Abstract
Transcription factors are believed to play a pivotal role in the activation and fine-tuning of plant defense responses, but little is known about the exact function of individual transcription factors in this process. We analyzed the role of the IId subfamily of WRKY transcription factors in the regulation of basal resistance to Pseudomonas syringae pv tomato (Pst). The expression of four members of the subfamily was induced upon challenge with virulent and avirulent strains of Pst. Mutant analyses revealed that loss of WRKY11 function increased resistance toward avirulent and virulent Pst strains and that resistance was further enhanced in wrky11 wrky17 double mutant plants. Thus, WRKY11 and WRKY17 act as negative regulators of basal resistance to Pst. Genome-wide expression analysis and expression studies of selected genes in single and double mutants demonstrated that both transcription factors modulate transcriptional changes in response to pathogen challenge. Depending on the target gene, WRKY11 and WRKY17 act either specifically or in a partially redundant manner. We demonstrate complex cross-regulation within the IId WRKY subfamily and provide evidence that both WRKY transcription factors are involved in the regulation of Pst-induced jasmonic acid–dependent responses. These results provide genetic evidence for the importance of WRKY11 and WRKY17 in plant defense.
INTRODUCTION
The defense responses that plants mount against invading microorganisms are orchestrated by a complex reprogramming of host cells, both at the infection site and in systemic tissues, and rely on major changes in gene expression (Somssich and Hahlbrock, 1998; Maleck et al., 2000; Nimchuk et al., 2003; Tao et al., 2003; De Vos et al., 2005). These responses are highly specific to be effective against each individual species within the panoply of potential plant pathogens, ranging from necrotrophic fungi to biotrophic oomycetes, or from bacteria colonizing the vascular system to root-feeding nematodes. But defense is also intimately connected to general plant physiological and developmental processes to benefit the plant and to minimize the costs associated with resistance (Heil, 2002; Brown, 2003). Thus, induction of the plant defense response does not occur through a linear pathway but by a complex signaling network interconnected by crosstalk to the networks regulating other plant functions (Thomma et al., 2001; Glazebrook et al., 2003; Katagiri, 2004). Two well-studied key mediators of plant resistance are the plant hormones salicylic acid (SA) and jasmonic acid (JA). Research on the plant Arabidopsis thaliana has led to a model in which SA primarily activates defense responses to biotrophic pathogens, such as Hyaloperonospora parasitica and Pseudomonas syringae, whereas JA mainly activates defense responses to necrotrophic pathogens such as Fusarium oxysporum (Thomma et al., 2001). Both signaling pathways have been shown to act antagonistically. Mutants affected in SA synthesis or SA signal transduction show enhanced susceptibility to Pseudomonas syringae pv tomato (Pst), whereas JA signal transduction mutants possess enhanced resistance to Pst (Delaney et al., 1995; Nawrath and Métraux, 1999; Kloek et al., 2001). In addition, JA inhibits SA-dependent responses and vice versa (Thomma et al., 1998).
The major transcriptional reprogramming associated with the plant defense response requires not only plant hormones but also the action of diverse transcription factors (Chen et al., 2002; Singh et al., 2002; Chen and Zhu, 2004; Eulgem, 2005). However, despite increasing circumstantial evidence implicating different families of transcriptional regulators in this process, little is known about the exact function of individual transcription factors. The WRKY class of transcriptional regulators appears to play a major role in the regulation of plant defense responses. Members of this family are characterized by the presence of one or two highly conserved WRKY domains. This 60–amino acid domain comprises the name-giving, absolutely conserved sequence motif WRKYGQK and a zinc-finger motif. Both conserved elements of the domain are necessary for the high binding affinity of WRKY proteins to the consensus sequence (C/T)TGAC(C/T), named the W-box (Eulgem et al., 2000; Maeo et al., 2001; Zhang and Wang, 2005). NMR analysis has shown that WRKY domains form a four-stranded β-sheet that is stabilized by a zinc binding pocket localized at one end of the β-sheet, and it is likely that the conserved WRKYGQK sequence directly binds the W-box consensus motif (Yamasaki et al., 2005).
There are 74 WRKY proteins in Arabidopsis that have been classified into three groups according to the number and the type of their WRKY domains (Eulgem et al., 2000). Group III members contain a single C-C-H-C zinc finger, group II proteins contain a single C-C-H-H zinc finger, and group I proteins contain two C-C-H-H zinc fingers. Group II is further divided into subgroups according to the level of homology within the WRKY domain and the presence of additional domains. For example, the proteins belonging to the IId subfamily possess three domains in common in addition to the WRKY domain: a putative nuclear localization signal, the so-called HARF domain of unknown function, and the C domain, which has been shown to mediate a calcium-dependent interaction with calmodulin (Park et al., 2005).
W-boxes are a major class of cis-acting elements that confer pathogen and elicitor inducibility, either on their own, when coupled to minimal promoters, or in the context of promoters of pathogen- or elicitor-responsive genes such as pathogenesis-related proteins, receptor protein kinases, or WRKY transcription factors (Rushton et al., 1996; Wang et al., 1998; Eulgem et al., 1999; Yang et al., 1999; Du and Chen, 2000; Robatzek and Somssich, 2002; Rushton et al., 2002). As shown by different transcriptome analyses, W-boxes are overrepresented in the promoters of Arabidopsis genes that are upregulated during resistance (R) gene–mediated resistance, basal defense, elicitor responses, and systemic acquired resistance (Maleck et al., 2000; Eulgem et al., 2004; Navarro et al., 2004; Zipfel et al., 2004). These data suggest that WRKY proteins are central regulators of the pathogen-induced active defense response. This notion is also supported by the finding that the majority of Arabidopsis WRKY genes are upregulated in defense responses or after treatment with defense-inducing elicitors or hormones (Chen et al., 2002; Kalde et al., 2003). In one expression profiling study, 49 of 72 tested Arabidopsis WRKY genes were induced in response to treatment with SA or Pst (Dong et al., 2003). The importance of WRKY factors in regulating the plant defense transcriptome is further underlined by studies using overexpression and gene-silencing approaches. Overexpression of the pathogen-induced WRKY18 gene in Arabidopsis leads to an amplification of developmentally regulated defense responses (Chen and Chen, 2002). Overexpression and antisense depletion of WRKY70 has provided compelling evidence for a central role of this factor in integrating signals from the antagonistic JA and SA signaling pathways (Li et al., 2004).
Apparent functional redundancy has severely hampered attempts to genetically define the roles of individual WRKY transcription factors in the regulation of plant defense (Ulker and Somssich, 2004; Eulgem, 2005). Recently, however, Li et al. (2006) demonstrated that loss of WRKY70 function impaired plant resistance toward the fungal pathogen Erysiphe cichoracearum, and Xu et al. (2006) established a function for the homologous genes WRKY18, WRKY40, and WRKY60 in resistance to Pst and Botrytis cinerea by direct genetic means.
In this study, the seven members of the Arabidopsis WRKY IId subfamily were analyzed for their role in resistance to Pst. We provide genetic evidence that two of them, WRKY11 and WRKY17, act as negative regulators of basal resistance to the bacterial pathogen. These two regulators show partial redundancy in the transcriptional reprogramming that occurs in response to pathogen challenge. In addition, WRKY11 and WRKY17 are shown to be involved in the regulation of Pst-induced responses that are JA-dependent. Finally, our results reveal complex cross-regulation within the IId WRKY subfamily.
