Abstract
A new pyranoindole class of small-molecule inhibitors was studied to understand viral resistance and elucidate the mechanism of inhibition in hepatitis C virus (HCV) replication. HCV replicon variants less susceptible to inhibition by the pyranoindoles were selected in Huh-7 hepatoma cells. Variant replicons contained clusters of mutations in the NS5B polymerase gene corresponding to the drug-binding pocket on the surface of the thumb domain identified by X-ray crystallography. An additional cluster of mutations present in part of a unique β-hairpin loop was also identified. The mutations were characterized by using recombinant replicon variants engineered with the corresponding amino acid substitutions. A single mutation (L419M or M423V), located at the pyranoindole-binding site, resulted in an 8- to 10-fold more resistant replicon, while a combination mutant (T19P, M71V, A338V, M423V, A442T) showed a 17-fold increase in drug resistance. The results of a competition experiment with purified NS5B enzyme with GTP showed that the inhibitory activity of the pyranoindole inhibitor was not affected by GTP at concentrations up to 250 μM. Following de novo initiation, the presence of a pyranoindole inhibitor resulted in the accumulation of a five-nucleotide oligomer, with a concomitant decrease in higher-molecular-weight products. The results of these studies have confirmed that pyranoindoles target the NS5B polymerase through interactions at the thumb domain. This inhibition is independent of GTP concentrations and is likely mediated by an allosteric blockade introduced by the inhibitor during the transition to RNA elongation after the formation of an initiation complex.
A wealth of literature has documented that RNA viruses or viruses that use RNA replicative intermediates have high degrees of genetic variability (5, 15). The high mutation rates found in these organisms are largely due to the lack of proofreading mechanisms and/or a high recombination frequency during cellular DNA replication. A DNA-dependent DNA polymerase complex allows an error rate of 10−9 mutations/nucleotide, whereas the error rates for an RNA-dependent DNA polymerase (reverse transcriptase) and RNA-dependent RNA polymerase (RdRp), such as those in human immunodeficiency virus (HIV) and hepatitis C virus (HCV), respectively, are on the order of 10−4 to 10−5 mutations/nucleotide (35, 37). The robust viral replication that leads to a large population size during viral infection in these RNA viruses and retroviruses contributes to the generation of quasispecies in infected patients (7, 12, 13).
During chemotherapy, the high rates of viral replication and the high frequency of mutation lead to the rapid generation of drug-resistant mutants. In the case of HIV, numerous mutations have been identified in patients treated with protease inhibitors as well as nucleoside and nonnucleoside reverse transcriptase inhibitors. The emergence of resistant viruses becomes one of the biggest challenges in developing effective antiviral therapies to eradicate these infections.
Hepatitis C virus is a positive-stranded RNA virus that possesses an RdRp, NS5B. Hitherto, a proofreading property has not been identified in this RNA replicase. As a consequence, quasispecies of viral variants have been found in HCV-infected patients (8, 13, 16). Recent research efforts have led to the discovery of many inhibitors that specifically target HCV protease and polymerase (18, 47). Among the HCV polymerase inhibitors, both nucleoside and nonnucleoside analogues have been reported (9, 14, 20, 28, 40, 42, 48). The nonnucleoside analogues, in general, bind to two regions of NS5B: near the catalytic site in the palm domain (33, 45) and near the interface with the C-terminal extension of the polymerase at the base thumb domain (3, 28, 48). The resistance profile and mechanism of action for the palm domain nonnucleoside RNA polymerase inhibitors (NNRPIs) have been reported (33). The studies reported here describe the generation of HCV replicon variants that showed reduced susceptibilities to a thumb domain NNRPI after prolonged passage of the cells containing an HCV replicon in the presence of a pyranoindole inhibitor.
HCV-570, a racemic mixture of pyranoindoles, has previously been shown to specifically inhibit HCV NS5B RdRp (17). The inhibitory activity of this racemate appears to reside in the R enantiomer (HCV-371) (20); no activity was found in the S enantiomer. The chiral structure-dependent inhibitory activity of HCV-371 is also consistent with the results of the cocrystal structure (R. Chopra, S. Sukits, and K. Svenson, unpublished data). In an attempt to understand the molecular interactions and the mechanisms of action of this series of inhibitors in a biological environment, clone A cells that contain an HCV genotype 1b replicon were used to select for replicon variants that showed reduced susceptibilities to these inhibitors. The HCV replicon is a subgenomic RNA that contains all essential elements and genes required for replication in the absence of structural genes (4, 26). The replicon also contains a foreign gene that encodes a drug-selectable marker (neomycin phosphotransferase) that allows the antibiotic (Geneticin, G418) selection of cells that contain a functional replicon. Treatment of cells with HCV polymerase inhibitors results in a dramatic reduction in the amount of HCV RNA, which, consequently, lowers the level of neomycin phosphotransferase production. Under this condition, administration of G418 to these cells facilitates the selection of replicon variants with reduced susceptibilities to the inhibitor. Key amino acids responsible for the resistance were identified by nucleotide sequence mapping and were characterized in replicon variants through the use of reverse genetics. The mechanism of inhibition of these thumb domain NNRPIs was studied in competition experiments with increasing concentrations of GTP and a de novo initiation experiment that examined the products of RNA synthesis. Insights into the mechanism of inhibition and HCV RNA initiation process are discussed.
