Abstract
Brassinosteroids (BRs) are biosynthesized from campesterol via several cytochrome P450 (P450)–catalyzed oxidative reactions. We report the functional characterization of two BR-biosynthetic P450s from Arabidopsis thaliana: CYP90C1/ROTUNDIFOLIA3 and CYP90D1. The cyp90c1 cyp90d1 double mutant exhibits the characteristic BR-deficient dwarf phenotype, although the individual mutants do not display this phenotype. These data suggest redundant roles for these P450s. In vitro biochemical assays using insect cell-expressed proteins revealed that both CYP90C1 and CYP90D1 catalyze C-23 hydroxylation of various 22-hydroxylated BRs with markedly different catalytic efficiencies. Both enzymes preferentially convert 3-epi-6-deoxocathasterone, (22S,24R)-22-hydroxy-5α-ergostan-3-one, and (22S,24R)-22-hydroxyergost-4-en-3-one to 23-hydroxylated products, whereas they are less active on 6-deoxocathasterone. Likewise, cyp90c1 cyp90d1 plants were deficient in 23-hydroxylated BRs, and in feeding experiments using exogenously supplied intermediates, only 23-hydroxylated BRs rescued the growth deficiency of the cyp90c1 cyp90d1 mutant. Thus, CYP90C1 and CYP90D1 are redundant BR C-23 hydroxylases. Moreover, their preferential substrates are present in the endogenous Arabidopsis BR pool. Based on these results, we propose C-23 hydroxylation shortcuts that bypass campestanol, 6-deoxocathasterone, and 6-deoxoteasterone and lead directly from (22S,24R)-22-hydroxy-5α-ergostan-3-one and 3-epi-6-deoxocathasterone to 3-dehydro-6-deoxoteasterone and 6-deoxotyphasterol.
INTRODUCTION
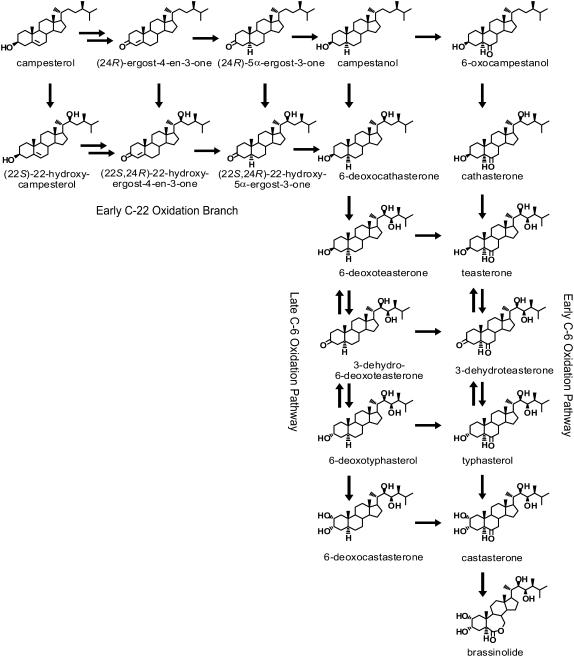
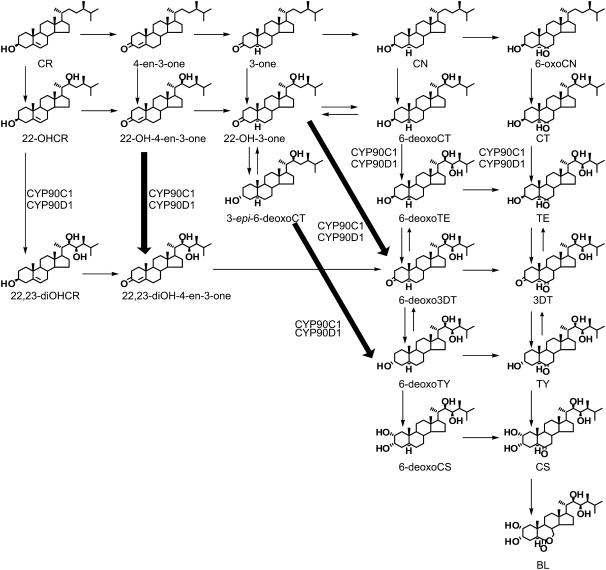
Brassinosteroids (BRs) are a group of plant steroid hormones that regulate plant growth and development. More than 50 naturally occurring BRs have been identified in a wide range of plant species (Bajguz and Tretyn, 2003; Fujioka and Yokota, 2003). The most abundant and widely occurring BRs in plants are C28 steroids, and among these, brassinolide (BL) is the most biologically active. The biosynthetic pathway of BL was elucidated by gas chromatography–mass spectrometry (GC-MS) identification of conversion products obtained from isotope-labeled intermediates fed to Catharanthus roseus cell suspensions and Arabidopsis thaliana seedlings (Sakurai, 1999; Fujioka et al., 2000; Noguchi et al., 2000). These studies show that campesterol (CR), the precursor of C28 BRs, is initially converted to campestanol (CN), which is then converted to BL via two parallel biosynthetic routes, the early and late C-6 oxidation pathways (Figure 1). In the course of early C-6 oxidation, CN is oxidized to 6-oxocampestanol (6-oxoCN), which is then successively converted to cathasterone (CT), teasterone (TE), 3-dehydroteaserone (3DT), typhasterol (TY), and castasterone (CS). In the late C-6 oxidation pathway, CN is first converted to 6-deoxocathasterone (6-deoxoCT) by C-22 hydroxylation and then further oxidized to 6-deoxoteasterone (6-deoxoTE), 3-dehydro-6-deoxoteasterone (6-deoxo3DT), 6-deoxotyphasterol (6-deoxoTY), 6-deoxocastasterone (6-deoxoCS), and CS, which is finally converted to BL. In all plant species analyzed to date, the 6-deoxo intermediates are predominant (Nomura et al., 2001), indicating that the late C-6 oxidation pathway is the main BR synthesis route. Fujioka et al. (2002) demonstrated the existence of an early C-22 oxidation branch, which was recently shown to be the main upstream BR synthesis route (Fujita et al., 2006).
Figure 1.
Pathways of BR Biosynthesis Deduced from in Vivo Metabolite Conversion Analyses.
The structural variability of BRs in the biosynthetic pathways results from the presence of several oxygen moieties at positions C-2, C-3, and C-6 of the steroid A/B rings and at side chain positions C-22 and C-23. These oxidative substitutions, except for the C-3 position, are introduced into steroids by cytochrome P450 monooxygenases (P450s) (Fujioka and Yokota, 2003). In Arabidopsis, the BR-biosynthetic mutants constitutive photomorphogenesis and dwarfism (cpd), dwarf4 (dwf4), and rotundifolia3 (rot3) were deficient in the P450s CYP90A1, CYP90B1, and CYP90C1, respectively (Szekeres et al., 1996; Choe et al., 1998; Kim et al., 1998). In tomato (Solanum lycopersicum), the dwarf (d) mutation causes a lesion in the Dwarf gene encoding CYP85 (Bishop et al., 1996), and the dumpy mutation has been suggested to affect an ortholog of Arabidopsis CPD/CYP90A1 (Koka et al., 2000). Rice (Oryza sativa) mutants BR-deficient dwarf1 and brassinosteroid-dependent1 (both designated brd1) have defective rice CYP85A (Hong et al., 2002; Mori et al., 2002); whereas other BR-deficient lines, such as ebisu dwarf (d2) and dwarf11 (d11), carry mutations in the P450 genes encoding CYP90D2 and CYP724B1, respectively (Hong et al., 2003; Tanabe et al., 2005).
The enzymatic functions of several BR-biosynthetic P450s were determined indirectly by analyzing the endogenous BR content in biosynthetic mutants or by measuring the efficiency of biosynthetic intermediates in rescuing their dwarf phenotypes. Using these approaches, the Arabidopsis P450s CPD/CYP90A1 and DWF4/CYP90B1 were identified as C-23 and C-22 hydroxylases, respectively (Szekeres et al., 1996; Choe et al., 1998). Recently, Kim et al. (2005a) reported that CYP90C1 and CYP90D1 are involved in distinct BR-biosynthetic steps, suggesting that CYP90C1 is responsible for TY C-2 hydroxylation to CS, while CYP90D1 likely participates in the oxidative C-3 epimerization of BRs. Analyses of the rice d2 and d11 mutants suggested that D2/CYP90D2 catalyzes the C-3 oxidation converting 6-deoxoTE to 6-deoxo3DT and TE to 3DT (Hong et al., 2003) and that the rice D11/CYP724B1 enzyme is involved in 6-deoxoTY and TY supply (Tanabe et al., 2005). The BR-biosynthetic pathways constitute a metabolic grid where enzymatic conversions, such as C-22 hydroxylation and C-6 oxidation, occur at several intermediate steps (Shimada et al., 2001; Fujioka et al., 2002). Broad substrate specificities of the BR-biosynthetic P450s can induce physiologically subordinate reaction routes in the mutants. Furthermore, in BR-deficient mutants, the expression of endogenous BR-biosynthetic P450s is upregulated (Bancos et al., 2002; Goda et al., 2002), potentially distorting the mutation effects on the endogenous BR content. As a result, the data obtained by indirect analyses are difficult to interpret, necessitating identification or confirmation of catalyzed reactions by biochemical assays using heterologously expressed enzymes.