RESULTS
Four Members of the WRKY IId Subfamily Are Induced in Response to Pseudomonas
Transcriptome analysis of hxc2 (Godard et al., 2000), an Arabidopsis mutant affected in specific resistance to Xanthomonas campestris pv campestris and basal resistance to Pst, revealed the downregulation of one member of the IId subfamily of WRKY proteins, WRKY11, in the mutant compared with wild-type plants. To evaluate the role of the IId subfamily of WRKY proteins (Figure 1A) in the regulation of Arabidopsis defense responses, the expression of all seven members of this subfamily was analyzed after challenge with virulent or avirulent strains of Pst DC3000 by quantitative real-time PCR (Q-RT-PCR). In untreated leaves or leaves treated with water, WRKY7, WRKY11, WRKY15, WRKY17, WRKY21, and WRKY39 are weakly expressed, whereas WRKY74 is barely detectable (data not shown). After inoculation with virulent Pst DC3000 or avirulent Pst DC3000 expressing the avirulence gene avrRpt2, the expression of WRKY7, WRKY11, WRKY15, and WRKY17 is strongly induced, whereas the expression of WRKY21, WRKY39, and WRKY74 is not altered significantly (Figure 1B; data not shown for WRKY74, which is below the detection limit). WRKY11, WRKY7, and WRKY17 show similar expression profiles, with a rapid and transient induction that peaks 2 h after inoculation. Induction in response to virulent bacteria is similar to the induction in response to avirulent bacteria, but slightly weaker. The expression profile of WRKY15 in response to Pst is different. In the incompatible interaction with Pst DC3000 (avrRpt2), induction is rapid and sustained, whereas in the compatible interaction with Pst DC3000, it is delayed but increases steadily between 6 and 12 h after inoculation.
Figure 1.
The Pathogen-Responsive Expression Pattern of WRKY IId Subfamily Members.
(A) The phylogenetic tree of the WRKY IId subfamily was generated using the neighbor-joining method after alignment of the entire amino acid sequences of the IId subfamily members WRKY7, WRKY15, WRKY11, WRKY17, WRKY21, WRKY39, and WRKY74 and the IIe subfamily member WRKY22 with ClustalW (see Supplemental Figure1 online). Distance in the tree corresponds to evolutionary distance. Bootstrap values are indicated at the nodes of the tree. Tree building with the maximum-parsimony, minimum-evolution, and UPGMA methods gave similar results.
(B) Transcript levels of the WRKY genes belonging to the IId subfamily were determined by Q-RT-PCR with cDNA generated from leaves of 4-week-old Col-0 plants inoculated with water (closed diamonds), Pst DC3000 (closed squares), and Pst DC3000 (avrRpt2) (open squares) at 107 colony-forming units (cfu)/mL. The expression values of individual genes were normalized using the expression level of β-Tubulin4 as an internal standard. Mean expression values were calculated from the results of two independent experiments. WRKY74 mRNA levels are not shown because they are below the level of detection. hpi, hours after inoculation; AU, arbitrary units.
Many pathogen-responsive genes are also induced by a wide range of abiotic stresses. This has been shown for some WRKY transcription factors, for which induction by wounding, drought, and cold has been documented (Hara et al., 2000; Chen et al., 2002; Lee et al., 2005; Taki et al., 2005). Therefore, it was important to evaluate the specificity of the induction of the WRKY IId gene members in the pathogen response. For this purpose, we analyzed the responsiveness of WRKY11, WRKY17, WRKY7, and WRKY15 to various abiotic stresses, including wounding, cold, high salt concentration, and drought, as well as to treatment with the stress hormone abscisic acid. None of these treatments led to a strong induction of any of the WRKY IId genes (data not shown for WRKY7 and WRKY15 or for drought and abscisic acid treatments for WRKY11 and WRKY17). We observed a weak induction (fourfold) of WRKY11 after cold treatment and salt treatment and a rapid and transient induction of WRKY11 between 30 min and 1 h after wounding (threefold) (see Supplemental Figure 1 online). However, these activations are considerably weaker than those observed in response to inoculation with Pst (>30-fold), indicating that WRKY11, WRKY17, WRKY7, and WRKY15 are specifically upregulated in response to pathogen challenge and are not general stress response genes.
wrky11 but Not wrky7 or wrky17 Mutants Show Enhanced Basal Resistance
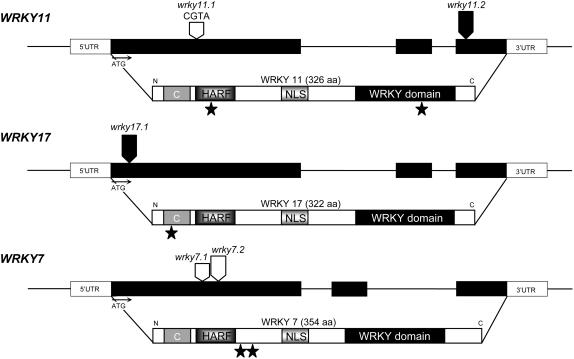
To study the function of the WRKY IId genes WRKY11, WRKY17, WRKY7, and WRKY15 in the regulation of defense responses, we searched Arabidopsis mutant collections for loss-of-function insertion mutants. For WRKY11, we identified two alleles that we named wrky11.1 and wrky11.2 (Figure 2). wrky11.1 was isolated from the ZIGIA En-insertion population (Wisman et al., 1998). This mutant carries a 4-bp insertion in the 5′ part of the coding sequence, resulting in a premature stop codon at position 121 in the predicted amino acid sequence of WRKY11 and thus a truncated gene product. By Q-RT-PCR, we detected the transcript at a slightly but significantly lower level than in wild-type plants (see Supplemental Figure 2 online). The second allele, wrky11.2, is from the GABI collection (Li et al., 2003; Rosso et al., 2003) and carries an insertion within the third and last exon, just after the position encoding the first Cys of the zinc finger motif (Figure 2). By Q-RT-PCR analysis, we verified disruption of the gene. Primer pairs positioned 3′ to the insertion site or around the insertion site did not amplify any product, whereas primer pairs in the 5′ part of the transcript amplified a product showing that a truncated mRNA is produced. As disruption of the zinc finger motif has been shown to completely abolish the W-box–specific DNA binding activity of WRKY transcription factors (Maeo et al., 2001), it is very likely that wrky11.2 is a true loss-of-function allele. For WRKY17, we identified one allele from the SALK collection (Alonso et al., 2003), which carries a T-DNA insertion in the 5′ part of the first exon (Figure 2). Q-RT-PCR analysis of the insertion line indicated that no wrky17 transcript is produced (data not shown). For WRKY7, we isolated two alleles, named wrky7.1 and wrky7.2 (Figure 2), from the ZIGIA En-insertion population. Both lines carry truncated En-1 transposons, inserted in the first exon of the gene. In both cases, this leads to premature stop codons, resulting in a truncated protein with no WRKY DNA binding domain. In the case of WRKY15, we could not identify any insertion mutant in the public mutant collections.
Figure 2.
Structures of the WRKY11, WRKY17, and WRKY7 Genes, Proteins, and Mutants.
The gene models show the intron/exon structures of the genes, the transcribed (black boxes) and untranscribed regions (UTR; white boxes), and the positions of En footprints (open arrows) and T-DNA insertions (closed arrows) in the different mutant alleles. The WRKY11, WRKY17, and WRKY7 proteins are shown with the N-terminal C domain, the HARF domain, the putative nuclear localization sequence (NLS), and the WRKY domain. Black stars indicate the predicted C termini of truncated mutant wrky proteins.
