Abstract
By PCR, we screened for qnr genes 112 clinical isolates of extended-spectrum β-lactamase-producing Escherichia coli collected from hospitals in France during 2004. For the first time, 7.7% of CTX-M-producing E. coli isolates presented a plasmid-mediated resistance to quinolones. All strains harbored a qnrA gene located on a sul1-type class 1 integron with similar structure to the In36 integron.
Recently, several studies revealed that CTX-M-type enzymes are becoming the most prevalent extended-spectrum β-lactamases (ESBL) in Escherichia coli isolates in different geographic areas (5, 29, 32, 35, 36, 38). Localized outbreaks of CTX-M-producing E. coli infection have been reported in the north of France (12, 21, 22). However, the link between the increase in ESBL-producing E coli prevalence and the emergence of CTX-M enzyme is not yet established. Moreover ESBL-producing strains generally displayed a high level of resistance to several other non-β-lactam antibiotic families, in particular, to quinolones. A plasmid-mediated quinolone resistance based on the Qnr production has been successively identified in the United States, in China, and recently in France. Interestingly, the qnr-containing plasmid was frequently associated with plasmid-mediated ESBLs (7, 16, 26, 31), but no search was focused on Qnr determinant detection in CTX-M-producing E. coli strains.
(This work was presented in part in 25th Réunion Interdiciplinaire de Chimiothérapie Anti-Infectieuse [RICAI], Paris, France, December 2005 [abstr. 222/520].)
To investigate the relationship between CTX-M-producing E. coli and qnr genes, we undertook a prospective study of ESBL-producing E. coli isolates from 1 January 2004 until 31 December 2004 in three university hospitals (Clermont-Ferrand, Montpellier, and Nîmes) and one community hospital (Perpignan) in the south and center of France. Only one ESBL-producing isolate per sampling type and patient was included in the study. Patients were deemed to have community disease if the first culture found positive for ESBL-producing E. coli was obtained within 48 h of admission. The genus and species were determined biochemically either with the Vitek 2-ID-GNB identification card or the API 20E system (bioMérieux, Marcy-l'Etoile, France). Susceptibility to antimicrobial agents was tested by using the agar disk diffusion assay on Mueller-Hinton agar. Strains were classified as susceptible, intermediate resistant, or resistant to the antibiotics tested according to the recommendations of the Antibiotic Susceptibility Testing Committee of the French Society for Microbiology (40). ESBL production was confirmed by the double-disk synergy test using not only ceftazidime and cefotaxime but also cefpodoxime disks (39). Isoelectric focusing was performed with polyacrylamide gels as previously described (10). The genes blaTEM, blaSHV, and blaCTX-M were detected by PCR using specific primers as previously reported (3, 10, 11, 24) and further identified by sequencing the PCR products. A macrorestriction analysis of chromosomal DNA was performed using pulsed-field gel electrophoresis (PFGE) according to a previous published procedure after XbaI restriction (New England Biolabs, Inc.) by using the contour-clamped homogeneous electric field system (Bio-Rad SA, Ivry-sur-Seine, France) (20). The PFGE patterns were analyzed with Gel compar computer software (Applied Math, Kortrijk, Belgium) and an unweighted-pair group method with the Dice coefficient of similarity. Isolates were considered to be within a cluster if the coefficient of similarity was >80% (42). Phylogenetic grouping of the E. coli isolates was determined by a PCR-based method developed by Clermont et al. (8). The fingerprinting analysis of ESBL-carrying plasmid DNA was performed with the clinical isolates and their electroporants, after digestion with HindIII endonuclease (New England Biolabs, Inc.) and electrophoresis on 0.8% (wt/vol) agarose gels at 100 V for 2 h. The qnrA, qnrB, and qnrS genes were screened by PCR as previously described (15, 16, 34, 43, 44) in all quinolone-resistant strains and electroporants. For PCR mapping of the integrons that contained the blaCTX-M and the qnr genes, PCR primers were used in combination and studied by sequencing the PCR products (26). A search for additional chromosome-encoded quinolone resistance determinants (gyrA, gyrB, parC, and parE genes) was performed by PCR (26). PCR products were sequenced to detect mutations.