MATERIALS AND METHODS
Materials.
All tissue culture reagents were purchased from Gibco/BRL and HyClone. Clone A cells (licensed from Apath, LLC) were derived from Huh-7 cells, a human hepatoma cell line. The clone A cells contain approximately 500 to 1,000 genome copies of the HCV genotype 1b replicon per cell when they are maintained in a subconfluent monolayer in the presence of 1 mg/ml G418. The sequence of the replicon in the clone A cells is similar to that of the genotype 1b Con 1 strain of HCV (GenBank accession no. AJ238799), with the exception of two mutations at NS3 (Q1112R) and NS5A (S2204I). Clone A cells were propagated in Dulbecco's minimal essential medium (Gibco/BRL) containing 10% fetal calf serum (FCS; HyClone) supplemented with 1% penicillin-streptomycin (GibcoBRL), 1% nonessential amino acids (Gibco/BRL), 1 mg/ml G418 sulfate (Geneticin; GibcoBRL), and 0.66 mM HEPES buffer (pH 7.5).
Plasmid pBB7, which contains HCV genotype 1b BB7 replicon cDNA, was also licensed from Apath, LLC. The coding sequence of pBB7 is similar to that of the genotype 1b Con 1 strain of HCV, except that there is 1-nucleotide mutation that results in an amino acid change of S2204I within NS5A. All other molecular biology reagents were obtained from the indicated suppliers.
Selection of drug-resistant HCV replicons in clone A cells.
Approximately 6 × 105 clone A cells per well were seeded in a six-well tissue culture dish in four replicates. The cells were treated multiple times over a 16-day period in medium containing 2% FCS without G418 and 30 μM HCV-570 dissolved in dimethyl sulfoxide (DMSO; final concentration of DMSO in the tissue culture medium, 0.5% [vol/vol]). HCV-570 is a racemic mixture that contains about 50% of the R enantiomer, HCV-371 (20). As a control, clone A cells were passaged in parallel in the same medium with 0.5% (vol/vol) DMSO and no compound. When the cells reached approximately 80% confluence (about 3 to 4 days), the cells were passaged in a 1:4 dilution and the old medium was replaced with fresh medium containing 30 μM HCV-570. An aliquot of the cells from each passage was collected for monitoring of the effect of the compound on viral replication. As the HCV viral load reduced to a plateau (day 16), fresh medium containing 30 μM HCV-570 and 0.5 mg/ml G418 was added to select for cells containing the resistant replicons. The concentration of G418 was subsequently increased to 1 mg/ml after four passages. During the course of selection, small colonies of cells resistant to the compound and antibiotics became visible and were pooled and expanded for subsequent analyses.
The susceptibilities of the replicon-containing cells to compounds were evaluated as described previously (20). Briefly, cells were treated with increasing concentrations of compounds in a medium containing 2% FCS and no G418 for 3 days at 37°C in 5% CO2. After incubation, the total RNA from the replicon-containing cells was isolated. The levels of HCV RNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA, and rRNAs were quantified by TaqMan reverse transcriptase PCRs. The amounts of HCV RNA, 18S rRNA, and GAPDH RNA in each sample were estimated by comparing the number of cycles during the exponential phase of the PCR amplification with those in the corresponding standard curves. The HCV RNA standards used for the construction of the standard curve were prepared by extracting the total RNA from the clone A cells. The RNA sample was sent to National Genetics Institute to quantify the HCV RNA. Total RNA extracted from clone A cells was quantified by measurement of the optical density at 260 nm and was used for construction of the standard curves of rRNA and GAPDH RNA. The concentrations of the compounds that inhibited 50% of the HCV RNA level (EC50) were determined by using MDL LSW data analysis software in Microsoft Excel. The amounts of HCV or GAPDH RNA in the samples were expressed as the amount of HCV RNA (number of copies) or GAPDH (ng) per μg of total RNA, respectively, by using rRNA as a marker for total RNA measurement.