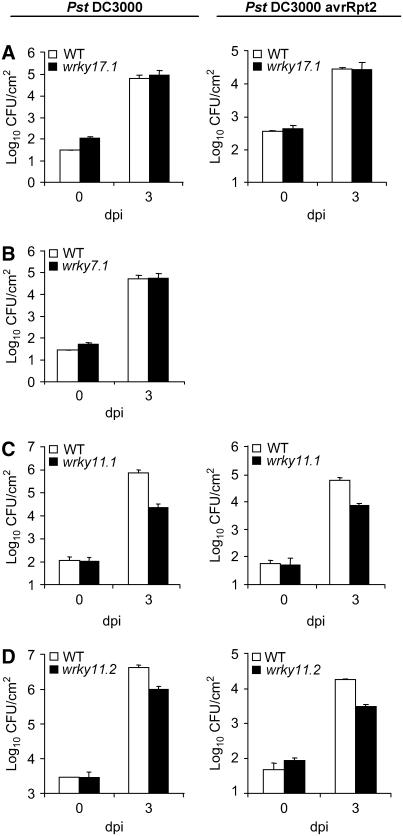
wrky17.1 and wrky7.1 plants behaved like wild-type plants in both incompatible and compatible interactions with the appropriate Pst strains. Bacterial colonization of wrky17.1 and wrky7.1 was not significantly different from the colonization of wild-type plants (Figures 3A and 3B), and the disease symptoms observed on mutant leaves after inoculation with virulent Pst DC3000 were indistinguishable from those of wild-type controls (Figure 4A for wrky17.1; data not shown for wrky7.1). On the other hand, both wrky11 alleles exhibited enhanced resistance to both virulent and avirulent Pst. They were less efficiently colonized and, depending on the experiment, 5- to 50-fold less bacteria were present 3 d after inoculation in wrky11 plants compared with wild-type plants (Figures 3C and 3D). Likewise, in the compatible interaction with Pst DC3000, we observed weaker disease symptoms on wrky11 leaves compared with wild-type leaves (Figure 4A for wrky11.1; data not shown for wrky11.2).
Figure 3.
Resistance of wrky17, wrky7, and wrky11 Mutants to Pst DC3000 and Pst DC3000 (avrRpt2).
Leaves of 4-week-old wild-type, wrky17.1 (A), wrky7.1 (B), wrky11.1 (C), and wrky11.2 (D) plants were syringe-infiltrated with a bacterial suspension of Pst DC3000 (105 cfu/mL) and Pst DC3000 (avrRpt2) (2 × 105 cfu/mL). Mean bacterial densities and se values calculated from the bacterial densities of six to eight individual plants are shown at 0 and 3 d after inoculation (dpi). These results are representative of three to six independent replications.
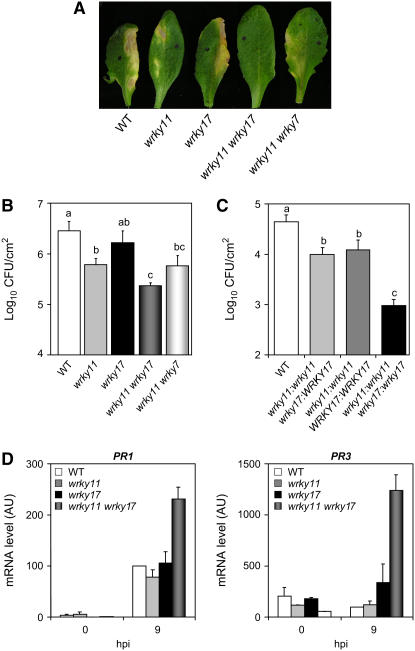
Figure 4.
Disease Phenotypes of wrky11.1 and wrky17.1 Single Mutant and wrky11.1 wrky17.1 and wrky11.1 wrky7.2 Double Mutant Plants.
(A) Representative leaves of wild-type, wrky11.1, wrky17.1, wrky11.1 wrky17.1, and wrky11.1 wrky7.2 plants 6 d after syringe inoculation with a bacterial suspension of Pst DC3000 at a density of 2 × 105 cfu/mL.
(B) Growth of Pst DC3000 in wild-type, wrky11.1, wrky17.1, wrky11.1 wrky17.1, and wrky11.1 wrky7.2 plants. Inoculation was performed with a bacterial suspension of 105 cfu/mL, and bacterial growth was determined at 3 d after inoculation. Mean bacterial densities and se values calculated from six to eight replicate plants are shown. According to Student's t test (P ≤ 0.05), means of cfu do not differ significantly if they are indicated with the same lowercase letter. These results are representative of four independent experiments.
(C) Cosegregation analysis using the progeny of a wrky11.1:wrky11.1 wrky17.1:WRKY17 plant. Wild-type plants and 60 plants from the progeny of an F2 line homozygous for the wrky11.1 mutation and heterozygous for the wrky17.1 mutation were infiltrated with a bacterial suspension of 105 cfu/mL. Bacterial growth was determined at 3 d after inoculation. In parallel, the plants were genotyped individually for the wrky17.1 mutation. The mean bacterial densities and se values were calculated from at least 14 replicate plants. According to Student's t test (P ≤ 0.05), means of cfu do not differ significantly if they are indicated with the same lowercase letter.
(D) Expression patterns of the defense-related genes PR1 and PR3 in wrky11.1 and wrky17.1 single and double mutants. The transcript levels of PR1 and PR3 were determined by Q-RT-PCR with cDNA generated from leaves of 4-week-old plants inoculated with Pst DC3000 at 107 cfu/mL. The expression values of the individual genes were normalized using the expression level of β-Tubulin4 as an internal standard and are expressed as percentages with respect to the value of wild-type plants at 9 h after inoculation (hpi), which is set at 100%. Mean expression values and se values were calculated from the results of three independent experiments.
Enhanced resistance is frequently observed in mutants with pleiotropic developmental and/or morphological perturbations and in these cases is associated with the constitutive expression of stress-responsive genes or defense marker genes. No morphological or developmental alterations under standard growth conditions were observed for wrky11 plants, as indeed for all other WRKY IId mutants tested. Using Q-RT-PCR, we checked the expression of the defense marker genes PR1, PR2, PR3, PR4, PR5, and Isochorismate Synthase (ICS) in wrky11.1 and wrky11.2 plants and found no altered levels either before or after inoculation (Figure 4D for PR1 and PR3; data not shown for PR2, PR5, ICS, and wrky11.2).
Together, these data suggest that WRKY11 is a negative regulator of resistance and that enhanced resistance in wrky11 mutants is not correlated with constitutive expression or with a stronger or more rapid induction of defense marker genes.
Double Mutant Analysis Reveals That WRKY11 and WRKY17 Act Redundantly as Negative Regulators of Basal Resistance
Analysis of single wrky mutant plants did not provide any information concerning the role of WRKY17 and WRKY7 in resistance to Pst, although both genes show pathogen-induced expression profiles very similar to WRKY11. This was somewhat unexpected, because WRKY17 shows extensive sequence homology with WRKY11 (72% amino acid identity), making it probable that both proteins have similar molecular activities.
To investigate the role of WRKY17 and WRKY7 and to evaluate their functional overlap with WRKY11, crosses between wrky11.1 and wrky17.1 and between wrky11.1 and wrky7.2 were performed, and double homozygous plants were isolated from the progeny. F2 lines homozygous for the wrky11.1 locus and heterozygous for the wrky17.1 locus were also obtained and used later for cosegregation analysis. The phenotypes of the double mutant plants after inoculation with Pst DC3000 and Pst DC3000 (avrRpt2) were analyzed. As shown in Figure 4A, wrky11.1 wrky17.1 plants displayed markedly reduced disease symptoms compared with wrky11.1 in the interaction with Pst DC3000, whereas wrky11.1 wrky7.2 responded similarly to wrky11.1. Consistent with these data, bacterial multiplication was reduced significantly in wrky11.1 wrky17.1 compared with wrky11.1 and the wrky11.1 wrky7.2 double mutant (Figure 4B). Three days after inoculation, bacterial density was 5- to 20-fold lower in wrky11.1 wrky17.1 compared with wrky11.1 and 20- to 100-fold lower compared with the wild type in both compatible (Figure 4B) and incompatible interactions (see Supplemental Figure 3 online). To confirm the enhanced resistance phenotype of wrky11.1 wrky17.1, we also analyzed the progeny of an F2 line that was homozygous for the wrky11.1 locus and heterozygous for the wrky17.1 locus. We determined the genotype for the wrky17.1 locus of 60 individual plants and the corresponding bacterial density 3 d after inoculation with Pst DC3000. Again, we found significantly reduced bacterial colonization in mutant plants that were homozygous for the wrky17.1 locus compared with mutant plants that were heterozygous or wild type (Figure 4C). From this result, we concluded that both WRKY11 and WRKY17 act as negative regulators of the plant defense response toward Pst.
