1-4∷Mu were determined by standard PCR using primers flanking the insertion events (for Gln1-3∷Mu, these were 5′-CCTTCCGTGGGTATTTCTTCAG-3′ and 5′-AGTGGCTTTGCAATCCGCTTC-3′, and for Gln1-4∷Mu, these were 5′-CTTCGTCCATAATGGCAATTATCTC-3′ and 5′-AGCAACCCACACCTGATCG-3′) and a primer specific for the Mu elements (5′-CTTCGTCCATAATGGCAATTATCTC-3′; Hanley et al., 2000). PCR fragments were sequenced using standard procedures (Hanley et al., 2000).
To study the genetic background of the material, a set of single sequence repeat (SSR) markers, chosen among the SSRs assigned on the IBM map (B73*Mo17 population; Maize Genome Database; http://www.maizegdb.org/), was genotyped on the three GS1-deficient mutants and the wild type. A total of 34 SSRs were genotyped, with an average of four SSRs per chromosome covering both the short and the long arm.
Two additional Gln1-3 Mu insertion events (gln1-3∷Mub and gln1-3∷Muc) were isolated at Biogemma using a resource of 27,500 maize lines in which an endogenous Mu element had been allowed to transpose at high frequency. This collection of mutants was built up using a Mu transposon stock kindly provided by B. Taylor (Commonwealth Scientific and Industrial Research Organization, Canberra, Australia). The Mu transposon stock was crossed with maize hybrids developed by Limagrain (Chappes, France) to increase the vigor of the plants and their adaptation to European environmental conditions. The screening for Mu insertion events was performed on the F1 generation and confirmed for their germinal status (heritability) on the F2 generation. Mutant screens were accomplished through a PCR-based approach using a Mu-specific primer called OmuA: 5′-CTTCGTCCATAATGGCAATTATCTC-3′. OMuA that binds to the edges of the element, and the so-called terminal inverted repeat, was used in combination with a PCR primer (GS F5, 5′-GCACTGAGAACAGAGATTCCC-3′; GS R6, 5′-GGTGTGGGATTGAGATTGAGGA-3′).
Because of the process used to make the maize mutant collection, each plant from the F2 generation for each family has a heterogeneous genetic background. Therefore, to minimize phenotypic variation between plants belonging to the same family, each mutant was crossed with a standard elite line adapted to the European climate. In that case, for each family investigated, the maize line FV2 (Hirel et al., 2001) was used and crossed with the mutant heterozygous for the insertion event. Thus, at each generation, there is a genetic segregation for the mutant allele; therefore, the presence of a Mu insertion was verified for each plant at each generation. After three rounds of crosses with the maize line FV2, heterozygous mutants were self-pollinated, and the phenotype of the kernel was scored directly on the ear.
Plant Transformation, Regeneration, and Characterization
Maize transformation of the inbred line A188 with Agrobacterium tumefaciens strain LBA4404 harboring a superbinary plasmid was performed essentially as described by Ishida et al. (1996). In particular, the composition of all media cited hereafter is detailed in this reference. The protocol was slightly modified concerning the selective marker, which was the NPTII gene instead of the bar gene. The superbinary plasmid used for transformation was the result of a recombination between plasmid pBIOS 445, harboring between the T-DNA borders a neomycin resistance cassette (NPTII gene flanked by an actin promoter and 3′Nos terminator) and the Gln1-3 full-length cDNA (Sakakibara et al., 1992) flanked by pCsVMV (Verdaguer et al., 1998) and 3′Nos terminator and the plasmid pSB1 harboring the virB and virG genes isolated from the supervirulent strain A281. The resulting recombinant plasmid used for transformation was pRec 445 (see Supplemental Figure 2 online).
Plant transformation was conducted using immature maize embryos isolated at 10 d after pollination according to the procedure described by Voisin et al. (2006). Transgenic plants were then cultivated in a glasshouse (18 to 24°C) and selfed or pollinated with line A188 to produce seeds.
A number of transgenic lines were selected and tested for the pattern of insertion of the pCsVMV-Gln1-3 construct. Following digestion of the genomic DNA with NcoI, hybridization was performed with Gln1-3 cDNA, Actin intron, and 3′Nos probes to determine the T-DNA copy number. In addition, two probes were designed on the plasmid pREc, 445 each overlapping the two right and left T-DNA borders, to check the eventual presence of plasmid sequence external to the T-DNA (see Supplemental Figure 2 online for the position of the probes). In two primary transformants (Lines 1 and 9), a single insertion event was detected without the presence of additional T-DNA sequences, apart from those flanking the insert containing the pCsVMV-Gln1-3 and the pActin-NPTII chimeric constructs (data not shown). These two primary transformants were crossed to the line FV2, which contained the unfavorable allele for Gln1-3 (Hirel et al., 2001). T1 transformants were then selected and backcrossed three times with the line FV2. In the T4 generation containing 50% transgenics, the 50% null segregants for Lines 1 and 9 were used as control plants and named WT1 and WT9, respectively.