Among the BR-biosynthetic P450s, only CYP85s of tomato, Arabidopsis, and rice are characterized by functional expression in yeast cells, which demonstrates that these enzymes catalyze a two-step C-6 oxidation converting 6-deoxoCS to CS (Bishop et al., 1999; Shimada et al., 2001, 2003; Hong et al., 2002). BR C-6 oxidases exist in two isoforms in Arabidopsis (CYP85A1 and CYP85A2) and tomato (CYP85A and CYP85A3). Nomura et al. (2005) and Kim et al. (2005b) demonstrated that CYP85A2 and CYP85A3 are bifunctional enzymes possessing both BR C-6 oxidase and BL synthase activities. Recently, we characterized functional CYP90B1, expressed in Escherichia coli, finding that the main reaction catalyzed by this enzyme is C-22 hydroxylation of CR to (22S)-22-hydroxycampesterol (22-OHCR) (Fujita et al., 2006). Sakamoto et al. (2006) recently reported that the rice double mutant deficient in both DWF4/CYP90B2 and D11/CYP724B1 has a severe dwarf phenotype and low BR levels. Biochemical characterization of CYP90B2 and CYP724B1 expressed in the baculovirus/insect cell system showed that these P450s are functionally redundant C-22 hydroxylases. Heterologous expression techniques proved instrumental in clarifying the roles of these enzymes; however, technical difficulties prevented their use for studying the functions of other BR-biosynthetic P450s.
The absence of CYP90C1 in the rot3 mutants results in a weak phenotype with reduced leaf elongation (Kim et al., 1998) but causes no appreciable alteration of the endogenous BR content (Kim et al., 2005a). Also, CYP90D1 deficiency does not have any effect on Arabidopsis morphology (Kim et al., 2005a). By contrast, a double null mutant devoid of both of these P450s enzymes has a severe dwarf stature and reduced BR content, suggesting that the closely related CYP90C1 and CYP90D1 function redundantly in BR biosynthesis (Kim et al., 2005a). Despite this expectation, GC-MS analysis of the endogenous BR content in the rot3-1 and cyp90d1 null mutants led Kim et al. (2005a) to the conclusion that these two enzymes play distinct roles, though these authors discussed some inconsistencies of their results, in particular those indicating redundancies of both the CYP90C1 and CYP90D1 functions in the respective mutants. Further support for distinct catalytic roles was from transgenic CYP90D1 overexpression in rot3-1 plants, which did not rescue the leaf phenotype of the mutant (Kim et al., 2005a). However, the authors gave no evidence for an increased level of the CYP90D1 protein or enzyme activity in the transgenic line, and the rot3-1 background already contained its endogenous CYP90D1 function. Therefore, these studies left unanswered questions regarding the relation between the CYP90C1 and CYP90D1 functions.
We describe genetic and biochemical characterization of the Arabidopsis CYP90C1 and CYP90D1 enzymes. We show that mutations in both the CYP90C1 and CYP90D1 genes result in a severe dwarf phenotype, which can be rescued by 22,23-dihydroxylated, but not 22-hydroxylated, BRs. When expressed in a baculovirus/insect cell system, CYP90C1 and CYP90D1 catalyzed C-23 hydroxylation of some 22-hydroxylated BRs, confirming that these P450s act as functionally redundant C-23 hydroxylases in BR biosynthesis. Kinetic studies showed that the early 22-hydroxylated intermediates are better CYP90C1 and CYP90D1 substrates than 6-deoxoCT. Based on these results, we propose a novel shortcut in BR biosynthesis, which allows direct conversion of early 22-hydroxylated intermediates to 6-deoxo3DT and 6-deoxoTY via C-23 hydroxylation.
RESULTS
Isolation of CYP90C1- and CYP90D1-Deficient Mutants
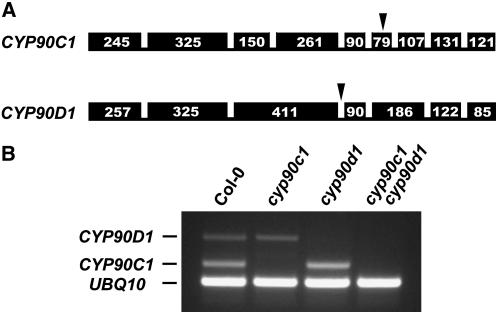
To gain insight into the functions of CYP90C1 and CYP90D1, we isolated mutants that carry T-DNA insertions within the genes encoding these P450 enzymes. The cyp90c1 and cyp90d1 mutants were identified in the Koncz collection of 90,000 T-DNA–tagged Arabidopsis Columbia (Col-0) lines following a PCR-based screen using combinations of T-DNA– and gene-specific primers (Ríos et al., 2002). Sequencing the genomic regions flanking the T-DNA borders determined that the insertion in CYP90C1 occurred in exon 6, 3220 bp downstream of the ATG codon, and in CYP90D1 between exons 3 and 4, 1596 bp downstrem of the ATG (Figure 2A). We crossed homozygous lines to generate a cyp90c1 cyp90d1 double mutant. RT-PCR assays performed with total RNA isolated from cyp90c1, cyp90d1, and cyp90c1 cyp90d1 seedlings clearly showed that the mutant lines do not contain detectable CYP90C1 or CYP90D1 transcript (Figure 2B).
Figure 2.
Null Mutations Caused by T-DNA Insertions in the CYP90C1 and CYP90D1 Genes.
(A) Exon-intron structures of the CYP90C1 and CYP90D1 genes. Bars represent exons, and numbers indicate exon sizes in nucleotide base pairs. Arrowheads show T-DNA insertion sites in the cyp90c1 and cyp90d1 mutant alleles.
(B) RT-PCR transcript analysis of the CYP90C1, CYP90D1, and UBQ10 transcripts in wild-type Col-0, cyp90c1, cyp90d1, and cyp90c1 cyp90d1 seedlings.
Characterization of the Mutant Phenotypes
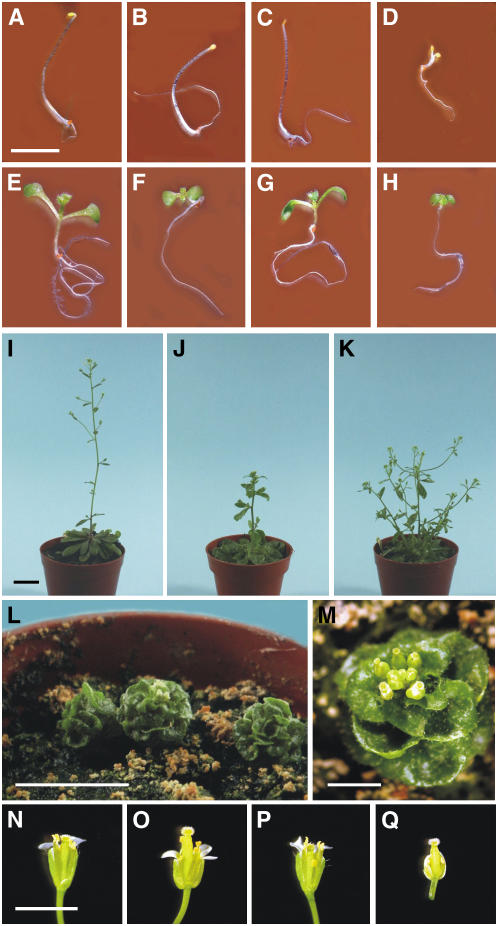
We characterized the cyp90c1, cyp90d1, and cyp90c1 cyp90d1 mutants to determine how their phenotypes are affected by the absence of CYP90C1 and/or CYP90D1. As anticipated, neither cyp90c1 nor cyp90d1 gave recognizable phenotypes at any developmental stage in heterozygous plants. In agreement with earlier reports describing reduced leaf elongation in rot3 mutants (Tsuge et al., 1996; Kim et al., 1998), both etiolated and light-grown seedlings of our cyp90c1 mutant had somewhat rounded cotyledons compared with those of the respective wild-type controls (Figures 3A, 3B, 3E, and 3F). By contrast, cyp90d1 seedlings were indistinguishable from the wild type (Figures 3C and 3G). The cyp90c1 cyp90d1 double mutant exhibited a dwarf phenotype. In etiolated seedlings, the hypocotyl length was <10% of wild-type control, and the cotyledons were opened. Light-grown double mutants had reduced hypocotyl length and short, epinastic cotyledons (Figures 3D and 3H).
Figure 3.
Morphology of the CYP90C1- and CYP90D1-Deficient Mutants.
(A) to (H) Seven-day-old etiolated ([A] to [D]) and light-grown ([E] to [H]) seedlings. Wild-type Col-0 ([A] and [E]), cyp90c1 ([B] and [F]) cyp90d1 ([C] and [G]), and cyp90c1 cyp90d1 ([D] and [H]). Bar = 5 mm.