To check whether the enhanced resistance phenotype of wrky11.1 wrky17.1 plants is correlated with a constitutive or increased activation of defense responses, we analyzed the expression of defense marker genes in the double mutants compared with single mutants and the wild type. No modification in the expression level of the marker genes was detectable in untreated plants. However, during the interaction with Pst DC3000, PR1 and PR3 were found to be significantly upregulated in the double mutant (Figure 4D).
Screening for Candidate Target Genes by Genome-Wide Expression Analysis in wrky11.1 and wrky17.1
Because both WRKY11 and WRKY17 can be assumed to function as transcription factors involved in the transcriptional reprogramming that occurs after pathogen recognition and contributes to the establishment of resistance, we were particularly interested in identifying their direct or indirect target genes. For this, a genome-wide expression analysis was undertaken using the Arabidopsis ATH1 GeneChip (Affymetrix), which contains 22,500 probe sets representing ∼24,000 genes. Three independent experiments were performed in which leaves of wild-type, wrky11.1, and wrky17.1 plants were inoculated with Pst DC3000 and harvested at 0, 2, 5, and 12 h after inoculation. Unfortunately, the wrky11.1 wrky17.1 double mutant was not yet available when this experiment was performed. Each individual sample from each of the three biologically independent experiments was used for the hybridization of one GeneChip. Thus, 36 hybridizations were performed, and expression data for each time point of the three independent experiments were obtained.
As a screen for candidate target genes, we used an analysis with MAS5.0 software (see Methods for details). In this way, 133 genes were identified as differentially expressed in wrky11.1 and 217 genes were identified as differentially expressed in wrky17.1 compared with the wild type (see Supplemental Figure 4 online; Supplemental Tables 1 and 2 online list the genes differentially expressed in wrky11.1 and wrky17.1). Only 11 differentially expressed genes were common to both wrky11.1 and wrky17.1 mutant lines. Among the genes differentially expressed in wrky11.1, we identified at different time points WRKY11, which we knew from previous experiments to be weakly but significantly downregulated in wrky11.1. This showed that we can indeed identify weakly differentially expressed genes by this method.
Functional Overlap between WRKY11 and WRKY17 in the Regulation of Target Genes Identified by Transcriptome Analysis
To validate some of the candidate genes, and to address the question of a potential functional overlap between WRKY11 and WRKY17, we compared in three independent biological experiments (different from those used for the microarray analysis) the expression of 10 differentially expressed genes in wild-type, wrky11.1, wrky17.1, and wrky11.1 wrky17.1 plants. We focused on genes with putative roles in signal perception and signal transduction, such as FRK1/SIRK (Asai et al., 2002; Robatzek and Somssich, 2002), RFO1 (Diener and Ausubel, 2005), CRK5 (Du and Chen, 2000; Chen et al., 2003), ATKC1 (Reintanz et al., 2002), and RRPK (for Related to Receptor Protein Kinases), because these classes of genes are overrepresented in the candidate gene lists (see Supplemental Table 3 online). In noninoculated wrky11.1 plants, a large number of resistance gene homologs (5 of 30 genes) and receptor-like protein kinases (5 of 30 genes) seem to be upregulated. A high enrichment for genes encoding protein kinases with putative functions in signal perception or signal transduction was observed among the genes upregulated in wrky11.1 at 5 h after inoculation (8 of 44 genes; see Supplemental Table 3A online) and in wrky17.1 at 2 h after inoculation (18 of 110 genes; see Supplemental Table 3C online). In addition, two JA-associated genes, LOX2 and AOS, were analyzed, because a high proportion of genes downregulated in wrky11.1 at 5 h (see Supplemental Table 3B online) have been described in the literature as being JA-responsive, including VSP1, VSP2, and COR1 (Benedetti et al., 1995, 1998; Tsuchiya et al., 1999), or are strongly JA-inducible according to transcriptome databases (see Supplemental Figure 5 online). Three of the genes, LOX2, AOS, and JMT, encode enzymes of JA biosynthesis or derivatization (Bell et al., 1995; Laudert et al., 1996; Seo et al., 2001). Finally, the expression of WRKY11 and two other WRKY genes with described functions in the regulation of plant defense, WRKY54 (Kalde et al., 2003) and WRKY70 (Li et al., 2004), was analyzed.
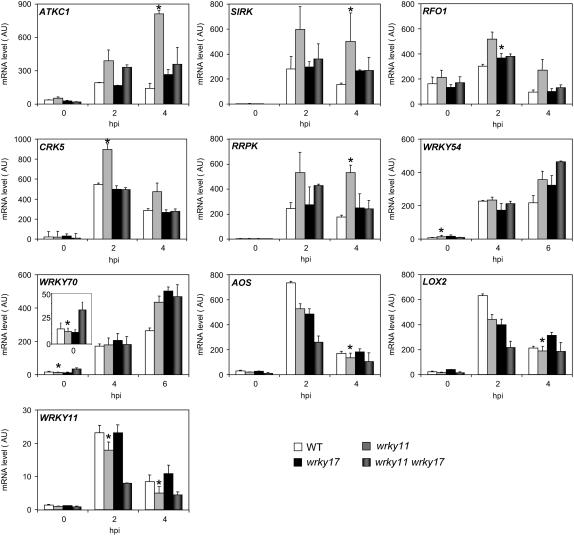
All of the genes analyzed showed significantly altered expression in the wrky single and/or double mutants by Q-RT-PCR analysis (Figure 5), confirming the trends observed in the transcriptome analysis. Genes upregulated or downregulated in the microarray experiment could be confirmed by Q-RT-PCR as, respectively, upregulated or downregulated. In some cases, however, differential expression was not observed in the corresponding single mutant but only in the double mutant (WRKY70) or at a different time point (LOX2 and AOS). These differences may be attributable to variations in the extent of functional complementation between the two WRKY homologs or to variations in the kinetics of gene induction. According to their expression profiles in the different mutants, the validated target genes could be classified into three groups. Group 1, the largest one, is characterized by a strong upregulation in inoculated wrky11.1 plants. This group contains ATKC1, FRK1/SIRK, RFO1, CRK5, and RRPK (Figure 5). The expression of these genes in wrky17.1 and wrky11.1 wrky17.1 plants is either not altered or only increased slightly compared with that in wild-type plants but is reduced significantly compared with that in wrky11.1 plants. These results suggest that WRKY17 acts in the same pathway as WRKY11 in the regulation of this group of genes, downstream of WRKY11 and in an opposite manner, because the loss of WRKY17 abolishes the loss-of-function effect of WRKY11. It also indicates that WRKY11 and WRKY17 have specific nonredundant functions in the regulation of these genes.
Figure 5.
Expression Analysis of Genes Potentially Regulated by WRKY11 and/or WRKY17.
Transcript levels of ATKC1, FRK1/SIRK, RFO1, CRK5, RRPK, WRKY54, WRKY70, LOX2, AOS, and WRKY11 were determined by Q-RT-PCR with cDNA generated from leaves of 4-week-old plants inoculated with Pst DC3000 at 107 cfu/mL. The expression values of the individual genes were normalized using the expression level of β-Tubulin4 as an internal standard. Mean expression values and se values were calculated from the results of three independent experiments. Stars indicate in which mutant and at which time point genes were identified as differentially expressed in the microarray analysis. hpi, h after inoculation.
The second group consists of WRKY54 and WRKY70 and is characterized by similar upregulation in wrky11.1, wrky17.1, and wrky11.1 wrky17.1 at 6 h after inoculation and by upregulation in untreated wrky11.1 wrky17.1 for WRKY70 (Figure 5). The synergistic effect of losing WRKY11 and WRKY17 function in noninfected plants suggests that they act in a partially redundant manner as negative regulators of WRKY70. Finally, the third group contains WRKY11 and the two JA-responsive genes AOS and LOX2. In wrky11.1 and wrky17.1 single mutants, the expression of these genes is either not altered or weakly but significantly downregulated (Figure 5), whereas in the wrky11.1 wrky17.1 double mutant, their expression is strongly downregulated. This finding suggests that WRKY11 and WRKY17 positively regulate the expression of this group of genes with partial redundancy.