Plant Material for Molecular and Physiological Studies
Seeds of the three GS1-deficient mutants, the corresponding wild type, the transgenic Lines 1 and 9, and the corresponding control plants WT1 and WT9 were first sown on coarse sand and after 1 week, when two to three leaves had emerged, were transferred either to hydroponic culture for root harvesting or to soil for leaf harvesting. For the hydroponic culture, 12 plants (three for the wild type and three for each mutant) were randomly placed on a 130-liter aerated culture unit. The experiment was performed in triplicate for each line, and plants were grown for 18 d in a growth chamber with a 16/8 light/dark period. A photosynthetic photon flux density of 400 μmol m−2 s−1 was provided by metal halide lamps. The relative humidity was maintained at 60% of saturation. Plants were harvested at the 10 to 11 leaf stage between 9 to 12 am and separated into young leaves (three youngest leaves) and roots. The root samples were immediately placed in liquid N2 and then stored at −80°C until further analysis.
The glasshouse experiment for the three mutants and the wild type were performed on May 15, 2004 and for the two transgenic overexpressing lines and the corresponding wild types on May 15, 2005. Three plants of the three mutants and six plants of the two transgenic lines were transferred to pots (diameter and height of 30 cm) containing clay loam soil, and at the 10 to 11 leaf stage, the three youngest fully expanded leaves were harvested and pooled for the VS samples. The leaf below the ear was harvested at later stages of plant development, including 15 and 55 d after flowering. The leaf, below the ear, was selected since it has been shown to provide a good indication of the source sink transition during grain filling (Hirel et al., 2005a, 2005b; Martin et al., 2005). No major variations in N metabolite content and enzyme activity within a single leaf blade have been observed until the latest stages of leaf development. Therefore, it has been concluded that the entire leaf blade can be used for measuring physiological traits related to N metabolism (Hirel et al., 2005b). Leaf samples were harvested between 9 and 12 am and frozen in liquid N2, ground to a homogenous powder, and stored at −80°C for use in subsequent RNA, protein, and metabolite analyses. In the hydroponic culture, plants were grown on a complete nutrient solution containing 10 mM NO3− as the sole N source (Coïc and Lesaint, 1971). The nutrient solution was replaced daily. In the glasshouse, plants were watered daily with a complete nutrient solution containing 10 mM NO3− as the sole N source (Coïc and Lesaint, 1971). For both growth methods, the complete nutrient solution contained 1.25 mM K+, 0.25 mM Ca2+, 0.25 mM Mg2+, 1.25 mM H2PO4−, 0.75 mM SO42−, 21.5 μM Fe2+ (Sequestrene; Ciba-Geigy), 23 μM B3+, 9 μM Mn2+, 0.3 μM Mo2+, 0.95 μM Cu2+, and 3.5 μM Zn2+.
Agronomic Trait Evaluation for Kernel Yield
Kernel yield, its components, and the N content of different parts of the plant at stages of development from silking to maturity were determined according to the method described by Martin et al. (2005). The plants were grown in the glasshouse on soil and watered daily with the medium described by Coïc and Lesaint (1971) as described above. Total N content was measured using the Dumas combustion method.
Metabolite Extraction and Analyses
Lyophilized plant material was used for metabolite extraction. NH4+ and amino acids were extracted with 2% 5-sulfosalicylic acid (10 mg DW mL−1; Ferrario-Méry et al., 1998). Total amino acid content and individual amino acid composition was determined by ion exchange chromatography on pooled samples extracted from equal DWs. Total free amino acids were determined by the Rosen colorimetric method using Leu as a standard (Rosen, 1957). The composition of individual amino acids was performed by ion exchange chromatography followed by detection with ninhydrin using the AminoTac JLC-500/V amino acid analyzer according to the instructions of the manufacturer (JEOL). Free NH4+ was determined by the phenol hypochlorite assay (Berthelot reaction), which provides reliable data for comparative studies when the concentration of NH4+ is low, although a more precise quantification can be obtained by other methods (Husted et al., 2000). Sucrose, glucose, fructose, and starch were extracted with 1 M HClO4 (1 mL per 5 to 10 mg DW of plant material) as described by Ferrario-Méry et al. (1998). The soluble sugars (glucose, fructose, and sucrose) were measured enzymatically using a commercially available kit assay (Boehringer).
Xylem and Phloem Sap Collection
For xylem sap collection, stems of plants were cut 2 cm above the root system, and the cut stem was rinsed with water and blotted dry. Root pressure bleeding sap (∼200 to 400 μL per plant) was collected with a micropipette, and the samples were immediately stored at −80°C. Amino acid analysis was performed as described previously.