(I) to (M) One-month-old wild-type Col-0 (I), cyp90c1 (J), cyp90d1 (K), and cyp90c1 cyp90d1 ([L] and [M]) plants. Bars = 30 mm in (I) to (L) and 5 mm in (M).
(N) to (Q) Flowers of wild-type (N), cyp90c1 (O), cyp90d1 (P), and cyp90c1 cyp90d1 (Q) plants. Bar = 5 mm.
Compared with the wild type, 1-month-old cyp90c1 plants formed a more compact rosette resulting from shortened petioles. In addition, the inflorescence stem was shorter and thicker, with larger cauline leaves (Figures 3I and 3J). At the same age, cyp90d1 plants showed reduced apical dominance with multiple, shortened primary inflorescence stems and elongated secondary inflorescence stems (Figure 3K). In contrast with the weak cyp90c1 and cyp90d1 phenotypes, cyp90c1 cyp90d1 double mutant plants were severely dwarfed, with a globular shape and curly, succulent leaves (Figures 3L and 3M).
The flowers of cyp90d1 and wild-type plants were indistinguishable; cyp90c1 flowers had shorter petals and anthers (Figures 3N to 3P). The pistil was much longer than the anthers in cyp90c1 plants, resulting in less efficient self-fertilization (data not shown). Due to the reduced petals, the stigma emerged early during flower bud formation. Compared with single mutants and the wild type, cyp90c1 cyp90d1 double mutant flowers were smaller and developed ∼1 week later. The petals were short and almost entirely hidden by the sepals. The stamens were also very short due to the rudimentary filaments (Figure 3Q). Double mutant flowers were male sterile because they never opened to release pollen, even though the capsule size was normal.
Heterologous Expression of CYP90C1 and CYP90D1 in Insect Cells
We characterized biochemical properties of CYP90C1 and CYP90D1 by expressing their cDNAs in insect cells using a baculovirus expression system. We confirmed expression in the microsomal fractions via SDS-PAGE analysis (see Supplemental Figure 1A online). Solubilized insect cell microsomal fractions expressing either CYP90C1 or CYP90D1 had a cytochrome P450-specific reduced-CO difference spectrum with a clear absorption peak at 450 nm (see Supplemental Figures 1B and 1C online). In the insect cell cultures, the expression levels of CYP90C1 and CYP90D1 proteins were 80 nmol P450/liter and 113 nmol P450/liter, respectively. However, mock-infected cells did not have a 450-nm peak (data not shown). These data indicate that insect cell–expressed CYP90C1 and CYP90D1 proteins are active P450s.
Catalytic Activities of CYP90C1 and CYP90D1
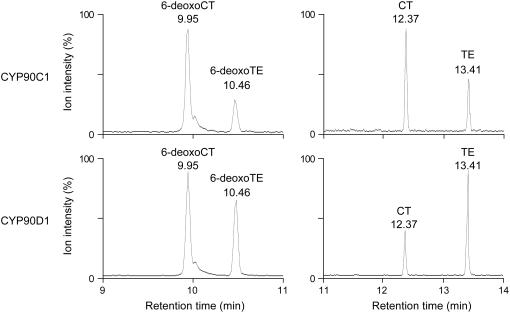
To determine the catalytic functions of CYP90C1 and CYP90D1, we performed in vitro conversion assays using the microsomal fractions containing CYP90C1 or CYP90D1, each BR-biosynthetic intermediates of the late C-6 oxidation pathway, and purified Arabidopsis NADPH-P450 reductase (Mizutani and Ohta, 1998), which was required to support electron transfer from NADPH to the P450s. After 2 h of incubation at 30°C, BRs were extracted from the reaction mixtures, converted to methaneboronate-trimethylsilyl derivatives, and then analyzed by GC-MS. Of the BR intermediates tested, CYP90C1 metabolized 6-deoxoCT to a product with the same GC retention time and mass spectrum as the authentic 6-deoxoTE standard (Figure 4). CYP90C1 also metabolized CT, the corresponding early C-6 oxidation intermediate, to TE. The other intermediates assayed in these reactions did not yield any detectable conversion product (see Supplemental Table 1 online). Although a recent report suggested that Arabidopsis CYP90C1 functions as a BR C-2 hydroxylase (Kim et al., 2005a), we did not detect conversion of 6-deoxoTY to 6-deoxoCS under our assay conditions. Our results suggest that CYP90C1 catalyzes C-23 hydroxylation, converting 6-deoxoCT to 6-deoxoTE and CT to TE.
Figure 4.
GC-MS Analysis of BR Metabolites Produced by Heterologously Expressed CYP90C1 and CYP90D1.
Selected ion chromatograms of the reaction products generated by CYP90C1 and CYP90D1 from 6-deoxoCT and CT, respectively.
We also performed conversion assays with CYP90D1 under the same conditions, using the same BR intermediates (see Supplemental Table 1 online). We found that CYP90D1 converted 6-deoxoCT and CT to 6-deoxoTE and TE, respectively (Figure 4), but the other BR intermediates were not metabolized. This indicates that CYP90D1 catalyzes the same C-23 hydroxylation reaction as CYP90C1.
Kinetic Studies with CYP90C1 and CYP90D1
Recently, we found that CYP90B1 preferentially catalyzes C-22 hydroxylation of CR to form 22-OHCR, indicating that the early C-22 oxidation branch is the predominant route of the BR-biosynthetic pathway in Arabidopsis (Fujita et al., 2006). Because 22-hydroxylated compounds of the BR pathway are likely potential substrates of C-23 hydroxylases, we examined the metabolism of 22-OHCR, (22S,24R)-22-hydroxyergost-4-en-3-one (22-OH-4-en-3-one), (22S,24R)-22-hydroxy-5α-ergostan-3-one (22-OH-3-one), 6-deoxoCT, and 3-epi-6-deoxoCT by CYP90C1 and CYP90D1 (see Supplemental Figure 2 online). These conversion assays revealed that both enzymes metabolized all of these 22-hydroxylated compounds to the corresponding 22,23-dihydroxylated products (Table 1). At 20 μM substrate concentration, CYP90C1 had the highest activity with 3-epi-6-deoxoCT, lower activity with 22-OH-3-one and 22-OH-4-en-3-one, and even weaker activity with 22-OHCR. 6-deoxoCT proved to be the worst substrate. CYP90D1 also was highly active on 3-epi-6-deoxoCT, 22-OH-3-one, and 22-OH-4-en-3-one, while 22-OHCR and 6-deoxoCT were poor substrates. The results indicate that CYP90C1 and CYP90D1 are C-23 hydroxylases with a broad specificity for 22-hydroxylated substrates.
Table 1.
GC-MS Data of Products Obtained in CYP90C1 and CYP90D1 Reconstitution Assays
| Enzyme | Substrate | Product | Retention Time (min) | Characteristic Ions m/z (Relative Intensity Percentage) |
|---|---|---|---|---|
| CYP90C1 | 22-OHCR | 22,23-OHCR | 10.34 | 529 (0.6), 438 (4.1), 243 (8.1), 129 (100) |
| synthetic | 10.37 | 529 (0.3), 438 (3.3), 243 (7.8), 129 (100) | ||
| 22-OH-4-en-3-one | 22,23-OH-4-en-3-one | 11.55 | 454 (2.1), 281 (3.1), 229 (25.9), 124 (100) | |
| synthetic | 11.53 | 454 (1.0), 281 (1.2), 229 (25.8), 124 (100) | ||
| 22-OH-3-one | 6-deoxo3DT | 10.72 | 456 (3.4), 283 (7.7), 231(100), 155 (52.5) | |
| synthetic | 10.70 | 456 (1.4), 283 (3.7), 231(100), 155 (52.3) | ||
| 3-epi-6-deoxoCT | 6-deoxoTY | 9.68 | 530 (0.3), 440 (8.0), 425 (18.8), 215 (100) | |
| synthetic | 9.68 | 530 (0.2), 440 (6.7), 425 (17.2), 215 (100) | ||
| 6-deoxoCT | 6-deoxoTE | 10.46 | 530 (0.8), 516 (4.8), 425 (7.4), 215 (100) | |
| synthetic | 10.46 | 530 (0.1), 516 (4.5), 425 (8.5), 215 (100) | ||
| CYP90D1 | 22-OHCR | 22,23-OHCR | 10.35 | 529 (0.1), 438 (4.1), 243 (7.3), 129 (100) |
| synthetic | 10.37 | 529 (0.3), 438 (3.3), 243 (7.8), 129 (100) | ||
| 22-OH-4-en-3-one | 22,23-OH-4-en-3-one | 11.51 | 454 (2.1), 281 (3.3), 229 (25.9), 124 (100) | |
| synthetic | 11.53 | 454 (1.0), 281 (1.2), 229 (25.8), 124 (100) | ||
| 22-OH-3-one | 6-deoxo3DT | 10.70 | 456 (1.7), 283 (6.3), 231(100), 155 (54.5) | |
| synthetic | 10.70 | 456 (1.4), 283 (3.7), 231(100), 155 (52.3) | ||
| 3-epi-6-deoxoCT | 6-deoxoTY | 9.68 | 530 (0.1), 440 (7.3), 425 (18.6), 215 (100) | |
| synthetic | 9.68 | 530 (0.2), 440 (6.7), 425 (17.2), 215 (100) | ||
| 6-deoxoCT | 6-deoxoTE | 10.46 | 530 (0.8), 516 (4.6), 425 (7.1), 215 (100) | |
| synthetic | 10.46 | 530 (0.1), 516 (4.5), 425 (8.5), 215 (100) |
To further investigate the specificities of CYP90C1 and CYP90D1, we determined their kinetic parameters using 22-hydroxylated intermediates. Of the compounds tested, CYP90C1 showed the highest dissociation constant (Km) and lowest catalytic efficiency (kcat/Km) with 6-deoxoCT. In the case of 3-epi-6-deoxoCT, the Km of CYP90C1 was 5.3-fold lower and the kcat value 22.6-fold higher than those for 6-deoxoCT, resulting in a 120-fold higher catalytic efficiency compared with 6-deoxoCT. The kinetic parameters determined with 22-OH-4-en-3-one and 22-OH-3-one were comparable to the values obtained with 3-epi-6-deoxoCT. However, 22-OHCR was a poor substrate similar to 6-deoxoCT (Table 2).