Because WRKY17 regulates WRKY11 expression, we asked whether additional cross-regulation exists among the IId subfamily members. Therefore, the expression of WRKY17, WRKY7, and WRKY15 was analyzed in wrky11.1, wrky11.2, wrky17.1, and wrky11.1 wrky17.1 mutants after inoculation with Pst DC3000 and Pst DC3000 (avrRpt2). Although WRKY7 and WRKY15 expression was not altered in any of the mutant plants (data not shown), WRKY17 was substantially upregulated in the wrky11.1 and wrky11.2 mutants compared with wild-type plants (Figure 6), indicating that WRKY11 negatively regulates the expression of WRKY17. Whether WRKY17 regulates its own expression could not be tested because of the lack of detectable WRKY17 transcripts in the wrky17.1 T-DNA insertion line.
Figure 6.
WRKY17 Expression Analysis in Wild-Type and Mutant Plants.
The transcript level of WRKY17 was determined by Q-RT-PCR with cDNA generated from leaves of 4-week-old wild-type (closed diamonds), wrky11.1 (closed squares), and wrky11.2 (open squares) plants inoculated with Pst DC3000 or Pst DC3000 (avrRpt2) at 107 cfu/mL. The expression values of the individual genes were normalized using the expression level of β-Tubulin4 as an internal standard. These results are representative of four independent replicates. hpi, h after inoculation.
DISCUSSION
Using mutant analysis, we were able to demonstrate that WRKY11 and WRKY17 act as negative regulators of basal resistance during compatible and incompatible interactions with Pst. This lends strong genetic support to the hypothesis that members of this gene family play important roles in the regulation of plant defense. The enhanced resistance conferred by the loss of WRKY11 and WRKY17 function reveals a certain specificity, because resistance to avirulent and virulent strains of X. campestris pv campestris is not altered in wrky11 single and wrky11 wrky17 double mutants (N. Journot-Catalino and T. Kroj, unpublished data). In addition, transcriptome analysis and expression studies of selected defense genes indicate that this is not attributable to constitutive expression of defense responses. Although we have been able to correlate increased resistance with the altered expression of a certain number of genes, we cannot as yet establish the precise relationship between the altered expression of these genes and enhanced resistance. Nevertheless, we have identified specific and redundant functions of WRKY11 and WRKY17 in the regulation of plant resistance, both during normal growth and upon pathogen challenge.
The Biological Significance of the Negative Regulation of Resistance by WRKY11 and WRKY17
At first sight, it may appear surprising that a negative regulator of defense responses is rapidly induced during the establishment of resistance. However, it must be borne in mind that the plant defense system can be deleterious to the host, as shown in mutants with constitutively activated defense responses or with hypersensitive response lesion–mimic phenotypes, which often show stunted growth and low fertility (Lorrain et al., 2003). In addition, the activation of defense and even the maintenance of surveillance by the expression of R genes is associated with significant costs for the plant (Heil, 2002; Brown, 2003). For these reasons, the plant defense system needs to be under tight, fine-tuned regulation to be efficient but also beneficial to the plant. Our study suggests that the biological function of WRKY11 and WRKY17 could be both to limit the expression of the pathogen surveillance system in nonstimulated plants and to attenuate the expression of plant defense responses upon challenge with pathogens, thus contributing to a balanced allocation of resources by limiting the activation of defense-related functions.
Another important role of such negative regulators of defense responses could be to ensure an equilibrated defense response, which is effective not only against Pst but also against the panoply of other potentially pathogenic microorganisms (Thomma et al., 2001). It has repeatedly been observed that enhanced resistance to one class of pathogens results in enhanced susceptibility to another class and that in many of these cases the mutually antagonistic action of SA and JA is disturbed (Kloek et al., 2001; Veronese et al., 2006). In favor of this idea, wrky11 plants show enhanced susceptibility to virulent strains of Ralstonia solanacearum (Y. Marco and X. Barlet, personal communication). Therefore, it will be interesting in the future to test the resistance and susceptibility of the different single and multiple wrky11, wrky17, and wrky7 mutants to a wider range of pathogens with varying physiologies and invasion strategies.
Redundant versus Specific Functions of WRKY11 and WRKY17
A frequent problem in the study of plant transcription factors is functional overlap between closely related homologs, attributable to similar molecular activities and to overlapping expression patterns. For this reason, knockouts in multiple transcription factors have often been necessary to produce informative phenotypes (Liljegren et al., 2000; Kroj et al., 2003; Zhang et al., 2003; Xu et al. 2006). We have also found strong evidence for functional overlap between WRKY11 and WRKY17. The resistance phenotype of the double mutant is not simply the sum of the phenotypes of the single mutants but reflects a synergistic interaction between mutations in WRKY11 and WRKY17. Analyzing the expression of single genes and of the entire genome indicated that no target genes are strongly deregulated in wrky11 or wrky17 and revealed only subtle changes in the expression levels of individual genes. This may also be attributable to partial compensation of one factor by its closest homolog.
However, our data also indicate specific functions for the two WRKY proteins. First, wrky11 single mutants show an enhanced resistance phenotype; second, the group 1 genes are upregulated in wrky11 in a WRKY17-dependent manner. Because WRKY17 is also upregulated in wrky11 mutant plants, the regulation of group 1 genes can be explained by a model in which WRKY11 and WRKY17 act in a sequential and partly nonoverlapping manner (Figure 7A).
Figure 7.
Working Model for WRKY11 and WRKY17 Action.
(A) Group 1 genes are regulated in a positive manner by WRKY17, which is itself negatively regulated by WRKY11. In wrky11, WRKY17 is upregulated, leading to stronger expression of the downstream genes. In wrky17 and wrky11 wrky17 plants, WRKY17 function may be replaced by another functionally similar WRKY factor that is independent of WRKY11. This leads to an expression level of group 1 genes that is unaltered or increased slightly compared with that in wild-type plants.
(B) WRKY11 and WRKY17 regulate in a partially redundant manner the Pst-induced expression of the central JA biosynthetic enzymes LOX2 and AOS and by this may indirectly modulate the level of pathogen-responsive JA production. In addition, they negatively regulate the expression of WRKY70, which is under the control of JA and acts as an integrator of the mutually antagonistic JA and SA pathways (Li et al., 2004). By these two interconnected mechanisms, WRKY11 and WRKY17 may induce the expression of JA-responsive genes, repress the expression of genes responsive to SA, and ultimately negatively regulate basal resistance.
A molecular view of specificity and redundancy in WRKY protein function has emerged from a recent study describing the elicitor-responsive binding of the parsley (Petroselinum crispum) WRKY1 protein to its own promoter and to the promoter of the defense gene Pc PR1-1 in vivo (Turck et al., 2004). Regulatory W-box elements seem to be constantly occupied by WRKY proteins, but this occupancy is changing dynamically in a stimulus-dependent manner. Different WRKY factors are proposed to compete with one another for individual WRKY binding sites, generating a dynamic equilibrium of W-box occupancy. This equilibrium seems to be regulated by posttranslational modifications of individual WRKY proteins and their de novo synthesis or degradation (Turck et al., 2004). In this conceptual framework, loss of function of one individual WRKY protein may lead to a shift in equilibrium, with the respective WRKY protein being replaced by certain homologs at various W-boxes sites. Depending on the trans-activating or repressing activity of these homologs, the transcriptional output of individual downstream target genes may be either positively or negatively affected.