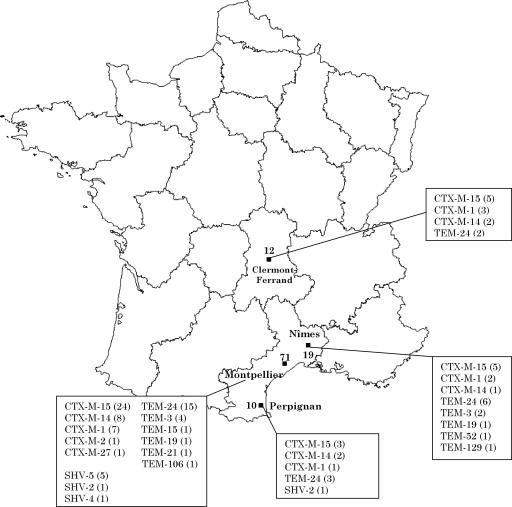
During the period studied, 112 ESBL-producing E. coli strains were isolated from 111 patients. The prevalence of the ESBL production in the E. coli isolates was <3% in the four centers. The distribution of the different types of ESBL according to the geographical origin of the corresponding strains is shown in Fig. 1. E. coli produced mainly CTX-M-ESBLs (58.0%), followed by TEM-type (34.8%) and SHV-type (7.2%) β-lactamases. CTX-M-15 was the most prevalent ESBL in our study (57.0% of CTX-M β-lactamases). We observed that 33.8% of the CTX-M-producing strains were isolated in outpatients compared to strains producing other ESBLs (6.4%), as previously noted by other authors (1, 30, 32, 35, 37, 45). PFGE revealed three independent clonal propagations of CTX-M-15-producing isolates in hospitals in Montpellier (19 isolates), Nîmes (4 isolates), and Perpignan (2 isolates) (Fig. 2). Twelve of these strains were shown to be of community origin, and seven patients infected by these strains had never been hospitalized. The clonal strains belonged to the B2 phylotyping group. No epidemiological link between patients could be demonstrated. The study on the CTX-M-15-producing plasmids revealed that all isolates had one or more plasmids. Electroporation of plasmid DNA from the clinical strains into E. coli DH5α successfully transferred the ESBL phenotype. Analysis of the resulting electroporants revealed the presence of large plasmids (>80 kb) (data not shown) and the same ESBL resistance pattern. Plasmids corresponding to the clonal CTX-M-15-producing E. coli strains yielded similar restriction patterns after digestion with HindIII, whereas those corresponding to the unrelated CTX-M-15-producing E. coli strains harbored different restriction patterns (data not shown).
FIG. 1.
Distribution of ESBL-producing Escherichia coli strains found in four hospitals in France.
FIG. 2.
Dendrogram of XbaI-digested genomic DNAs from all of the CTX-M-15-producing E. coli strains isolated in four hospitals in France. MTP, Montpellier; CF, Clermont-Ferrand; NEC, Nîmes; and PEC, Perpignan. Strains were clustered by the unweighted-pair group method using arithmetic averages (UPGMA). The scale indicates the percentage of genetic similarity. Max. tol., percent maximum tolerance of the curve that matches the bands; Min. surf., percent minimum surface area of a band. The four strains harboring the qnrA gene are noted with asterisks.