Table 2.
Kinetic Parameters of CYP90C1- and CYP90D1-Catalyzed Reactions with 22-HydroxyBRs
|
Km
|
kcat
|
kcat/Km
|
||
|---|---|---|---|---|
| Enzymes | Substrate | (μM) | (min−1) | (min−1 μM−1) |
| CYP90C1 | 22-OHCR | 19.4 ± 1.18 | 0.11 ± 0.01 | 0.0057 ± 0.0003 |
| 22-OH-4-en-3-one | 4.94 ± 0.02 | 1.40 ± 0.05 | 0.29 ± 0.02 | |
| 22-OH-3-one | 8.61 ± 0.34 | 2.75 ± 0.04 | 0.32 ± 0.01 | |
| 3-epi-6-deoxoCT | 6.61 ± 0.38 | 3.01 ± 0.22 | 0.45 ± 0.018 | |
| 6-deoxoCT | 35.9 ± 1.35 | 0.14 ± 0.004 | 0.0038 ± 0.0002 | |
| CYP90D1 | 22-OHCR | 18.1 ± 1.01 | 0.12 ± 0.01 | 0.0068 ± 0.0008 |
| 22-OH-4-en-3-one | 0.77 ± 0.03 | 1.01 ± 0.07 | 1.32 ± 0.09 | |
| 22-OH-3-one | 0.73 ± 0.04 | 1.27 ± 0.05 | 1.73 ± 0.14 | |
| 3-epi-6-deoxoCT | 1.06 ± 0.03 | 1.10 ± 0.04 | 1.04 ± 0.006 | |
| 6-deoxoCT | 16.9 ± 1.12 | 0.29 ± 0.01 | 0.017 ± 0.001 |
The substrate specificity of CYP90D1 resembled that of CYP90C1. The Km and kcat values measured with 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT were similar, giving 60- to 100-fold higher catalytic efficiency than those detected with 6-deoxoCT, and the weakest substrate was 22-OHCR. Comparing the two enzymes, the Km values of CYP90D1 for 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT were 6- to 11-fold lower, and the kcat values were 1.4- to 3-fold lower than those of CYP90C1 (Table 2).
Our in vitro experiments determined that 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT were preferential substrates of CYP90C1 and CYP90D1, whereas 22-OHCR and 6-deoxoCT were poor substrates. These data suggest that in vivo the two enzymes can use similar substrates. It is noteworthy that C-23 hydroxylation of 3-epi-6-deoxoCT and 22-OH-3-one directly supplies 6-deoxoTY and 6-deoxo-3DT, respectively, which are downstream of 6-doxoTE in the BR-biosynthetic pathway (Figure 1). In addition, CYP90C1 and CYP90D1 convert 22-OHCR and 22-OH-4-en-3-one to the novel biosynthetic intermediates (22R,23R)-22,23-dihydroxycampesterol (22,23-diOHCR) and (22R,23R)-22,23-dihydroxy-campest-4-en-3-one (22,23-diOH-4-en-3-one), respectively.
Endogenous BR Levels in cyp90c1 cyp90d1 and Wild-Type Arabidopsis Plants
To investigate the effects of BR C-23 hydroxylase deficiency, we determined endogenous BR levels of 1-month-old cyp90c1 cyp90d1 plants by quantitative GC-MS analysis (Table 3). The amounts of 6-deoxoCT and 6-deoxoTE, which are early intermediates of the late C-6 oxidation pathway, were comparable to those of the wild-type control. However, we observed a progressive reduction in the levels of 6-deoxo3DT (23% of the control) and 6-deoxoTY (5% of the control), while 6-deoxoCS and CS were near or below the detection limit in the mutant. BL and the early C-6 oxidation intermediates CT, TE, 3DT, and TY were not detectable in either the wild-type or double mutant plants.
Table 3.
Endogenous BR Levels of Col-0 and cyp90c1 cyp90d1 Plants (ng kg−1 FW)
| BRs | Wild Type | cyp90c1 cyp90d1 | Wild Type | cyp90c1 cyp90d1 |
|---|---|---|---|---|
| 1st Analysis 27.1 g (FW) 54.4 g (FW) | 2nd Analysis 20.0 g (FW) 44.2 g (FW) | |||
| CT | ND | ND | ND | ND |
| TE | ND | ND | 3.5 | Trace |
| 3DT | ND | ND | 47 | 10 |
| TY | ND | ND | Trace | ND |
| 22-OHCR | 326 | 305 | 180 | 215 |
| 22-OH-4-en-3-one | ND | ND | ND | ND |
| 22-OH-3-one | 412 | 380 | 950 | 448 |
| 6-deoxoCT | 1502 | 1057 | 772 | 456 |
| 3-epi-6-deoxoCT | 913 | 625 | 697 | 427 |
| 22,23-diOHCR | ND | ND | ND | ND |
| 22,23-diOH-4-en-3-one | ND | ND | ND | ND |
| 6-deoxoTE | 154 | 186 | 40 | 20 |
| 6-deoxo3DT | 266 | 62 | 255 | 25 |
| 6-deoxoTY | 911 | 45 | 988 | ND |
| 6-deoxoCS | 394 | ND | 1318 | 3 |
| CS | 177 | ND | 141 | ND |
| BL | ND | ND | ND | ND |
ND, not detectable; FW, fresh weight.
Because our biochemical results indicated that early 22-hydroxylated BR precursors are preferential substrates of CYP90C1 and CYP90D1, we also determined the levels of these intermediates (Table 3). In wild-type plants, the levels of 22-OH-3-one and 3-epi-6-deoxoCT, the preferred CYP90C1 and CYP90D1 substrates, were similar to 6-deoxoCT. Compared with these intermediates, the amount of 22-OHCR was severalfold lower. GC-MS analysis could not detect 22,23-diOHCR or 22,23-diOH-4-en-3-one, the 23-hydroxylated derivatives of 22-OHCR and 22-OH-4-en-3-one, in either the wild-type or mutant plants.
The cyp90c1 cyp90d1 plants are deficient in 6-deoxo3DT and 6-deoxoTY; therefore, we expected that the C-23 hydroxylation substrates 22-OH-3-one, 3-epi-6-deoxoCT, and perhaps 6-deoxoCT may accumulate in this mutant. However, our data revealed that the inactivation of CYP90C1 and CYP90D1 did not increase the levels of 22-OH-3-one, 3-epi-6-deoxoCT, and 6-deoxoCT, which were slightly lower in the double mutant than those measured in wild-type plants (Table 3).
Phenotypic Rescue of the cyp90c1 cyp90d1 Mutant by BR Intermediates
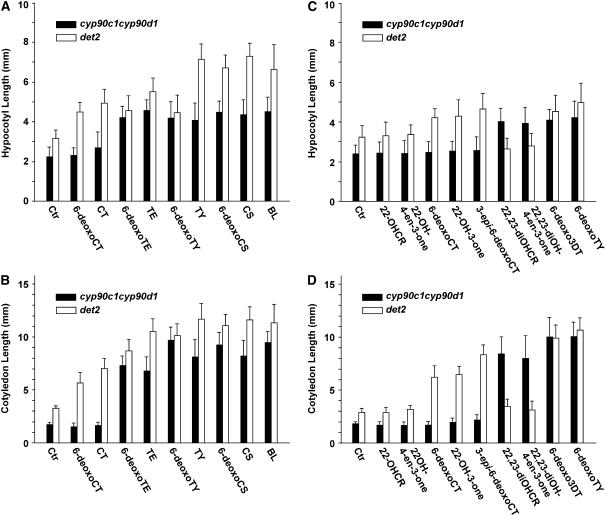
To determine whether the dwarf phenotype of the cyp90c1 cyp90d1 mutant is caused by BR deficiency, we performed feeding rescue assays with seedlings that received BL or one of the following precursors: 6-deoxoCT, CT, 6-deoxoTE, TE, 6-deoxoTY, TY, 6-deoxoCS, and CS at 100 nM each. In 7-d-old etiolated seedlings, BL treatment caused an approximately twofold hypocotyl elongation, but the seedlings remained shorter than the steroid 5α-reductase–deficient de-etiolated 2 (det2) BR-biosynthetic mutant used as control (Li et al., 1996). We found that 6-deoxoTE, TE, 6-deoxoTY, TY, 6-deoxoCS, and CS at 100 nM concentration were as efficient as BL in rescuing the elongation of the cyp90c1 cyp90d1 hypocotyl. By contrast, CT elicited only a weak growth response, and 6-deoxoCT had no appreciable effect on hypocotyl length (Figure 5A).