Phloem exudates were obtained using the technique described by King and Zeevaart (1974). The leaves were cut off and recut under water before rapid immersion in the collection buffer. For each experiment, fully expanded leaves of three individual plants (the wild type and the three mutants) were placed separately in a solution of 10 mM HEPES and 10 mM EDTA (adjusted to pH 7.5 with NaOH) in a humid chamber (relative humidity >90%) and in the dark. Exudates were collected during 6 h from 10 am to 4 pm. The fresh weights of the leaves were then measured and the exudates stored at −80°C. Phloem exudates (in the EDTA solution) were adjusted to pH 2.1 and centrifuged to remove debris and EDTA, which precipitate at that pH.
Enzymatic Assays and Ion Exchange Chromatography
Enzymes were extracted from frozen leaf material that had been previously stored at −80°C. All extractions were performed at 4°C. GS was measured according to the method of Lea et al. (1999) for the transferase reaction and O'Neal and Joy (1973) for the synthetase reaction.
To determine the relative proportions of plastidic and cytosolic GS activities in the wild type and the mutants, frozen leaf material harvested at the VS was ground to a fine powder in liquid N2 and then extracted in a buffer containing 100 mM TEA, 1 mM EDTA, 10 mM MgSO4, 5 mM Glu, 10% (v/v) ethylene glycol, and 6 mM DTT, pH 7.6. Extracts were then centrifuged at 10,000g for 15 min at 4°C. Separation on a Mono Q anion exchange column (Amersham Pharmacia Biotech) attached to an HPLC (DX 500; Dionex) was performed as described by Habash et al. (2001), except that a 0.1 to 0.7 M NaCl linear gradient was used for elution and 1-mL fractions were collected.
Gel Electrophoresis, Protein Gel Blot Analysis, and Protein Sequencing
Proteins were extracted from frozen leaf powder in cold extraction buffer containing 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM MgCl2, 0.5% (w/v) PVP, 0.1% (v/v) 2-mercaptoethanol, and 4 mM leupeptin and separated by SDS-PAGE (Laemmli, 1970). The percentage of polyacrylamide in the gels was 10%, and equal amounts of protein (10 μg) were loaded onto each track. Proteins were electrophoretically transferred to nitrocellulose membranes for protein gel blot analysis. GS1 and GS2 polypeptides were detected using polyclonal antisera raised either against GS2 of tobacco (Nicotiana tabacum; Hirel et al., 1984) or GS1 from Phaseolus vulgaris root nodules (Cullimore and Miflin, 1984). Soluble protein was determined using a commercially available kit (Coomassie protein assay reagent; Bio-Rad) using BSA as a standard.
Protein Identification by LC-MS/MS
For protein identification, a preparative gel containing 250 μg of protein was prepared as described by Mechin et al. (2004) and stained with colloidal Coomassie Brilliant Blue. Spots were picked out and the acrylamide pieces were collected in 96-well microplates. In-gel digestion was performed with the Progest system (Genomic Solution) according to a standard trypsin protocol. Gel pieces were washed twice by successive separate baths of 10% acetic acid, 40% ethanol, and acetonitrile (ACN). They were then washed twice with successive baths of 25 mM NH4CO3 and ACN. Digestion was subsequently performed for 6 h at 37°C with 125 ng of modified trypsin (Promega) dissolved in 20% methanol and 20 mM NH4CO3. The peptides were extracted successively with 2% trifluoroacetic acid (TFA) and 50% ACN and then with ACN. Peptide extracts were dried in a vacuum centrifuge and suspended in 20 μL of 0.05% TFA, 0.05% HCOOH, and 2% ACN.
HPLC was performed on an Ultimate LC system combined with a Famos autosampler and a Switchos II microcolumn switch system (Dionex).
A 4-μL sample was loaded at 5 μL/min−1 on a μ-Precolumn cartridge (stationary phase: C18 PepMap 100, 5 μm; column: 300 μm i.d., 5 mm; Dionex) and desalted with 0.05% TFA, 0.05% HCOOH, and 2% ACN. After 2.5 min, the precolumn cartridge was connected to the separating PepMap C18 column (stationary phase: C18 PepMap 100, 3 μm; column: 75 μm i.d., 150 mm; Dionex). Buffers were 0.1% HCOOH, 3% ACN (A) and 0.1% HCOOH and 95% ACN (B). The peptide separation was achieved with a linear gradient from 5 to 30% B for 25 min at 200 nL min−1. Including the regeneration step at 100% B and the equilibration step at 100% A, one run took 45 min.
Eluted peptides were analyzed online with a LCQ Deca XP+ ion trap (Thermo Electron) using a nanoelectrospray interface. Ionization (1.2 to 1.6 kV ionization potential) was performed with liquid junction and a noncoated capillary probe (10 μm i.d.; New Objective). Peptide ions were analyzed using Xcalibur 1.3 with the following data-dependent acquisition steps: (1) full MS scan (mass-to-charge ratio (m/z) 400 to 1900, centroid mode), (2) ZoomScan on a selected precursor (scan at high resolution in profile mode on a m/z window of 4), and (3) MS/MS (Qz = 0.22, activation time = 50 ms, and collision energy = 40%; centroid mode). Steps 2 and 3 were repeated for the two major ions detected in step 1, or step 3 was performed for the three major ions. Dynamic exclusion was set to 30 s.