In the selected ESBL-producing E. coli strains, 64.3% of the isolates were resistant to nalidixic acid and 56.3% were resistant to ciprofloxacin. The comparison of susceptibility profiles observed for CTX-M- and other ESBL-producing strains indicates that CTX-M-producing strains had more associated resistances than the other ESBL-producing strains. A total of 73.8% of CTX-M-producing E. coli strains were resistant to quinolones versus 51.1% of TEM- and SHV-producing E. coli strains (P < 0.01). Similarly, 66.2% of CTX-M-producing isolates were resistant to ciprofloxacin versus 42.6% for TEM- and SHV-producing isolates (P < 0.01). With the aim of investigating an association between CTX-M production and resistance to quinolones, we screened for qnr genes and transfer of quinolone resistance. We detected the qnrA gene in 7.7% of CTX-M-producing E. coli strains, including four epidemiologically unrelated isolates (MECJ, MECT, MECA6, and NEC39) producing CTX-M-15 and strain NEC34 producing CTX-M-1. No TEM- or SHV-producing strains had qnr genes. In these five qnr-positive strains, MICs of nalidixic acid and ciprofloxacin varied between 64 to 256 μg/ml and 4 to 16 μg/ml, respectively. Except for strain NEC39, the resistance to nalidixic acid was cotransferred with the ESBL-type resistance phenotype for the qnrA-containing strains and the qnrA gene was detected by PCR in the transformants (Table 1). The clinical strains and electroporants harbored a similar qnrA gene to that originally identified in a Klebsiella pneumoniae isolate in United States (43) with a single functionally silent nucleotide change, CTA→CTG, at position 537. The qnrA gene was located on a sul1-type class 1 integron. The structures of the integrons were identical in the five strains and were homologous to the In36 integron (orf513 qnr ampR qacEΔ1 sul1) identified in E. coli isolates from Shanghai (44). The detection of an associated chromosomal quinolone resistance revealed the presence of GyrA in all five strains, with a mutation at codon 83 (Ser→Tyr) and a parC mutation in one strain (NEC39) at codon 80 (Ser→Ile).
TABLE 1.
Characteristics of qnr-positive strainsa
| Strain | MIC (mg/liter)
|
Mutation(s) in topoisomerase genes | Associated ESBL | Phylotyping group | |||
|---|---|---|---|---|---|---|---|
| Clinical strain
|
Electroporant
|
||||||
| Nalidixic acid | Ciprofloxacin | Nalidixic acid | Ciprofloxacin | ||||
| MECJ | >256 | 4 | 8 | 0.25 | gyrA (Ser83→Tyr) | CTX-M-15 | A |
| MECT | >256 | 8 | 8 | 0.125 | gyrA (Ser83→Tyr) | CTX-M-15 | D2 |
| MECA6 | 64 | 8 | 4 | 0.125 | gyrA (Ser83→Tyr) | CTX-M-15 | D1 |
| NEC39 | >256 | 16 | gyrA (Ser83→Tyr) parC (Ser80→Ile) | CTX-M-15 | D1 | ||
| NEC34 | >256 | 16 | 8 | 0.5 | gyrA (Ser83→Tyr) | CTX-M-1 | A |
The aim of this study was to establish the dissemination of and a link between ESBL-producing E. coli isolates and quinolone plasmid resistance. The selected strains represented an actual evolution of E. coli in most parts of the world: increasing prevalence of ESBL-producing E. coli, emergence and diffusion of CTX-M-producing strains (4, 6, 9, 14, 18, 27-29, 38, 41), diffusion of CTX-M-15 (2, 5, 13, 17, 19, 20, 22, 23, 27, 29), and community outbreak of clonally CTX-M isolates (32). The strains were particularly interesting because the majority of CTX-M-producing strains displayed a high level of resistance to quinolones. This high level of resistance associated with CTX-M production was previously described (14), but the actual causes of this association remain not well known. Recently, a plasmid-mediated quinolone resistance determinant named Qnr has been described as leading to a low level of resistance to quinolones (43). Different studies throughout the world showed that the Qnr determinant occurred in between 0.3 and 48% of the strains (15, 26, 33, 43, 44). We found a high rate of qnrA genes (7.7%) associated with CTX-M-type ESBL, in comparison to 0.3% found in a first French study (26). However, we found that four out five strains transferred the two resistances in electroporant strains, suggesting colocalization of qnrA and blaCTX-M genes on the same plasmid. The majority of qnr-positive strains were associated with multidrug resistance and ESBL- or cephalosporinase-producing strains (15, 43). Interestingly, we have described how in the five strains, the qnrA gene is embedded in a complex sul1-type class 1 integron known to integrate numerous multidrug resistance genes as previously suggested (44). The integrons are frequent in strains of Enterobacteriaceae and in strains with multidrug resistance (25). This genetic support induces the transfer between bacteria of the same plasmid or integron and then the transmission and dissemination of these strains. The origin of the plasmid supporting the Qnr region has been detected in a waterborne species, Shewanella algae (31). Our study suggests that the plasmid-mediated Qnr-based mechanism of quinolone resistance could be emerging in France in CTX-M-producing strains. To date, no genetic link has been found between the two emerging mechanisms of resistance. However, recently qnrB has been detected on plasmids also encoding CTX-M-15 in Klebsiella pneumoniae isolates in India (16). Interestingly, an associated chromosomal quinolone resistance was detected in all our strains. This observation explains the higher level of resistance to quinolones in our isolates compared with those observed for qnr-positive transconjugants (Table 1).