Figure 5.
Rescue of Hypocotyl and Cotyledon Elongation in the cyp90c1 cyp90d1 Mutant by BRs.
Hypocotyl elongation of etiolated seedlings (A) and cotyledon elongation of light-grown seedlings (B) treated by intermediates (100 nM) of the early and late C-6 oxidation pathways. Hypocotyl elongation of etiolated seedlings (C) and cotyledon elongation of light-grown seedlings (D) treated by intermediates (100 nM) of the early C-22 oxidation pathway. Seedlings of det2 were used as control. Ctr, control; 22-OHCR, 22-hydroxycampesterol; 3-epi-6-deoxoCT, 3-epi-6-deoxocathasterone; 22,23-diOHCR, 22,23-dihydroxycampesterol. Panels show representative experiments. Values are averages of 50 seedlings; error bars indicate se.
Because the CYP90C1/ROT3 function is important in leaf elongation, we also measured the BR rescue of cotyledon elongation in 7-d-old green seedlings grown under normal light/dark photoperiods. Compared with the untreated control, a 48-h treatment with 100 nM BL resulted in an approximately fivefold cotyledon elongation in the cotyledons of the cyp90c1 cyp90d1 seedlings (Figure 5). In the cotyledon assay, the same concentration of 6-deoxoCT and CT did not have detectable effect, but all other BR intermediates induced at least threefold elongation (Figure 5B). Whereas the absolute det2 hypocotyl length exceeded those of the double mutant in each treatment, their BR-induced relative elongation was weaker. In contrast with the cyp90c1 cyp90d1 mutant, det2 seedlings showed a clear elongation response to all tested BR intermediates, although 6-deoxoCT, CT, and 6-deoxoTE were less effective than the other compounds (Figure 5A).
We also examined the efficiency of several 22-hydroxylated intermediates and their 23-hydroxylated derivatives in rescuing the phenotypes of cyp90c1 cyp90d1 and the det2 control (Figures 5C and 5D). The hypocotyl and cotyledon lengths of cyp90c1 cyp90d1 plants were not appreciably affected by 22-OHCR, 22-OH-4en-3-one, 22-OH-3-one, 6-deoxoCT, and 3-epi-6-deoxoCT. By contrast, exogenous application of the 22,23-dihydroxylated compounds 22,23-diOHCR, 22,23-diOH-4-en-3-one, and 6-deoxo3DT efficiently rescued both hypocotyl and cotyledon elongation. As expected, det2 seedling phenotype was rescued by all intermediates with saturated A and B rings, namely, 6-deoxoCT, 22-OH-3-one, 3-epi-6-deoxoCT, and 22,23-diOH-3-one. These intermediate rescue analyses indicate that C-23 hydroxylation of BRs is impaired in the cyp90c1 cyp90d1 mutant. The results support the proposed C-23 hydroxylase function of CYP90C1 and CYP90D1 determined from our enzymatic experiments. Furthermore, the rescue data also indicate a potential metabolic flow from 22,23-diOHCR and 22,23-diOH-4-en-3-one toward bioactive BRs in vivo.
DISCUSSION
CYP90C1 and CYP90D1 Are BR C-23 Hydroxylases
In this report, we provide biochemical evidence that CYP90C1 and CYP90D1 are BR C-23 hydroxylases. This finding is well supported by intermediate feeding experiments, showing stimulation of hypocotyl and cotyledon elongation of the cyp90c1 cyp90d1 mutant by 23-hydroxylated BRs. Based on similar BR intermediate feeding rescue of the cpd mutant, CPD/CYP90A1 has been proposed to function as C-23 hydroxylase in Arabidopsis (Szekeres et al., 1996). However, the severe dwarf phenotype of cpd plants that have wild-type CYP90C1 and CYP90D1 enzymes suggests that CYP90A1 must have an enzymatic function different from C-23 hydroxylation. To address this contradiction, we investigated the enzymatic function of baculovirus/insect cell–expressed CYP90A1. Our preliminary data indicate that CYP90A1 acts earlier than CYP90C1 and CYP90D1 in the early C-22 oxidation pathway and performs a function different from C-23 hydroxylation (T. Ohnishi and M. Mizutani, unpublished results).
Previously, Kim et al. (2005a) reported that only the Arabidopsis mutant lacking both CYP90C1 and CYP90D1 has a severe BR-deficient phenotype. Nevertheless, based on intermediate rescue and GC-MS analysis data, these authors proposed distinct functions for these two P450s. They suggested that CYP90C1 may catalyze the C-2 hydroxylation reaction leading to 6-deoxoCS and CS, whereas CYP90D1 would act upstream of this conversion step in the BR pathway. However, as Kim et al. (2005a) pointed out, their rescue and BR analysis data were somewhat inconclusive and difficult to assess, principally because single mutations of either CYP90C1 or CYP90D1 gave rise to only minor effects on the endogenous BRs and the phenotype. An apparent controversy is that their rot3-1 cyp90d1 double mutant was rescued by all intermediates downstream of TE in the early C-6 oxidation pathway and 6-deoxoCT in the late C-6 oxidation pathway, although both of these BRs are upstream of the C-2 hydroxylation step that, according to these authors, should be blocked by the rot3-1 mutation. In agreement with Kim et al. (2005a), our GC-MS analyses also detected a considerable amount of 23-hydroxylated 6-deoxoTE in the double mutant plants lacking both CYP90C1 and CYP90D1 (Table 3). This indicates that another Arabidopsis enzyme(s) may also have weak steroid C-23 hydroxylase activity. Nevertheless, our biochemical data clearly indicate redundant C-23 hydroxylase functions for CYP90C1 and CYP90D1, in good agreement with the finding that only 23-hydroxylated compounds rescue the elongation defect of the cyp90c1 cyp90d1 double mutant.
The BR-deficient d2 mutants of rice carry lesions in the D2 gene encoding CYP90D2, a P450 with significant sequence identity to CYP90C1 (46%) and CYP90D1 (54%) (Hong et al., 2003). Endogenous BR measurement and intermediate feeding analyses of d2 plants led to the conclusion that D2/CYP90D2 catalyzes C-3 oxidation of 6-deoxoTE and TE to 6-deoxo3DT and 3DT, respectively (Hong et al., 2003). However, the interpretation of their data is difficult because the rice genome contains a second CYP90D gene, CYP90D3, which is highly homologous with CYP90D2 (Hong et al., 2003). Thus, these two CYP90D enzymes have potentially redundant functions; therefore, CYP90D2 may have BR C-23 hydroxylase activity similar to that of Arabidopsis ROT3/CYP90C1 and CYP90D1.
Novel C-23 Hydroxylation Shortcuts
Our enzyme kinetic studies obtained with CYP90C1 and CYP90D1 indicated that 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT are preferred substrates of C-23 hydroxylases in vitro. These 22-hydroxylated compounds lie in the early C-22 oxidation branch, in which CR is first 22-hydroxylated to 22-OHCR and further converted to 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT (Fujioka et al., 2002). The authenticity of this early C-22 oxidation branch is supported by feeding studies by Fujioka et al. (2002) as well as the identification of 22-OHCR, 22-OH-3-one, and 3-epi-6-deoxoCT in Arabidopsis (Fujioka et al., 2002; this study). Furthermore, our recent observations indicate that Arabidopsis CYP90B1 preferentially catalyzes C-22 hydroxylation of CR to form 22-OHCR (Fujita et al., 2006). Thus, BR biosynthesis is likely initiated primarily from 22-OHCR through the early C-22 hydroxylation route in Arabidopsis.
Based on our results, we propose a novel route of the BR-biosynthetic pathway where the primary substrates of C-23 hydroxylation are early 22-hydroxylated intermediates (Figure 6). In the late C-6 oxidation pathway, 6-deoxo3DT and 6-deoxoTY are synthesized from CR via six and seven steps, respectively. The synthesis occurs in the following sequence: CR → 4-en-3-one → 3-one → CN → 6-deoxoCT → 6-deoxoTE → 6-deoxo3DT → 6-deoxoTY (Figure 1). Our results indicate that 6-deoxo3DT and 6-deoxoTY may be directly derived from 22-OH-3-one and 3-epi-6-deoxoCT by C-23 hydroxylation and synthesized from CR in only four and five steps, respectively (Figure 6, thick arrows). The possibility of the direct conversion from 3-epi-6-deoxoCT to 6-deoxoTY has already been predicted by Fujioka et al. (2002) on the basis of their chemical structures. This synthetic route of BL from CR is shorter by two enzymatic steps because a C-3 reduction step (3-one → CN) and a C-3 oxidation step (6-deoxoTE → 6-deoxo3DT) are bypassed. We therefore designate the reaction routes leading from the early 22-hydroxylated intermediates to 6-deoxo3DT and 6-deoxoTY as C-23 hydroxylation shortcuts (Figure 6).