To better understand the specific and redundant functions of WRKY11 and WRKY17, it is indispensable to identify direct target genes of the two factors. We found a significant overrepresentation of W-boxes in the promoters of candidate genes upregulated in wrky11 plants (P < 10−5 in the −1.5-kb region according to an analysis with ATHENA [O'Connor et al., 2005]), suggesting that several of them may be regulated directly by WRKY11 and/or WRKY17. For certain genes, such as FRK1/SIRK, additional evidence exists indicating direct regulation by WRKY proteins binding to defined W-boxes within their promoters (Asai et al., 2002; Robatzek and Somssich, 2002). In vitro binding studies and promoter analysis are a prerequisite to identify direct target genes of WRKY11 and WRKY17. By this means, WRKY11 binding to several of the W-boxes in the FRK1/SIRK promoter has been demonstrated (I. Ciolkowski and I. Somssich, unpublished data). Ultimately, however, defining in vivo target genes using methods such as chromatin immunoprecipitation (Orlando, 2000; Hanlon and Lieb, 2004) will be essential to decipher the dynamics of WRKY protein–DNA interactions under physiological conditions at individual promoter sites and to define the extent to which WRKY11 and WRKY17 directly act as transcriptional repressors or activators of target genes on a genome-wide basis.
Function of WRKY11 and WRKY17 in the Regulation of Defense Responses
In Pst-inoculated plants, WRKY11 attenuates the induction of a range of pathogen-responsive genes, notably protein kinases with putative functions in signal perception and signal transduction. Protein kinases from different classes play important roles in the activation of plant defense (Nürnberger and Scheel, 2001; Romeis, 2001), and their overexpression leads to enhanced resistance (Chen et al., 2003). Thus, enhanced resistance in wrky11 and wrky11 wrky17 could in part result from the amplification of signal transduction attributable to the upregulation of signaling elements.
In addition to their function as negative regulators, WRKY11 and WRKY17 act also as positive regulators of gene expression. Among the genes downregulated in wrky11 after pathogen challenge, JA-responsive genes are particularly abundant, suggesting that WRKY11 and maybe also WRKY17 positively modulates JA signaling, either upstream of JA production or downstream of JA. Expression analysis of two key enzymes of JA biosynthesis, LOX2 and AOS, in wrky11 and wrky17 single and double mutant plants showed that both are regulated positively and redundantly by WRKY11 and WRKY17. LOX2 has been demonstrated to be essential for JA production in the wound response (Bell et al., 1995) and may have a similar function in pathogen responses. AOS, which catalyzes the first step of the octadecanoid pathway leading to jasmonates, has been proposed to be a key regulatory element in the production of jasmonates (Laudert et al., 1996; Laudert and Weiler, 1998; Park et al., 2002). Together with the fact that the expression of WRKY11 and WRKY17 is not affected by JA (transcriptome database at Genevestigator [Zimmermann et al., 2004]; see Supplemental Figure 4 online), these results imply a model in which WRKY11 and WRKY17 act upstream of JA by positively regulating the Pst-stimulated accumulation of jasmonates (Figure 7B).
The timing of WRKY11 and WRKY17 expression is consistent with the timing of the induction of the JA biosynthetic enzymes AOS and LOX2 and with the timing of JA accumulation, which occurs within the first hour of the interaction with Pst (De Vos et al., 2005). This model can explain the enhanced-resistance phenotype of wrky11 and wrky11 wrky17 and the upregulation of the SA-dependent marker gene PR1 in the double mutant, because JA negatively influences SA action and basal resistance to Pst (Kloek et al., 2001; Glazebrook et al., 2003; Devoto et al., 2005). Another link between WRKY11 and WRKY17 action and JA signaling is the negative regulation of WRKY70 by both proteins. WRKY70 has been shown to act as a negative regulator of the expression of JA-responsive genes downstream of JA and as a positive regulator of SA-induced genes such as PR1 (Li et al., 2004, 2006). Because the promoter of WRKY70 contains no W-boxes, its expression is presumably not regulated directly by WRKY proteins. However, JA has been shown to negatively regulate WRKY70 transcript levels, whereas SA regulates WRKY70 transcription positively (Li et al., 2004). Upregulation of WRKY70 in wrky11 and wrky17 single and double mutants could thus result from reduced JA levels and contribute to the downregulation of JA-responsive genes. Future experiments will be required to validate the roles of WRKY11 and WRKY17 in positively regulating the JA pathway. In particular, determining the concentrations of JA and other jasmonates in the wild type, wrky11 and wrky17 single mutants, and the wrky11 wrky17 double mutant will be of particular interest.
WRKY11 and WRKY17 positively regulate WRKY11 expression in a partly redundant manner, whereas WRKY11 negatively regulates the expression of WRKY17 (Figure 7A). As the promoters of both genes are highly enriched for W-boxes (Dong et al., 2003), it is conceivable that direct WRKY interactions are involved. The complex pattern of autoregulation and cross-regulation observed in the IId subfamily seems to be a general characteristic of the WRKY family. For example, WRKY6 has been demonstrated to negatively regulate its own expression and the expression of the closely related gene WRKY42 (Robatzek and Somssich, 2002), whereas the group III WRKY protein WRKY54 has been shown to influence in a complex manner the expression of other group III WRKY genes in response to different pathogens and pathogen-related treatments (Kalde et al., 2003). In addition, cross-regulation between distantly related WRKY genes also exists, as exemplified in this study, which shows that WRKY11 and WRKY17 negatively regulate the expression of WRKY70 and WRKY54. The emerging picture appears to be that of a highly interconnected network of WRKY transcription factors modulating WRKY gene expression and thereby fine-tuning the plant defense system.
In summary, we have been able to demonstrate a particular function for individual WRKY genes in the regulation of plant defense. Apart from the role that our study establishes for WRKY11 and WRKY17 in the negative regulation of basal resistance, we show that specific functions can be attributed to individual WRKY proteins. However, this study has also revealed the existence of considerable functional overlap between closely related WRKY transcription factors and, as a consequence, the subtle and quantitative nature of single mutant phenotypes. Finally, it illustrates the existence of a network of WRKY transcription factors that regulate and integrate signaling through the antagonistic SA and JA signal transduction pathways.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used in this study are in the Columbia background. As a wild-type control, we used Col-0 (Nottingham Arabidopsis Stock Centre [NASC] accession number N1093). Plants were grown in Jiffy pods in a growth chamber at 22°C, with a 9-h light period and a light intensity of 190 μmol·m−2·s−1. All experiments were performed with 4- to 5-week-old plants.
Bacterial Strains, Plant Inoculation Procedures, and Bacteria Growth Measurements
Pseudomonas syringae pv tomato (Pst) strains were grown at 29°C on King B's medium supplemented with 50 μg/mL rifampicin for Pst DC3000 and 50 μg/mL rifampicin and 10 μg/mL tetracycline for Pst DC3000 (avrRpt2) (Whalen et al., 1991). Plant inoculations and in planta bacterial growth analysis were performed essentially as described previously (Lorrain et al., 2004) using six to eight replicate plants per experiment. For the determination of disease symptoms and in planta bacterial growth, we used an inoculum of 105 cfu/mL for Pst DC3000 and of 2 × 105 cfu/mL for Pst DC3000 (avrRpt2). For gene expression analysis, we used an inoculum of 107 cfu/mL.
Identification of Insertion Mutants and Generation of Double Mutants
The mutant lines wrky7.1, wrky7.2, and wrky11.1 were derived from insertion mutants identified by a PCR-based screen of an En-1 insertion population (Wisman et al., 1998). The use of gene-specific and En-1–specific primers led to the identification of the line 5AAH57 with an En-1 insertion in WRKY11 and the lines 5AAO102 and 6AAL50 with En-1 insertions in WRKY7. The En-1 insertions were confirmed by DNA gel blot analysis, and the insertion sites were determined by sequencing. Footprint mutants were identified with gene-specific primers flanking the original En-1 insertion site. The individual lines 6AAL50.3 and 5AAO102.12, which contain fragments of the original En-1 transposon, and 5AAH57.3, which carries a 4-bp insertion, were backcrossed at least five times to eliminate additional En-1 insertions, leading to the isolation of the wrky7.1, wrky7.2, and wrky11.1 lines.