In conclusion, CTX-M β-lactamases were associated with quinolone/fluoroquinolone resistance, and in some cases, this association was linked to the Qnr determinant. The frequency of CTX-M strains in weakened patients and their community character invite examination of the epidemiological evolution of these strains and the necessity to inform the medical profession of these results.
Acknowledgments
We are very grateful to A. Gouby for help with this work and Josiane Campos for technical assistance.
This work was supported by Université de Montpellier 1 (BQR, BQ 68, and 88), La Ville de Nîmes, Le CHU de Nîmes, and La Région Languedoc Roussillon.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Arpin, C., V. Dubois, L. Coulange, C. André, I. Fischer, P. Noury, F. Grobost, J.-P. Brochet, J. Jullin, B. Dutilh, G. Larribet, I. Lagrange, and C. Quentin. 2003. Extended-spectrum β-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrob. Agents Chemother. 47:3506-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R., C. Dutour, J. L. M. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brigante, G., F. Luzzaro, M. Perilli, G. Lombardi, A. Coli, G. M. Rossolini, G. Amicosante, and A. Toniolo. 2005. Evolution of CTX-M-type beta-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int. J. Antimicrob. Agents 25:157-162. [DOI] [PubMed] [Google Scholar]
- 6.Chanawong, A., F. H. M'Zali, J. Heritage, J.-H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, T. K., Y. W. Chu, M. Y. Chu, C. H. Ma, R. W. Yung, and K. M. Kam. 2005. Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. J. Antimicrob. Chemother. 56:586-589. [DOI] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and the French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Champs, C., C. Chanal, D. Sirot, R. Baraduc, J. P. Romaszko, R. Bonnet, A. Plaidy, M. Boyer, E. Carroy, M. C. Gbadamassi, S. Laluque, O. Oules, M. C. Poupart, M. Villemain, and J. Sirot. 2004. Frequency and diversity of class A extended-spectrum beta-lactamases in hospitals of the Auvergne, France: a 2 year prospective study. J. Antimicrob. Chemother. 54:634-639. [DOI] [PubMed] [Google Scholar]
- 11.Dutour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez, J. R., L. Martinez-Martinez, R. Canton, T. M. Coque, A. Pascual, and the Spanish Group for Nosocomial Infections (GEIH). 2005. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Antimicrob. Agents Chemother. 49:2122-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J., Y.-M. Lim, Y.-S. Jeong, and S.-Y. Seol. 2005. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 extended-spectrum β-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob. Agents Chemother. 49:1572-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lartigue, M. F., L. Poirel, C. Heritier, V. Tolun, and P. Nordmann. 2003. First description of CTX-M-15-producing Klebsiella pneumoniae in Turkey. J. Antimicrob. Chemother. 52:315-316. [DOI] [PubMed] [Google Scholar]
- 20.Lavigne, J.-P., N. Bouziges, C. Chanal, A. Mahamat, S. Michaux-Charachon, and A. Sotto. 2004. Molecular epidemiology of Enterobacteriaceae isolates producing extended-spectrum β-lactamases in a French hospital. J. Clin. Microbiol. 42:3805-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavollay, M., K. Mamlouk, T. Frank, A. Akpabie, B. Burghoffer, S. Ben Redjeb, R. Bercion, V. Gautier, and G. Arlet. 2006. Clonal dissemination of a CTX-M-15 β-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 50:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leflon-Guibout, V., C. Jurand, S. Bonacorsi, F. Espinasse, M. C. Guelfi, F. Duportail, B. Heym, E. Bingen, and M.-H. Nicolas-Chanoine. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 24.Mabilat, C., S. Goussard, W. Sougakoff, R. C. Spencer, and P. Courvalin. 1990. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum beta-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23:27-34. [DOI] [PubMed] [Google Scholar]