Figure 6.
Proposed Pathway of BR Biosynthesis.
The flow of intermediates is indicated by arrows. The C-23 hydroxylation shortcuts from 22-OH-4-3-one to 22,23-diOH-4-en-3-one, from 22-OH-3-one to 6-deoxo3DT, and from 3-epi-6-deoxoCT to 6-deoxoTY are highlighted by thick arrows. The conversion steps catalyzed by CYP90C1 and CYP90D1 are indicated.
The observed CYP90C1 and CYP90D1 substrate preferences are indicative of the functional importance of the shortcuts. In vitro C-23 hydroxylation of 6-deoxoCT by CYP90C1 and CYP90D1 has a much lower catalytic efficiency than those measured with 22-OH-3-one and 3-epi-6-deoxoCT, even though the endogenous content of these three compounds is comparable in Arabidopsis. Furthermore, whereas the levels of the 22,23-dihydroxylated products 6-deoxo-3DT and 6-deoxoTY in the cyp90c1 cyp90d1 double mutant are severely reduced, the relatively high abundance of endogenous 6-deoxoTE (Table 3; Kim et al., 2005a) suggests that this intermediate is not easily metabolized to bioactive BRs. In addition, 22-OH-3-one and 3-epi-6-deoxoCT feeding readily stimulates the elongation of hypocotyls of det2 seedlings (Fujioka et al., 2002; Figure 5). In vivo evidence for the subordinate role of the biosynthetic route via 6-oxoCN was provided by the analysis of BR intermediate pools in the cyp85a1 cyp85a2 double mutant (Kwon et al., 2005). In this late C-6 oxidation-deficient mutant, early C-6 oxidation is fully functional, allowing normal synthesis of 6-oxoCN. Whereas the cyp85a1 cyp85a2 plants are BR deficient, they contain higher than wild-type levels of 6-oxoCN, indicating that this compound is inefficiently used in BR synthesis. This is consistent with in vitro conversion assays that detected no C-22 hydroxylation of 6-oxoCN by CYP90B1 (Fujita et al., 2006). Similarly, CN is a very poor substrate for C-22 hydroxylation by CYP90B1 and CYP724B (Fujita et al., 2006; Ohnishi et al., 2006), and 6-deoxoCT is a poor substrate for C-23 hydroxylation by CYP90C1 and CYP90D1 (Table 2). Fujioka et al. (1997) have observed that CN and 6-oxoCN show extremely low activity in rescuing the det2 phenotype, though these compounds are downstream of the predicted DET2 function. Taken together, these data suggest that the side chain hydroxylation steps from CN and 6-oxoCN in the late and early C-6 oxidation pathways are physiologically less significant. Hence, the reactions via the C-23 hydroxylation shortcuts likely constitute a more important route in the BR-biosynthetic pathway.
Although we found that in vitro 22-OH-4-en-3-one is also a preferred substrate of CYP90C1 and CYP90D1, this intermediate and its product, 22,23-diOH-4-en-3-one, were not detectable by GC-MS analysis in wild-type Arabidopsis (Fujioka et al., 2002; this study). However, 22,23-diOH-4-en-3-one showed growth stimulation in the hypocotyl and cotyledon elongation assays with the cyp90c1 cyp90d1 mutant, indicating a likely intermediary role. The low levels of these intermediates may be due to their efficient conversion to downstream intermediates. Interestingly, 22-OH-4-en-3-one was detected in det2 seedlings and also in cultured cells of C. roseus (Fujioka et al., 2002); therefore, these compounds are probably involved in 6-deoxo3DT production in vivo. By contrast, 22-OHCR is not a favorable substrate for CYP90C1 and CYP90D1 in vitro; therefore, it is less conceivable that 22-OHCR is converted to 22,23-diOH-4-en-3-one via 22,23-diOHCR.
Intriguingly, we found that the disruption of C-23 hydroxylation in cyp90c1 cyp90d1 plants does not result in the accumulation of the expected potential CYP90C1 and CYP90D1 substrates. Our GC-MS analyses of all known BR intermediates revealed that the total amount of these compounds in the double mutant is only ∼30 to 50% of that of the wild type (Table 3), although BR deficiency strongly induces the expression of BR-biosynthetic genes. This suggests that the cyp90c1 cyp90d1 mutations may block the whole BR-biosynthetic pathway. Most BR-related enzymes, such as CYP85A, CYP90A1, CYP90B1, CYP90C1, CYP90D1, and DET2, are likely localized on the endoplasmic reticulum membrane; therefore, the colocalization of these enzymes may facilitate multienzyme complex formation and metabolic channeling in BR synthesis (Winkel, 2004; Jorgensen et al., 2005). The absence of both the CYP90C1 and CYP90D1 proteins in the double mutant may disrupt such interaction between the BR-related enzymes. In addition, the reduction of the total BR levels in the mutant suggests unknown mechanisms capable of removing excess amounts of unused BR intermediates. Rouleau et al. (1999) identified an inducible steroid sulfotransferase (BNST3) in Brassica napus that preferentially sulfonates early BR-biosynthetic intermediates, such as 24-epiCT, at the C-22 hydroxy group. Another possibility is that the pools of 22-OH-3-one and 3-epi-6-deoxoCT may be stabilized by reversible glucosylation or acylation, which have been suggested to regulate TE and BL levels (Asakawa et al., 1996; Poppenberger et al., 2005).
Physiological Functions of CYP90C1 and CYP90D1
Mutations of Arabidopsis DET2, DWF4, and CPD genes cause BR deficiency and dwarfism, indicating that these genes code for nonredundant BR-biosynthetic functions (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996; Azpiroz et al., 1998; Choe et al., 1998). However, two Arabidopsis genes, CYP85A1 and CYP85A2, encode BR C-6 hydroxylase enzymes (Shimada et al., 2001, 2003), and only mutants defective in both genes show the characteristics of the BR dwarf phenotype (Kwon et al., 2005; Nomura et al., 2005). CYP85A1 and CYP85A2 expression displays differential organ-specific and developmental regulation, which can be important for flexible control over bioactive BR synthesis (Shimada et al., 2003; Castle et al., 2005). Our functional analyses of CYP90C1 and CYP90D1 indicate that in Arabidopsis C-23 hydroxylation is also performed by redundant enzymes. These data are consistent with observations that deficiency in either CYP90C1 or CYP90D1 does not result in severe BR deficiency, while the lack of both enzymes does (Kim et al., 2005a).
While CYP90C1 and CYP90D1 show similar biochemical properties, they are classified within different P450 subfamilies because they show only 47% identity in their amino acid sequences. Recently, we have reported that CYP90Bs and CYP724Bs from rice and tomato, which share only 40 to 45% amino acid sequence identity, redundantly function as C-22 hydroxylases in the BR-biosynthetic pathway (Ohnishi et al., 2006; Sakamoto et al., 2006). The conserved biochemical functions, despite the low sequence identities of these enzymes, are quite remarkable for P450s and suggest the evolutionary importance of regulating the side chain hydroxylation steps in BR biosynthesis. The moderate level of sequence identity between CYP90C1 and CYP90D1 indicates that they may have appeared as a result of relatively ancient gene duplication. A different type of duplication occurred in the rice genome that resulted in genes coding for two CYP90D proteins with higher (66%) identity and a lack of a member of the CYP90C subfamily (Hong et al., 2003). It is therefore possible that members of the CYP90D subfamily represent more ancient forms of C-23 hydroxylases, and CYP90Cs may have evolved from this group following the divergence of dicot and monocot plants.
Normalized cDNA microarray data available at the Genevestigator database (https://www.geneveastigator.ethz.ch; Zimmermann et al., 2004) show twofold to fivefold higher levels of the CYP90C1 than the CYP90D1 transcript in most Arabidopsis organs and at most developmental stages. In mature plants, the highest CYP90C1 expression is detectable in developing petals, whereas the CYP90D1 transcript is most abundant in the inflorescence stem where its level exceeds that of the CYP90C1 mRNA. We found that the cyp90c1 mutant develops severely shortened petals, whereas cyp90d1 mutants have reduced apical dominance. These organ-specific effects may be due to local limitations of BR synthesis at the C-23 hydroxylation step and are in line with earlier reports indicating inefficient in planta BR transport (Bishop et al., 1996; Symons and Reid, 2004; Montoya et al., 2005).
METHODS
Chemicals
CR was purchased from Tama Biochemical, and BL and CS were purchased from Fuji Chemical Industries. CT, 6-deoxoTE, TE, 6-deoxoTY, TY, 6-deoxoCS, and CS used in the feeding experiments were from the Fujioka lab (RIKEN). Other BR compounds were chemically synthesized in our laboratory (see Supplemental Protocol 1 online).