A database search was performed with Bioworks 3.1 (Thermo Electron). Trypsin digestion was as specified. Cys carboxyamidomethylation and Met oxidation were set to static and variable modifications, respectively. Precursor mass and fragment mass tolerance were 2 and 1, respectively. The assembled unique transcripts of the Z. mays Plant Genome Database (68,579 entries, release 149a; http://plantgdb.org/) were used.
Identified tryptic peptides were filtered according to their cross-correlation score (Xcorr), >1.7, 2.2, and 3.3 for mono-, di-, and tricharged peptides, respectively. A minimum of two peptides was required. In the case of identification with only two MS/MS spectra, similarity between the experimental and the theoretical MS/MS spectra was visually confirmed. Annotation of the identified putative unique transcript sequence was performed by protein BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) against the nonredundant database.
Extraction of Total RNA and RNA Gel Blot Analysis
Total RNA extraction was performed on pooled root and leaf samples of three individual plants. RNA gel blot analysis was performed as described previously (Hirel et al., 2005a). The five maize cytosolic GS probes and one plastidic GS-specific probe (Sakakibara et al., 1992) were used for mRNA detection. The GS loci mapped by Hirel et al. (2001) corresponds to the GS genes named pGS122, pGS134, pGS107, pGS112 (cytosolic GS), and pGS202 (plastidic GS) by Sakakibara et al. (1992) and GS1-1, GS1-2, GS1-4, GS1-3, and GS2 by Li et al. (1993). After hybridization, filters were used to expose x-ray film at −80°C. The identification of the genes encoding GS in maize has proved somewhat complex, partially due to the numbering systems used by the different research groups. In this article, Gln1-1, Gln1-2, Gln1-3, and Gln1-4 encoding cytosolic GS1 and Gln2 encoding plastidic GS2 will be used to denote the five GS genes of maize that have been shown to be located on chromosomes 1top (Gln1 locus), 1bottom (Gln2 locus), 5 (Gln4 locus), 4 (Gln3 locus), and 10 (Gln5 locus), respectively (Gallais and Hirel, 2004). The further maize cytosolic gene Gln1-5 (Gln6 locus) is located on chromosome 9 (bin 9.06; M. Falque and B. Hirel, unpublished results) and corresponds to pGS117 (Sakakibara et al., 1992) and GS1-5 (Li et al., 1993).
Immunolocalization Studies
Leaf pieces (2 to 3 mm2) were fixed in freshly prepared 1.5% (w/v) paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, for 4 h at 4°C. For immunolocalization, material was dehydrated in an ethanol series (final concentration 90% [v/v] ethanol) and embedded in London Resin white resin (Polysciences). Polymerization was performed in gelatin capsules for 10 h at 54°C. For immunotransmission electron microscopy studies, ultrathin sections were mounted on 400-μm mesh nickel grids and allowed to dry at 37°C. Sections were first incubated with 5% (v/v) normal goat serum in T1 buffer (0.05 M Tris-HCl buffer containing 2.5% [w/v] NaCl, 0.1% [w/v] BSA, and 0.05% [v/v] Tween 20, pH 7.4) for 1 h at room temperature and then for an additional 6 h at room temperature with the GS antiserum diluted 100 times in T1 buffer. Sections were then washed three times with T1 buffer and incubated for 2 h at room temperature with 10 nm of colloidal gold goat anti-rabbit immunoglobulin complex (Sigma-Aldrich) diluted 70 times in T1 buffer. After several washes, grids were treated with 5% (w/v) uranyl acetate in water and observed with a Philips CM12 electron microscope (Philips) at 100 kV. Negative controls were conducted by substituting the serum containing GS antibodies with preimmune rabbit serum. For light microscope immunological studies, thin sections of 1 μm were floated on drops of sterile water on slides and treated at room temperature in 1% periodic acid for 1 h and then in 0.5 M NH4Cl for 1 h. Sections were then washed three times with distilled water and incubated for 1 h at room temperature in T1 buffer containing 10% BSA. The labeling procedure was the same as that used for immunotransmission electron microscopy. Labeling was silver enhanced as described by the supplier (British Biocell), and sections were back-stained with 1% (w/v) fuschin before microscopy observations under bright field plus epipolarized light.