- 25.Machado, E., R. Cantón, F. Baquero, J.-C. Galan, A. Rollán, L. Peixe, and T. M. Coque. 2005. Integron content of extended-spectrum-β-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 49:1823-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markovska, R., I. Schneider, E. Keuleyan, and A. Bauernfeind. 2004. Extended-spectrum beta-lactamase (ESBL) CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in Sofia, Bulgaria. Clin. Microbiol. Infect. 10:752-755. [DOI] [PubMed] [Google Scholar]
- 28.Moland, E. S., J. A. Black, A. Hossain, N. D. Hanson, K. S. Thomson, and S. Pottumarthy. 2003. Discovery of CTX-M-like extended-spectrum β-lactamases in Escherichia coli isolates from five U.S. states. Antimicrob. Agents Chemother. 47:2382-2383. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moubareck, C., Z. Daoud, N. I. Hakimé, M. Hamzé, N. Mangeney, H. Matta, J. E. Mokhbat, R. Rohban, D. K. Sarkis, and F. Doucet-Populaire. 2005. Countrywide spread of community- and hospital-acquired extended-spectrum β-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J. Clin. Microbiol. 43:3309-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munday, C. J., J. Xiong, C. Li, D. Shen, and P. M. Hawkey. 2004. Dissemination of CTX-M type beta-lactamases in Enterobacteriaceae isolates in the People's Republic of China. Int. J. Antimicrob. Agents 23:175-180. [DOI] [PubMed] [Google Scholar]
- 31.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 32.Pitout, J. D. D., D. B. Gregson, D. L. Church, S. Elsayed, and K. B. Laupland. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary health region. J. Clin. Microbiol. 43:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., J.-M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 56:1118-1121. [DOI] [PubMed] [Google Scholar]
- 35.Pournaras, S., A. Ikonomidis, D. Sofianou, A. Tsakris, and A. N. Maniatis. 2004. CTX-M-type beta-lactamases affect community Escherichia coli treatment, Greece. Emerg. Infect. Dis. 10:1163-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinteros, M., M. Radice, N. Gardella, M. M. Rodriguez, N. Costa, D. Korbenfeld, E. Couto, G. Gutkind, and the Microbiology Study Group. 2003. Extended-spectrum β-lactamases in Enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob. Agents Chemother. 47:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Baño, J., M. D. Navarro, L. Romero, L. Martinez-Martinez, M. A. Muniain, E. J. Perea, R. Pérez-Cano, and A. Pascual. 2004. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J. Clin. Microbiol. 42:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero, L., L. Lopez, J. Rodriguez-Bano, J. Ramon Hernandez, L. Martinez-Martinez, and A. Pascual. 2005. Long-term study of the frequency of Escherichia coli and Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 11:625-631. [DOI] [PubMed] [Google Scholar]
- 39.Sirot, J. 1996. Detection of extended-spectrum plasmid-mediated beta-lactamases by disk diffusion. Clin. Microbiol. Infect. 2:S35-S39. [DOI] [PubMed] [Google Scholar]
- 40.Soussy, C. J., G. Carret, J. D. Cavallo, et al. January 2005. Antibiotic Susceptibility Testing Committee of the French Society for Microbiology. Report. [Online.] http://www.sfm.asso.fr.
- 41.Spanu, T., F. Luzzaro, M. Perilli, G. Amicosante, A. Toniolo, G. Fadda, and the Italian ESBL Study Group. 2002. Occurrence of extended-spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrob. Agents Chemother. 46:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]