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Col-0 and all mutant lines were grown from surface-sterilized seeds on Murashige and Skoog (MS) medium (Duchefa) supplemented with 1% (w/v) sucrose and 0.2% (w/v) Phytagel (Sigma-Aldrich). Plants were raised in controlled environment chambers (SANYO Electronic) under alternating 12-h-white-light (60 μmol/m2/s) and 12-h-dark photoperiods at 22°C. T-DNA–tagged lines were maintained on selective medium containing 15 mg/L hygromycin (Calbiochem). For crosses and seed production, plants were grown in soil under controlled greenhouse conditions, at 22 ± 2°C air temperature and under 8-h-light/16-h-dark periods for the first 3 weeks and then 16-h-light/8-h-dark photoperiods during flower induction and reproductive development.
Mutant Isolation
Mutants deficient in the CYP90C1 (At4g36380) and CYP90D1 (At3g13730) gene functions were isolated from the T-DNA insertion mutant collection, in Col-0 background, in C.K.'s laboratory (Max Plank-Institut für Züchtungsforschung) using a PCR-based screening technique (Ríos et al., 2002). The gene-specific primers were 5′-GGTGATCGGAGAAACCCTAAACTTCATC-3′ and 5′-GATCTTCAAGTGAGATCGGAGAAGCAC-3′ for CYP90C1 and 5′-GTCATCTTCAACAAGATCAACGGTCTCAG-3′ and 5′-GTAATTTGTTCTTCATATGCACCGTTGGGAAG-3′ for CYP90D1. Homozygous lines of the cyp90c1 (Koncz collection No. 18430) and cyp90d1 mutants (Koncz collection No. 8777) were obtained, following two rounds of backcrosses with wild-type plants, from selfed heterozygous parents showing 3 to 1 segregation of the T-DNA–associated hygromycin resistance. Following crosses between the cyp90c1 and cyp90d1 lines, infertile cyp90c1 cyp90d1 double mutant plants were obtained as dwarf segregants from progenies of parents homozygous for the cyp90c1 allele and heterozygous for the cyp90d1 allele. During mutant line maintenance, genotypes were verified by PCR using conditions and the CYP90C1- and CYP90D1-specific primers described by Bancos et al. (2002).
Transcript Analysis
Total RNA was isolated from 0.5 g of 7-d-old green seedlings using TRI Reagent (Sigma-Aldrich). Template cDNA was synthesized from 5 μg RNA with the Ready-To-Go T-Primed First-Strand kit (Pharmacia Biotech). Transcripts of CYP90C1, CYP90D1, and the internal control POLYUBIQUITIN 10 (At4g05320) were detected in the same PCR reactions. Primers and PCR conditions were as described earlier (Bancos et al., 2002), except 30 cycles were used. PCR products were analyzed on 2% agarose gel and visualized with ethidium bromide.
Phenotypic Rescue by BR Intermediates
For the hypocotyl elongation test, seeds were plated on solidified MS medium with 1% sucrose. After an initial 12-h light period, etiolated seedlings were raised in continuous dark for 7 d. Because homozygous cyp90c1 cyp90d1 double mutant seeds could not be obtained, homozygous dwarf seedlings were collected and transferred under green safety light to Petri plates containing the same medium, with or without 100 nM of the BR intermediate tested. Homozygous det2 seedlings were treated the same way. Following transfer and an additional 7-d incubation in the dark, 50 mutant seedlings were collected from each plate, and hypocotyl lengths were measured using enlarged images.
For cotyledon elongation assays, seedlings were grown from seeds on solidified MS medium with 1% sucrose, under regular light/dark cycles. After 7 d, 50 mutant seedlings were transferred into Erlenmeyer flasks containing liquid MS medium plus 1% sucrose, with or without 100 nM of the BR intermediate tested. The cultures were incubated under the same light and temperature conditions on an orbitary shaker (50 rpm) for 2 d and then the seedlings were collected, and cotyledon lengths were measured as described above.
Determination of the Endogenous BR Content
Plant materials ranging from 30 to 60 g fresh weight were aseptically grown for 1 month and collected in liquid nitrogen and subsequently lyophilized. In the first analysis, basic analysis of BRs was performed as described by Nomura et al. (2001), except that silica gel chromatography was omitted. However, analysis of 22-hydroxy-BRs (22-OHCR, 22-OH-4-en-3-one, 22-OH-3-one, 6-deoxoCT, and 3-epi-6-deoxoCT) and 22,23-dihydroxy-BRs (22,23-diOHCR and 22,23-diOH-4-en-3-one) were performed using a different set of plant materials as described by Fujioka et al. (2002). Here, the eluates from silica gel columns with chloroform and methanol:chloroform (5:95) were used to analyze 22-hydroxy-BRs and 22,23-dihydroxy-BRs, respectively.
The second analysis was done accordingly to Nomura et al. (2001), although partitionings were done between 90% methanol and hexane, and silica gel and charcoal chromatography was not conducted. Furthermore, the following modifications were done in GC-MS analysis of HPLC fractions. Combined HPLC fractions 42 to 46 were expected to contain 22,23-diOH-4-en-3-one, 6-deoxo3DT, 22,23-diOHCR, and 6-deoxoTE. In the GC-MS analysis, [2H6] 6-deoxo3DT (M+, mass-to-charge ratio [m/z] 462) was used to quantify 6-deoxo3DT (M+, m/z 456) and 22,23-diOH-4-en-3-one (M+, m/z 454), while [2H6] 6-deoxoTE (M+, m/z 536) was used to quantify 6-deoxoTE (M+, m/z 530) and 22,23-diOHCR (M+, m/z 528). Fraction 47 was expected to contain 6-deoxoTY, and [2H6] 6-deoxoTY (M+, m/z 536) was used to quantify 6-deoxoTY (M+, m/z 530). Combined fractions 48 to 51 were expected to contain 22-OHCR, 22-OH-4-en-3-one, 22-OH-3-one, and 6-deoxoCT, and in the GC-MS analysis, [2H6] 6-deoxoCT (fragment ion at m/z 193) was used to quantify all these 22-hydroxylated BRs (fragment ion at m/z 187). Combined fractions 52 and 53 were expected to contain 3-epi-6-deoxoCT. Because these fractions did not contain an internal standard, a portion was mixed with a portion of combined fractions 48 to 51, and [2H6] 6-deoxoCT (fragment ion at m/z 193) was used to quantify 3-epi-6-deoxoCT (fragment ion at m/z 187) by GC-MS. GC-MS was performed as described by Nomura et al. (2001) using the Shimadzu QP2010 gas chromatograph mass spectrometer fitted with a DB-5 column (0.25 mm i.d. × 15 m; 0.25-m film thickness; J and W Scientific). Prior to analysis, BRs were converted to either methaneboronate, methaneboronate-trimethylsilyl ether (TY, TE, 6-deoxoTY, 6-deoxoTE, and 22,23-diOHCR), trimethylsilyl ether (CT, 6-deoxoCT, 3-epi-6-deoxoCT, 22-OHCR, 22-OH-4-en-3-one, and 22-OH-3-one), or methaneboronate (BL, CS, 6-deoxoCS, and 22,23-diOH-4-en-3-one).
Heterologous Expression in the Baculovirus/Insect Cell System
CYP90C1 and CYP90D1 cDNAs were cloned as BamHI-XhoI fragments in pFastBac1 vector (Invitrogen) and then were used for generating the corresponding recombinant Bacmid DNAs by transformation of Escherichia coli strain DH10Bac (Invitrogen). Preparation of the CYP90C1 and CYP90D1 cDNA-containing recombinant baculovirus DNAs and transfection of Sf9 (Spodoptera frugiperda 9) cells were performed according to the instructions of the manufacturer (Invitrogen). Heterologous expression of CYP90C1 and CYP90D1 proteins in Sf9 cells and spectrophotometric analysis were performed as described (Saito et al., 2004).
Measurement of CYP90C1 and CYP90D1 Activities by GC-MS
Microsomal fractions of the insect cells expressing CYP90C1 and CYP90D1 were obtained from the infected cells (300 mL of suspension-cultured cells). Infected cells were washed with PBS buffer and suspended in buffer A consisting of 20 mM potassium phosphate, pH 7.25, 20% (v/v) glycerol, 1 mM EDTA, and 1 mM DTT. The cells were sonicated and cell debris was removed by centrifugation at 10,000g for 15 min. The supernatant was further centrifuged at 100,000g for 1 h, and the pellet was homogenized with buffer A to provide the microsomal fractions. The microsomal fractions were stored at −80°C before the enzyme assay.