In Situ Hybridization
Paraffin-embedded leaf and root material was cut with a microtome (RM2145; Leica Microsystems), and semithin sections of 10 μm were floated on drops of sterile water on SuperFrost plus gold slides. Paraffin was removed in Histo-Clear (Agar Scientific). Sections were then dehydrated in an ethanol series (100%, 95%, 85%, 50%, and 30%), treated with 0.2 M HCl for 10 min, washed in PBS (150 mM NaCl and 10 mM phosphate buffer, pH 7.4), treated with proteinase K (1 μg/mL−1) for 10 min at 37°C, washed with 0.2% glycine in PBS, and postfixed with 4% paraformaldehyde in PBS buffer. After two washes in PBS buffer, the sections were treated with 1% acetic anhydride in 0.1 M triethanolamine, pH 8.0, and then washed again in PBS buffer. Sections were finally dehydrated in an ethanol series (30%, 50%, 85%, 95%, and 100%) and hybridized with digoxigenin-labeled RNA probes.
Dehydrated sections were air dried and incubated for 12 h at 57°C with digoxigenin-labeled RNA probes. For this purpose, 40 ng of probe was diluted in 100 μL of hybridization buffer (0.3 M NaCl, 5 mM EDTA, 10 mM Tris-HCl, pH 6.8, 50% deionized formamide, 10% dextran sulfate, 1 mg/mL−1 tRNA, and 1× Denhardt's solution). Sections were subsequently washed twice for 60 min in washing buffer (2× SSC [300 mM NaCl and 30 mM sodium citrate, pH 7.0] and 50% formamide) at 57°C and then washed in NTE (500 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) at 37°C, twice for 5 min each. Sections were incubated in NTE with 20 μg/mL−1 of RNase A at 37°C for 30 min and then washed in NTE at room temperature twice for 5 min and finally washed in washing buffer at 57°C for 60 min.
Sections were blocked for 1 h with buffer 1 (100 mM Tris-HCl, pH 7.5, and 150 mM NaCl), then for 1 h with 0.5% Blocking Reagent (Roche Diagnostic), then for 1 h with buffer 1 containing 1% BSA and 0.3% Triton, and finally incubated for 1 h at room temperature with antidigoxigenin alkaline phosphatase conjugate immunoglobulin (Roche Diagnostic) diluted 1:3000 in buffer 1 containing Triton and BSA. The sections were washed in buffer 1 containing 0.3% Triton (4 × 20 min), then for 5 min with buffer 1 alone, and finally for 5 min with buffer 2 (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2).
Development of label was performed by incubation of sections for ∼16 h in buffer 2 containing 10% polyvinylalcohol, 150 μg/mL−1 nitroblue tetrazolium, and 75 μg/mL−1 bromo-4-chloro-3-indolyl phosphate. Observation was performed using a bright-field light microscope (Eclipse E800; Nikon).
A 843-bp fragment of the coding region of Gln1-3 between the HincII and PvuII sites cloned into the SmaI site of the pSPT18 vector (Boehringer) was used to make digoxigenin-labeled riboprobes for in situ hybridization. Digoxygenin-labeled riboprobes were generated using digoxigenin/RNA SP6/T7 labeling (Boehringer). The full-length probes were then hydrolyzed with 100 mM carbonate buffer, pH 10.2, for 55 min at 60°C. The resulting probes of ∼200 bp were then precipitated by 0.25% acetic acid, 80 mM Na acetate, and 70% ethanol and resuspended in water. For the analysis of the wild type and the three GS-deficient mutants, both sense and antisense probes were used in parallel hybridizations with comparable tissue sections.
Sequencing of the Gln1-3 and Gln1-4 Genes
Sequence analysis of the flanking regions of Mu1 and Mu5 elements in plants identified from screening the two Mu populations indicated that they had inserted into the Gln1-3 and Gln1-4 genes, respectively. ESTs for these two genes were first reported by Li et al. (1993) and used to design primers for genomic DNA sequencing of line B73. Our results show that in the two maize GS genes, the intron/exon structure was highly conserved except for the last exon, which was 3 bp shorter in Gln1-4.
Statistics
For metabolite analyses, measurement of enzyme activities, and protein concentrations, results are presented as mean values for four plants with standard errors (se = sd/√n, where sd is the standard deviation, and n is the number of samples).
Accession Numbers
Sequence data for maize Gln1-3 and Gln1-4 can be found in the GenBank/EMBL data libraries under accession numbers x65928 and x65929, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Ear of Two Independent Mu Insertions within the 5′-Untranslated Region of Gln1-3.
Supplemental Figure 2. Map of the pREc 445 Plasmid Containing the Gln1-3 cDNA under the Control of the CsVMV Promoter.
Supplemental Figure 3. Transmission Electron Microscopic Immolocalization of GS in Leaf Sections of Maize Plants Overexpressing Gln1-3.