The wrky11.2 and wrky17.1 mutant lines were derived by backcrossing from the T-DNA insertion lines GABI-KAT 184D06 (Li et al., 2003) and SALK_176337 (Alonso et al., 2003), respectively. The position of the insertions was confirmed by PCR and sequencing, using the primers 5′-GGAGCCGGAGTTGTCACTTT-3′ (gene-specific) and 5′-CCCTTTAGGGTTCCGATTTAGTGCT-3′ (left border of the T-DNA) for wrky17.1 and the primers 5′-CCACCGTCTAGTGTAACACTCGAT-3′ (gene-specific) and 5′-GGGCTACACTGAATTGGTAGCTC-3′ (left border of the T-DNA) for wrky11.2.
The double mutant lines wrky11.1 wrky17.1 and wrky11.1 wrky7.1 were generated by crossing backcrossed homozygote mutant lines and determining the genotype of plants from the progeny by PCR analysis. The 4-bp footprint in wrky11.1 generates a new Rsa1 restriction site that allowed the generation of a cleaved-amplified polymorphic sequence marker (primers 5′-GGAGAAACTCTCTGAGTGCTCAT-3′ and 5′-GCTCCGAGATCACTGACTT-3′; digest of the 482-bp fragment by Rsa1).
Phylogenetic Analysis
Sequence alignment was performed with ClustalW and manually adjusted to get maximal alignment for the shared C domain, the putative nuclear localization signal, and the WRKY domain (Eulgem, 2000). Phylogenetic analysis was performed with the MEGA3 software package using neighbor-joining, UPGMA, maximum-parsimony, or minimum-evolution methods with 1000 bootstrap trials and complete gap deletion.
RNA Extraction and Q-RT-PCR Analysis
Material for RNA analysis was ground in liquid nitrogen, and total RNA was isolated using the Nucleospin RNA plant kit (Macherey-Nagel) according to the manufacturer's recommendations. Reverse transcription was performed using 1 μg of total RNA and SuperScript reverse transcriptase II (Invitrogen). Quantitative PCR was run on a Lightcycler system (Roche Diagnostics) according to the manufacturer's recommendations with the following conditions: 1 cycle of 9 min at 95°C, and 45 cycles of 5 s at 95°C, 10 s at 65°C, and 20 s at 72°C. β-Tubulin4 was used as an internal standard. The primer sets used in the different experiments are listed in Supplemental Table 4 online. The specificity of the amplifications was verified by analysis of the PCR products on agarose gels to ensure that only a single band was present and by melting curve analysis at the end of each experiment. Efficiency of the amplification was verified by the analysis of standard curves.
Microarray Experiments and Data Analysis
Samples were harvested and RNA was prepared as described for Q-RT-PCR analysis. To verify the induction of basal defense responses, the expression of the genes PR1, ICS, and WRKY11 was analyzed in the different experiments by Q-RT-PCR. Despite variations in kinetics and the amplitude of induction, expression patterns for the tested genes in the three experiments were very similar to one another and corresponded to the patterns observed in previous experiments. Processing of RNA, ATH1 GeneChip (Affymetrix) hybridization, and raw data collection were performed as described (Redman et al., 2004).
Using MAS5.0 statistical algorithms (Affymetrix; http://www.affymetrix.com/support/technical/whitepapers/sadd_whitepaper.pdf), GeneChip fluorescence intensity data were used to calculate for each probe set signal intensities (scaled to an average signal intensity of 100) and signal log ratios for pairwise slide-to-slide comparisons. In addition, MAS5.0 nonparametric rank tests (Liu et al., 2002) were used to determine whether a gene was detectable (detection call a for absent or p for present) and whether the signal log ratios represented a genuine change in mRNA level (change call i for increased, d for decreased, or nc for no change). Signal log ratios and change calls were determined for each mutant sample compared with its corresponding wild-type sample from the same biological experiment and the same time point.
Probe sets had to meet two criteria to be selected for further analysis. Those that were considered upregulated in a mutant at a given time point had to be present in the corresponding three replicate mutant samples (detection call p with an associated P < 0.05) and induced in all three independent biological experiments (change call i with an associated P < 0.006). Those that were considered downregulated in a mutant at a given time point had to be present in the corresponding three replicate wild-type samples (detection call p with an associated P < 0.05) and decreased in all three independent biological experiments (change call d with an associated P > 0.994).
To analyze whether after inoculation with Pst genes were upregulated or downregulated in wild-type plants (see Supplemental Tables 1 and 2 online), transcriptome data were analyzed by an alternative approach using Bioconductor packages (Gentlemen et al., 2005). Intensities were calculated from raw data by the robust multiple-array average expression measure (Irizarry et al., 2003). Subsequently, for time points 2, 5, and 12 h, a pairwise comparison between the intensities of the three replicates of each time point and the three replicates of time 0 was performed to identify significantly differentially expressed genes. This analysis was performed with the LIMMA package using an empirical Bayes linear modeling approach (Smyth, 2004, 2005) and by correcting obtained P values for multiple testing according to Benjamini and Hochberg (1995).
For functional classification of the differentially expressed genes, GO annotations at The Arabidopsis Information Resource (Berardini et al., 2004; Harris et al., 2004), FUN-CAT annotations at the Munich Information Center for Protein Sequences (Ruepp et al., 2004), and MapMan version 1.6.0 (Thimm et al., 2004) were used. Analysis of promoter regions was performed with ATHENA (O'Connor et al., 2005) and AGRIS (Davuluri et al., 2003; Palaniswamy et al., 2006). Public transcriptome data were analyzed using Genevestigator (Zimmermann et al., 2004).
Accession Numbers
Locus identifiers of the genes from this article are as follows: At4g24240 (WRKY7), At2g23320 (WRKY15), At4g31550 (WRKY11), At2g24570 (WRKY17), At2g30590 (WRKY21), At3g04670 (WRKY39), At5g28650 (WRKY74), At5g44340 (β-Tubulin4), At2g14610 (PR1), At3g57260 (PR2), At3g12500 (PR3), At3g04720 (PR4), At1g75040 (PR5), At1g74710 (ICS), At2g19190 (FRK1/SIRK), At2g40750 (WRKY54), At3g56400 (WRKY70), At4g23130 (CRK5), At1g79670 (RFO1), At3g45140 (LOX2), At5g42650 (AOS), At4g32650 (ATKC1), and At3g46280 (RRPK). Microarray data have been deposited in the NASCArrays database under the experiment reference number NASCARRAYS-393.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Genes Differentially Expressed in wrky11.1 Plants.
Supplemental Table 2. Genes Differentially Expressed in wrky17.1 Plants.
Supplemental Table 3. Selected Genes Differentially Expressed in wrky11 and wrky17.
Supplemental Table 4. Primers Used for Q-RT-PCR.
Supplemental Figure 1. Alignment of the WRKY IId Subfamily Proteins.
Supplemental Figure 2. Expression Pattern of WRKY11 and WRKY17 in Response to Abiotic Stresses.
Supplemental Figure 3. WRKY11 Expression in wrky11.1.
Supplemental Figure 4. Resistance of wrky11.1 and wrky17.1 Single and Double Mutants to Pst DC3000 (avrRpt2).
Supplemental Figure 5. Venn Diagrams of Genes Upregulated or Downregulated in wrky11 and wrky17.
Supplemental Figure 6. JA Responsiveness of Genes Downregulated in wrky11 at 5 h after Inoculation.