CYP90C1 and CYP90D1 activities were reconstituted by mixing the CYP90C1- and CYP90D1-containing microsomes with purified Arabidopsis NADPH-P450 reductase (Mizutani and Ohta, 1998). The reaction mixture consisted of 100 mM potassium phosphate, pH 7.25, 50 pmol/mL recombinant P450 protein, 0.1 unit/mL NADPH-P450 reductase, 1 mM NADPH, and 20 μM of BR intermediate compounds. Reactions were initiated by addition of NADPH and were performed at 30°C for 30 min. The reaction products were extracted three times with a half volume of ethyl acetate. The organic phase was collected and evaporated. The residue was treated with 2 mg/mL methaneboronic acid in pyridine at 80°C for 30 min and then with 10 mL of N-methyl-N-trimethylsilyltrifluoroacetamide at 80°C for 30 min. The derivatized products were analyzed by GC-MS as described (Fujita et al., 2006).
Kinetic Analysis of CYP90C1 and CYP90D1 Activities by HPLC
CYP90C1 and CYP90D1 activities were assayed using BRs at concentrations ranging from 0.5 to 48 μM. Reactions were initiated by the addition of NADPH and performed at 30°C for 10 min. The reaction products were extracted three times with equal volumes of ethyl acetate. The organic phase was collected and evaporated. The residue was treated with 1 mg/mL 9-phenanthreneboronic acid in pyridine at 80°C for 30 min (Gamoh et al., 1989), and 20 μL of the sample was subjected to HPLC: column, SunFire C18 column (20-mm length × 4.5-mm i.d.; Waters); solvent, 40% (v/v) acetonitrile in water; flow rate, 2.5 mL/min; detection, a fluorescence detector with an excitation wavelength at 305 nm and an emission wavelength at 375 nm. Retention times of authentic samples were 2.96 min for 22,23-diOHCR, 2.82 min for 22,23-diOH-4-en-3-one, 4.04 min for 6-deoxo3DT, 3.60 min for 3-epi-6-deoxoTE, and 4.46 min for 6-deoxoTY. The kinetic constants were calculated from triplicate data sets. Km and Vmax were calculated from a double reciprocal plot of the initial velocity (v0) versus substrate concentration.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At3g13730 (CYP90D1), At3g30180 (CYP85A2), At3g50660 (DWF4/CYP90B1), At4g36380 (ROT3/CYP90C1), At5g05690 (CPD/CYP90A1), and At5g38970 (CYP85A1). Further sequence data can be found in the GenBank/EMBL data libraries under accession numbers AF000307 (Brassica napus BNST3), AB084385 (Oryza sativa BRD1/CYP85), AB158759 (O. sativa D11/CYP724B1), AB206579 (O. sativa DWF4/CYP90B2), AC130732 (O. sativa CYP90D3), AP003244 (O. sativa D2/CYP90D2), AB190445 (Solanum lycopersicum CYP85A3), and U54770 (S. lycopersicum Dwarf/CYP85).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Protocol 1.
Supplemental Table 1. GC-MS Data of Products Obtained in CYP90C1 and CYP90D1 Reconstitution Assays.
Supplemental Figure 1. Heterologous Expression of the CYP90C1 and CYP90D1 Proteins in Baculovirus/Insect Cell System.
Supplemental Figure 2. Relative Activities of C-23 Hydroxylation Reactions with 22-Hydroxylated BRs.
Supplementary Material
Acknowledgments
We thank S. Fujioka (RIKEN, Wako, Japan) and S. Takatsuto (Joetsu University, Joetsu, Japan) for providing BRs used in the rescue experiments. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 18580091 to M.M.). This work was also supported in part by the Hungarian Scientific Research Fund (Grant T 42639 to M.S.) and by a scientific exchange program between the Deutsches Zentrum für Luft und Raumfahrt e.V. and the Hungarian Science and Technology Foundation (project No. HUN99/027 to C.K. and M.S.). T.O. is a research fellow of the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaharu Mizutani (mizutani@scl.kyoto-u.ac.jp).
Online version contains Web-only data.
References
- Asakawa, S., Abe, H., Nishikawa, N., Natsume, M., and Koshioka, M. (1996). Purification and identification of new acyl-conjugated teasterones in lily pollen. Biosci. Biotechnol. Biochem. 60 1416–1420. [Google Scholar]
- Azpiroz, R., Wu, Y.W., LoCascio, J.C., and Feldmann, K.A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz, A., and Tretyn, A. (2003). The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62 1027–1046. [DOI] [PubMed] [Google Scholar]
- Bancos, S., Nomura, T., Sato, T., Molnár, G., Bishop, G.J., Koncz, C., Yokota, T., Nagy, F., and Szekeres, M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D.G. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle, J., Szekeres, M., Jenkins, G., and Bishop, G.J. (2005). Unique and overlapping expression patterns of Arabidopsis CYP85 genes involved in brassinosteroid C-6 oxidation. Plant Mol. Biol. 57 129–140. [DOI] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., and Peto, C. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Li, J., Choi, Y.H., Seto, H., Takatsuto, S., Noguchi, T., Watanabe, T., Kuriyama, H., Yokota, T., Chory, J., and Sakurai, A. (1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Noguchi, T., Watanabe, T., Takatsuto, S., and Yoshida, S. (2000). Biosynthesis of brassinosteroids in cultured cells of Catharanthus roseus. Phytochemistry 53 549–553. [DOI] [PubMed] [Google Scholar]
- Fujita, S., Ohnishi, T., Watanabe, B., Yokota, T., Takatsuto, S., Fujioka, S., Yoshida, S., Sakata, K., and Mizutani, M. (2006). Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29, sterols. Plant J. 45 765–774. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., Takatsuto, S., and Yoshida, S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54 137–164. [DOI] [PubMed] [Google Scholar]
- Gamoh, K., Omote, K., Okamoto, N., and Takatsuto, S. (1989). High-performance liquid chromatography of brassinosteroids in plants with derivatization using 9-phenanthreneboronic acid. J. Chromatogr. A. 469 424–428. [Google Scholar]
- Goda, H., Shimada, Y., Asami, T., Fujioka, S., and Yoshida, S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32 495–508. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, K., Rasmussen, A.V., Morant, M., Nielsen, A.H., Bjarnholt, N., Zagrobelny, M., Bak, S., and Moller, B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8 280–291. [DOI] [PubMed] [Google Scholar]
- Kim, G.T., Fujioka, S., Kozuka, T., Tax, F.E., Takatsuto, S., Yoshida, S., and Tsukaya, H. (2005. a). CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 41 710–721. [DOI] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.W., Hwang, J.Y., Kim, Y.S., Joo, S.H., Chang, S.C., Lee, J.S., Takatsuto, S., and Kim, S.K. (2005. b). Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17 2397–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S. (2000). A putative role for the tomato genes DUMPY and CURL3 in brassinosteroid biosynthesis and response. Plant Physiol. 122 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, M., Fujioka, S., Jeon, J.H., Kim, H.B., Takatsuto, S., Yoshida, S., An, C.S., and Choe, S. (2005). A double mutant for the CYP85A1 and CYP85A2 genes of Arabidopsis exhibits a brassinosteroid dwarf phenotype. J. Plant Biol. 48 237–244. [Google Scholar]
- Li, J., Nagpal, P., Vitart, V., McMorris, T.C., and Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272 398–401. [DOI] [PubMed] [Google Scholar]
- Mizutani, M., and Ohta, D. (1998). Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya, T., Nomura, T., Yokota, T., Farrar, K., Harrison, K., Jones, J.G.D., Kaneta, T., Kamiya, Y., Szekeres, M., and Bishop, G.J. (2005). Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 42 262–269. [DOI] [PubMed] [Google Scholar]
- Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., Okabe, K., Kajiwara, H., Satoh, K., Yamamoto, K., Hirochika, H., and Kikuchi, S. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Kushiro, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yamaguchi, S. (2005). The last reaction producing brassinolide is catalyzed by cytochrome P450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 280 17873–17879. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Sato, T., Bishop, G.J., Kamiya, Y., Takatsuto, S., and Yokota, T. (2001). Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57 171–178. [DOI] [PubMed] [Google Scholar]
- Ohnishi, T., Watanabe, B., Sakata, K., and Mizutani, M. (2006). CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 70 2071–2080. [DOI] [PubMed] [Google Scholar]
- Poppenberger, B., Fujioka, S., Soeno, K., George, G.L., Vaistij, F.E., Hiranuma, S., Seto, H., Takatsuto, S., Adam, G., Yoshida, S., and Bowles, D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32 243–253. [DOI] [PubMed] [Google Scholar]
- Rouleau, M., Marsolais, F., Richard, M., Nicolle, L., Voigt, B., Adam, G., and Varin, L. (1999). Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 274 20925–20930. [DOI] [PubMed] [Google Scholar]
- Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., and Mizutani, M. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24 105–109. [DOI] [PubMed] [Google Scholar]
- Sakurai, A. (1999). Brassinosteroid biosynthesis. Plant Physiol. Biochem. 37 351–361. [Google Scholar]
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y., Goda, H., Nakamura, A., Takatsuto, S., Fujioka, S., and Yoshida, S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons, G.M., and Reid, J.B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 135 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182. [DOI] [PubMed] [Google Scholar]
- Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., Yoshimura, A., Kitano, H., Matsuoka, M., Fujisawa, Y., Kato, H., and Iwasaki, Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122 1589–1600. [DOI] [PubMed] [Google Scholar]
- Winkel, B.S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55 85–107. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.