Supplementary Material
Acknowledgments
Seeds of the maize GS mutants were obtained through the public gene knockout service offered by K.J.E.'s group funded by the Biotechnology and Biological Science Research Council (see www.cerealsdb.uk.net for further information). We thank Stephanie Boutet for the amino acid analysis and Jacques Durand for providing the Gln1-3 and Gln1-4 gene sequences. We also thank François Gosse for technical assistance in the glasshouse experiments. This work was in part funded by the GENOPLANTE and by the European Community framework VI (SUSTAIN, contact QLCK5-CT2001-01461) research programs.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Alain Murigneux (alain.murigneux@biobemma.com), Keith J. Edwards (k.j.edwards@bristol.ac.uk), and Bertrand Hirel (hirel@versailles.inra.fr).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Becker, T.W., Carrayol, E., and Hirel, B. (2000). Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: Localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211 800–806. [DOI] [PubMed] [Google Scholar]
- Below, F., Christensen, L.E., Redd, A.J., and Hageman, R.H. (1981). Availability of reduced N and carbohydrate for ear development of maize. Plant Physiol. 68 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Below, F.E. (1987). Growth and productivity of maize under nitrogen stress. In Developing Drought- and Low N-Tolerant Maize, G.O. Emerades, M. Bänzinger, H.R. Mickelson, and H.R. Pena-Valdivia, eds (El Batán, Mexico: CIMMYT), pp. 243–240.
- Below, F.E., Cazetta, J.O., and Seebauer, J.R. (2000). Carbon/nitrogen interactions during ear and kernel development of maize. In Physiology and Modelling Kernel Set in Maize, M.E. Westgate and K.J. Boote, eds (Madison, WI: Crop Science Society of American and American Society of Agronomy), pp. 15–24.
- Brears, T., Liu, C., Knight, T.J., and Coruzzi, G.M. (1993). Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol. 103 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière, N., Dubois, F., Limami, A., Lelandais, M., Roux, Y., Sangwan, R., and Hirel, B. (1999). Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11 1995–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, H., Lopes-Cardoso, I., Lima, L., Melo, P., and Cullimore, J.V. (2003). Nodule specific modulation of glutamine synthetase in transgenic Medicago truncatula leads to inverse alteration in asparagines synthetase expression. Plant Physiol. 133 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coïc, Y., and Lesaint, C. (1971). Comment assurer une bonne nutrition en eau et en ions minéraux en horticulture. Hortic. Française 8, 11–14.
- Coque, M., Bertin, P., Hirel, B., and Gallais, A. (2006). Genetic variation and QTLs for 15N natural abundance in a set of maize recombinant inbred lines. Field Crops Res. 97 310–321. [Google Scholar]
- Cren, M., and Hirel, B. (1999). Glutamine synthetase in higher plants: Regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 40 1187–1193. [Google Scholar]
- Cullimore, J.V., and Miflin, B.J. (1984). Immunological studies on glutamine synthetase using antisera raised against the two forms of the enzyme from Phaseolus vulgaris root nodules. J. Exp. Bot. 35 581–587. [Google Scholar]
- Esquivel, M.G., Ferreira, R.B., and Teixeira, A.R. (2000). Protein degradation in C3 and C4 plants subjected to nutrient starvation; particular reference to ribulose bisphosphate carboxylase/oxygenase and glycolate oxidase. Plant Sci. 153 15–23. [Google Scholar]
- Ferrario-Méry, S., Valadier, M.H., and Foyer, C. (1998). Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiol. 117 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais, A., Coque, M., Quilléré, I., Prioul, J.L., and Hirel, B. (2006). Modelling postsilking N-fluxes in maize (Zea mays) using 15N-labelling field experiments. New Phytol. 172 696–707. [DOI] [PubMed] [Google Scholar]
- Gallais, A., and Hirel, B. (2004). An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 55 295–306. [DOI] [PubMed] [Google Scholar]
- Good, A.G., Shrawat, A.K., and Muench, D.G. (2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9 597–605. [DOI] [PubMed] [Google Scholar]
- Habash, D.Z., Massiah, A.J., Rong, H.L., Wallsgrove, R.M., and Leigh, R.A. (2001). The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Biol. 138 83–89. [Google Scholar]
- Hanley, S., Edwards, D., Stevenson, D., Haines, S., Hegarty, M., Schuch, W., and Edwards, K.J. (2000). Identification of transposon-tagged genes by the random sequencing of Mutator-tagged DNA fragments from Zea mays. Plant J. 23 557–566. [DOI] [PubMed] [Google Scholar]
- Harrison, J., Pou de Crecenzo, M.A., Sené, O., and Hirel, B. (2003). Does lowering glutamine synthetase activity in nodules modify nitrogen metabolism and growth of Lotus japonicus L.? Plant Physiol. 133 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel, B., Andrieu, B., Valadier, M.H., Renard, S., Quillere, I., Chelle, M., Pommel, B., Fournier, C., and Drouet, J.L. (2005. b). Physiology of maize II: Identification of physiological markers representative of the nitrogen status of maize (Zea mays) leaves during grain filling. Physiol. Plant. 124 178–188. [Google Scholar]
- Hirel, B., Bertin, P., Quillere, I., Bourdoncle, W., Attagnant, C., Dellay, C., Gouy, A., Cadiou, S., Retailliau, C., Falque, M., and Gallais, A. (2001). Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 125 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel, B., and Lea, P.J. (2001). Ammonia assimilation. In Plant Nitrogen, P.J. Lea and J.F. Morot-Gaudry, eds (Berlin: Springer-Verlag), pp. 79–99.