Supplementary Material
Acknowledgments
We thank Susanna Rivas, Nemo Peeters, and David Barker for helpful discussions and critical reading of the manuscript. Matthew Hannah and Sébastien Déjean are acknowledged for advice and help with statistical analysis. Didier Aldon and Fabienne Magnan are acknowledged for providing abiotic stress experiments. This work was funded by the Genoplante Program Functional Analysis of Arabidopsis Genome (Grant AF 2001060). N.J.-C. was supported by a grant from the French Ministry of National Education and Research.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Imre E. Somssich (somssich@mpiz-koeln.mpg.de) and Thomas Kroj (kroj@toulouse.inra.fr).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, C.E., Costa, C.L., Turcinelli, S.R., and Arruda, P. (1998). Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the Coi1 mutant of Arabidopsis. Plant Physiol. 116 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, C.E., Xie, D., and Turner, J.G. (1995). Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. [Ser. A] 57 289–300. [Google Scholar]
- Berardini, T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.K. (2003). A cost of disease resistance: Paradigm or peculiarity? Trends Genet. 19 667–671. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K., Du, L., and Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53 61–74. [DOI] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.J., and Zhu, T. (2004). Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 9 591–596. [DOI] [PubMed] [Google Scholar]
- Davuluri, R.V., Sun, H., Palaniswamy, S.K., Matthews, N., Molina, C., Kurtz, M., and Grotewold, E. (2003). AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos, M., Van Oosten, V.R., Van Poecke, R.M., Van Pelt, J.A., Pozo, M.J., Mueller, M.J., Buchala, A.J., Metraux, J.P., Van Loon, L.C., Dicke, M., and Pieterse, C.M. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18 923–937. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Ellis, C., Magusin, A., Chang, H.S., Chilcott, C., Zhu, T., and Turner, J.G. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58 497–513. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., and Ausubel, F.M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51 21–37. [DOI] [PubMed] [Google Scholar]
- Du, L., and Chen, Z. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA- binding proteins in Arabidopsis. Plant J. 24 837–847. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005). Regulation of the Arabidopsis defence transcriptome. Trends Plant Sci. 10 71–78. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Weigman, V.J., Chang, H.S., McDowell, J.M., Holub, E.B., Glazebrook, J., Zhu, T., and Dangl, J.L. (2004). Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 135 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentlemen, R., Carey, V., Dudoit, S., Irizarry, R., and Huber, W. (2005). Bioinformatics and Computational Biology Solutions Using R and Bioconductor. (New York: Springer).
- Glazebrook, J., Chen, W., Estes, B., Chang, H.S., Nawrath, C., Metraux, J.P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34 217–228. [DOI] [PubMed] [Google Scholar]
- Godard, F., Lummerzheim, M., Saindrenan, P., Balagué, C., and Roby, D. (2000). Hxc-2, an Arabidopsis mutant with altered hypersensitive response to Xanthomonas campestris pv. campestris. Plant J. 24 749–762. [DOI] [PubMed] [Google Scholar]
- Hanlon, S.E., and Lieb, J.D. (2004). Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr. Opin. Genet. Dev. 14 697–705. [DOI] [PubMed] [Google Scholar]
- Hara, K., Yagi, M., Kusano, T., and Sano, H. (2000). Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. 263 30–37. [DOI] [PubMed] [Google Scholar]
- Harris, M.A., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32 D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M. (2002). Ecological costs of induced resistance. Curr. Opin. Plant Biol. 5 345–350. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Kalde, M., Barth, M., Somssich, I.E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe Interact. 16 295–305. [DOI] [PubMed] [Google Scholar]
- Katagiri, F. (2004). A global view of defense gene expression regulation—A highly interconnected signaling network. Curr. Opin. Plant Biol. 7 506–511. [DOI] [PubMed] [Google Scholar]
- Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26 509–522. [DOI] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Laudert, D., Pfannschmidt, U., Lottspeich, F., Hollander-Czytko, H., and Weiler, E.W. (1996). Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 31 323–335. [DOI] [PubMed] [Google Scholar]
- Laudert, D., and Weiler, E.W. (1998). Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15 675–684. [DOI] [PubMed] [Google Scholar]
- Lee, B.H., Henderson, D.A., and Zhu, J.K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Brader, G., Kariola, T., and Tapio Palva, E. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46 477–491. [DOI] [PubMed] [Google Scholar]
- Li, J., Brader, G., and Palva, E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Rosso, M.G., Strizhov, N., Viehoever, P., and Weisshaar, B. (2003). GABI-Kat SimpleSearch: A flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics 19 1441–1442. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. [DOI] [PubMed] [Google Scholar]
- Liu, W.M., Mei, R., Di, X., Ryder, T.B., Hubbell, E., Dee, S., Webster, T.A., Harrington, C.A., Ho, M.H., Baid, J., and Smeekens, S.P. (2002). Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18 1593–1599. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Lin, B., Auriac, M.C., Kroj, T., Saindrenan, P., Nicole, M., Balagué, C., and Roby, D. (2004). VASCULAR ASSOCIATED DEATH1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S., Vailleau, F., Balagué, C., and Roby, D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8 263–271. [DOI] [PubMed] [Google Scholar]
- Maeo, K., Hayashi, S., Kojima-Suzuki, H., Morikami, A., and Nakamura, K. (2001). Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 65 2428–2436. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26 403–410. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt, B.F., III, and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., and Scheel, D. (2001). Signal transmission in the plant immune response. Trends Plant Sci. 6 372–379. [DOI] [PubMed] [Google Scholar]
- O'Connor, T.R., Dyreson, C., and Wyrick, J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21 4411–4413. [DOI] [PubMed] [Google Scholar]
- Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25 99–104. [DOI] [PubMed] [Google Scholar]
- Palaniswamy, S.K., James, S., Sun, H., Lamb, R.S., Davuluri, R.V., and Grotewold, E. (2006). AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.J., Shin, R., Park, J.M., Lee, G.J., You, J.S., and Paek, K.H. (2002). Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol. Biol. 48 243–254. [DOI] [PubMed] [Google Scholar]
- Park, C.Y., Lee, J.H., Yoo, J.H., Moon, B.C., Choi, M.S., Kang, Y.H., Lee, S.M., Kim, H.S., Kang, K.Y., Chung, W.S., Lim, C.O., and Cho, M.J. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579 1545–1550. [DOI] [PubMed] [Google Scholar]
- Redman, J.C., Haas, B.J., Tanimoto, G., and Town, C.D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38 545–561. [DOI] [PubMed] [Google Scholar]
- Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., Palme, K., and Hedrich, R. (2002). AtKC1, a silent Arabidopsis potassium channel alpha-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. USA 99 4079–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T. (2001). Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 4 407–414. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Ruepp, A., Zollner, A., Maier, D., Albermann, K., Hani, J., Mokrejs, M., Tetko, I., Guldener, U., Mannhaupt, G., Munsterkotter, M., and Mewes, H.W. (2004). The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Reinstadler, A., Lipka, V., Lippok, B., and Somssich, I.E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K., Foley, R.C., and Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 1–26. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentlemen, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420.
- Somssich, I., and Hahlbrock, K. (1998). Pathogen defence in plants—A paradigm of biological complexity. Trends Plant Sci. 3 86–90. [Google Scholar]
- Taki, N., et al. (2005). 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Penninckx, I.A., Broekaert, W.F., and Cammue, B.P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13 63–68. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T., Ohta, H., Okawa, K., Iwamatsu, A., Shimada, H., Masuda, T., and Takamiya, K. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96 15362–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Zhou, A., and Somssich, I.E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16 2573–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker, B., and Somssich, I.E. (2004). WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7 491–498. [DOI] [PubMed] [Google Scholar]
- Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., Dietrich, R.A., Hirt, H., and Mengiste, T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Yang, P., Fan, B., and Chen, Z. (1998). An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with the plant defence response. Plant J. 16 515–522. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman, E., Hartmann, U., Sagasser, M., Baumann, E., Palme, K., Hahlbrock, K., Saedler, H., and Weisshaar, B. (1998). Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 95 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, K., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17 944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Chen, C., Wang, Z., Fan, B., and Chen, Z. (1999). A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J. 18 141–149. [Google Scholar]
- Zhang, Y., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.