- Hirel, B., and Lemaire, G. (2005). From agronomy and ecophysiology to molecular genetics for improving nitrogen use efficiency in crops. In Efficient Nitrogen Use in Crop Production. A. Basra and S. Goyal, eds (Binghamton, NY: Haworth's Food Product Press), pp. 213–257.
- Hirel, B., Martin, A., Terce-Laforque, T., Gonzalez-Moro, M.B., and Estavillo, J.M. (2005. a). Physiology of maize I: A comprehensive and integrated view of nitrogen metabolism in a C4 plant. Physiol. Plant. 124 167–177. [Google Scholar]
- Hirel, B., Weatherley, C., Cretin, C., Bergounioux, C., and Gadal, P. (1984). Multiple subunit composition of chloroplastic glutamine synthetase of Nicotiana tabacum L. Plant Physiol. 74 448–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted, S., Hebbern, C.A., Mattson, M., and Schjoerring, J.K. (2000). A critical experimental evaluation of methods for determination of NH4+ in plant tissue, xylem sap and apoplastic fluid. Physiol. Plant. 109 167–179. [Google Scholar]
- Ishida, Y., Saito, H., Ohta, S., Hiei, Y., Komari, T., and Kumashiro, T. (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14 745–750. [DOI] [PubMed] [Google Scholar]
- Jeuffroy, B., Ney, B., and Ourry, A. (2002). Integrated physiological and agronomic modelling of N capture and use within the plant. J. Exp. Bot. 53 809–823. [DOI] [PubMed] [Google Scholar]
- Kichey, T., Heumez, E., Pocholle, P., Pageau, K., Vanacker, H., Dubois, F., Le Gouis, J., and Hirel, B. (2006). Combined agronomic and physiological aspects of nitrogen management in wheat (Triticum aestivum L.). Dynamic and integrated views highlighting the central role for the enzyme glutamine synthetase. New Phytol. 169 265–278. [DOI] [PubMed] [Google Scholar]
- King, R.W., and Zeevaart, J.A.D. (1974). Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol. 53 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, T.J., and Langston-Unkefer, P.J. (1988). Enhancement of symbiotic dinitrogen fixation by a toxin-releasing plant pathogen. Science 241 951–954. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Wong, P., Chan, H.K., Yam, K.M., Chen, L., Chow, C.M., and Coruzzi, G.M. (2003). Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 132 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor, D.W. (2002). Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J. Exp. Bot. 53 773–787. [PubMed] [Google Scholar]
- Lea, P.J., Blackwell, R.D., Chen, F.L., and Hecht, U. (1999). Enzymes of ammonia assimilation. In Methods in Plant Biochemistry, Vol. 3, P.J. Lea, ed (London: Academic Press), pp. 257–267.
- Li, M.G., Villemur, R., Hussey, P.J., Silflow, C.D., Gantt, J.S., and Snustad, D.P. (1993). Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol. Biol. 23 401–440. [DOI] [PubMed] [Google Scholar]
- Lima, L., Seabra, A., Melo, P., Cullimore, J., and Carvalho, H. (2005). Phosphorylation and subsequent interaction with 14-3-3 proteins regulates plastid glutamine synthetase in Medicago truncatula. Planta 223 558–567. [DOI] [PubMed] [Google Scholar]
- Limami, A.M., Rouillon, C., Glevarec, G., Gallais, A., and Hirel, B. (2002). Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol. 130 1860–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.S., Cortadellas, N., Kichey, T., Dubois, F., Habash, D.Z., and Araus, J.L. (2006). Wheat nitrogen metabolism during grain filling: Comparative role of glumes and the flag leaf. Planta 125 165–181. [DOI] [PubMed] [Google Scholar]
- Martin, A., Belastegui-Macadam, X., Quilleré, I., Floriot, M., Valadier, M.H., Pommel, B., Andrieu, B., Donnison, I., and Hirel, B. (2005). Nitrogen management and senescence in two maize hybrids differing in the persistence of leaf greenness. Agronomic, physiological and molecular aspects. New Phytol. 167 483–492. [DOI] [PubMed] [Google Scholar]
- May, B.P., Liu, H., Vollbrecht, E., Senior, L., Rabinowicz, P.D., Roh, D., Pan, X., Stein, L., Freeling, M., Alexander, D., and Martienssen, R. (2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. USA 100 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechin, V., Balliau, T., Chateau-Joubert, S., Davanture, M., Langella, O., Negroni, L., Prioul, J.L., Thevenot, C., Zivy, M., and Damerval, C. (2004). A two-dimensional proteome map of maize endosperm. Phytochemistry 65 1609–1618. [DOI] [PubMed] [Google Scholar]
- Miflin, B.J., and Habash, D.Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53 979–987. [DOI] [PubMed] [Google Scholar]
- Muhitch, M.J. (2003). Distribution of the glutamine synthetase isozyme GS(p1) in maize (Zea mays). J. Plant Physiol. 160 601–605. [DOI] [PubMed] [Google Scholar]
- Muhitch, M.J., Liang, H., Rastogi, R., and Sollenberger, K.G. (2002). Isolation of a promoter sequence from the glutamine synthetase1–2 gene capable of conferring tissue-specific gene expression in transgenic maize. Plant Sci. 163 865–872. [Google Scholar]
- Nazoa, P., Vidmar, J.J., Tranberger, T.J., Mouline, K., Damiani, I., Tillard, P., Zhuo, D., Glass, A.D.M., and Touraine, B. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52 689–703. [DOI] [PubMed] [Google Scholar]
- Oaks, A. (1992). A re-evaluation of nitrogen assimilation in roots. Bioscience 42 103–110. [Google Scholar]
- Oaks, A. (1994). Efficiency of nitrogen assimilation in C3 and C4 cereals. Plant Physiol. 106 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal, D., and Joy, K.W. (1973). Glutamine synthetase of pea leaves. I. Purification, stabilisation and pH optima. Arch. Biochem. Biophys. 159 113–122. [DOI] [PubMed] [Google Scholar]
- Riedel, J., Tishner, R., and Mäck, G. (2001). The chloroplastic glutamine synthetase (GS2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 213 396–401. [DOI] [PubMed] [Google Scholar]
- Rosen, H. (1957). A modified ninhydrin colorimetric analysis for amino acids. Arch. Biochem. Biophys. 67 10–15. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Kawabata, S., Hase, T., and Sugiyama, T. (1992). Differential effect of nitrate and light on the expression of glutamine synthetase and ferredoxin-dependent glutamate synthase in maize. Plant Cell Physiol. 33 1193–1198. [Google Scholar]
- Sakakibara, H., Shimizu, H., Hase, T., Yamazaki, Y., Takao, T., Shimonishi, Y., and Sugiyama, T. (1996). Molecular identification and characterization of cytosolic isoforms of glutamine synthetase in maize roots. J. Biol. Chem. 271 29561–29568. [DOI] [PubMed] [Google Scholar]
- Seebauer, J.R., Moose, S.P., Fabbri, B.J., Crossland, L.D., and Below, F.E. (2004). Amino acid metabolism in maize earshoots. Implications for assimilate preconditioning and nitrogen signaling. Plant Physiol. 136 4326–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry, P.R., and Jones, H.D. (2005). Trangenic wheat: Where do we stand after 12 years? Ann. Appl. Biol. 147 1–14. [Google Scholar]
- Suarez, M.F., Avila, C., Gallardo, F., Canton, F., Garcia-Gutierez, A., Claros, M.G., and Canovas, F. (2002). Molecular and enzymatic analysis of ammonium assimilation in woody plants. J. Exp. Bot. 53 891–904. [DOI] [PubMed] [Google Scholar]
- Tabuchi, M., Sugiyama, T., Ishiyama, K., Inoue, E., Sato, T., Takahashi, H., and Yamaya, T. (2005). Severe reduction in growth and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase 1;1. Plant J. 42 641–655. [DOI] [PubMed] [Google Scholar]
- Taira, M., Valtersson, U., Burkhardt, B., and Ludwig, R.A. (2004). Arabidopis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplats. Plant Cell 16 2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercé-Laforgue, T., Dubois, F., Ferrario-Mery, S., Pou de Crecenzo, M.A., Sangwan, R., and Hirel, B. (2004). Glutamate dehydrogenase of tobacco (Nicotiana tabacum L.) is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol. 136 4308–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, O., Yoshimura, Y., and Sentoku, N. (2005). Variation in the activity of some enzymes of photorespiratory metabolism in C-4 grasses. Ann. Bot. (Lond.) 96 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer, B., de Kochko, A., Fux, C.I., Beachy, R.N., and Fauquet, C. (1998). Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 37 1055–1067. [DOI] [PubMed] [Google Scholar]
- Voisin, A.S., Reidy, B., Parent, B., Rolland, G., Redondo, E., Gerentes, D., Tardieu, F., and Muller, B. (2006). Are ABA, ethylene or their interaction involved in the response of leaf growth to soil water deficit? An analysis using naturally occurring variation or genetic transformation of ABA production in maize. Plant Cell Environ. 29 1829–1840. [DOI] [PubMed] [Google Scholar]
- Wong, H.K., Chan, H.K., Coruzzi, G.M., and Lam, H.M. (2004). Correlation of ASN2 gene expression with ammonium metabolism in Arabidopsis. Plant Physiol. 134 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya, T., Obara, M., Nakajima, H., Sasaki, T., Hayakawa, T., and Sato, T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53 917–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.