Isolation and sequencing of the NS5B gene from replicon-containing cells.
Total cellular RNA was extracted from the clone A cells by using an RNeasy kit purchased from QIAGEN. The NS5B-containing cDNA was generated in a two-step reverse transcription (RT)-PCR. The first-strand cDNA was generated by RT with a 3′-end primer for the NS5B gene (primer 5Bout3 [5′-GGATGGCCTATTGGCCTGGAGTG-3′]). A reaction mixture containing 0.1 to 0.3 μg of the total cellular RNA, 2 pmol of primer, and 10 nmol of each deoxynucleoside triphosphate was mixed, and the mixture was heated at 65°C for 5 min and placed on ice to facilitate primer annealing to the template RNA. Ten microliters of the RNA-primer mixture was added to 9 μl of the SuperScript II reaction mixture, which contained 10 mM dithiothreitol, 5 μM MgCl2, and 40 units of RNaseOut RNase inhibitor. After incubation of the reaction mixture (19 μl) at 42°C for 2 min, the RT reaction was initiated by adding 1 μl of SuperScript II reverse transcriptase (50 units), followed by incubation at 42°C for 50 min. The reaction was terminated by heating at 70°C for 15 min, followed by digestion with RNase H at 37°C for 20 min. To amplify the NS5B gene, 2 to 4 μl of the RT reaction product was mixed with 20 pmol of each of the primers (primer 5Bout5A [5′-ACGGGTCTTGGTCTACCGTAAG-3′] and primer 5Bout3 [as described above]), 10 nmol of each deoxynucleoside triphosphate, 2.5 units of Pfu Turbo polymerase, and 1× buffer provided by the supplier (Stratagene). The reaction (final volume, 50 μl) was carried out at 95°C for 30 s, followed by 25 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 2 min. The PCR products were evaluated by agarose gel electrophoresis. The band at 1.8 kb was excised, and the cDNA fragment was extracted from the gel. The cDNA was ligated with the PCR4Blunt TOPO vector (Zero Blunt TOPO PCR cloning kit; Invitrogen). The resulting recombinant DNA plasmid was transformed into One Shot electro- or chemical-competent Escherichia coli cells purchased from Invitrogen. The presence of the HCV NS5B insert in the plasmids was verified by EcoRI digestion. Plasmids containing the HCV NS5B inserts were subjected to nucleotide sequencing by using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (version 3.0; Applied Biosystems). The sequencing reactions were set up in a 96-well PCR plate in a final volume of 20 μl. The reaction mixture consisted of 1 μl of the Terminator ready reaction mix, 3.5 μl of 5× sequencing buffer, 3.2 pmol of primer, and 500 ng of plasmid DNA. The sequencing reaction was conducted under the conditions described in the manufacturer's instructions. The sequenced products were purified by using a DyeEx 96 kit (QIAGEN), dried, denatured with formaldehyde, and separated by electrophoresis with an ABI Prism 3700 DNA sequencer. Sequence data were analyzed by using Sequencher (version 4.0).
Generation of replicon cell lines with NS5B mutations.
The nucleotide sequence corresponding to amino acid residues 1 to 570 of the HCV NS5B protein was amplified by PCR with plasmid pBB7 as the template and primers 5B-For and 5B-D216Xhis (Table 1). An initiating methionine codon and an NdeI site (indicated in boldface in Table 1) were added to the 5′-end primer, primer 5B-For; and an XhoI site (indicated in boldface in Table 1) was added to the 3′-end primer, primer 5B-D216Xhis. The amplified DNA was cloned into the pET-21b vector, which had previously been digested with NdeI and XhoI (Novagen) to generate the wild type, pET-(BB7)5BΔ21CHis. This plasmid was used as the template for the generation of plasmids containing mutant NS5B. Site-directed mutagenesis was accomplished with a QuikChange site-directed mutagenesis kit, according to the manufacturer's recommendations (Stratagene). The sequences of the primers and the oligonucleotide pairs used to generate cDNA encoding single amino acid changes in the NS5B protein are indicated in Table 1. Additionally, as a negative control, a mutation which abolishes the catalytic activity of NS5B by changing the GDD at amino acid residues 317 to 319 to GAA to generate pET-5B-GDD was constructed. The sequences of the NS5B region in all plasmid constructs were verified.
TABLE 1.
Primer sets used for the NS5B site-directed mutagenesis
| Primera | Sequenceb |
|---|---|
| 5B-For | 5′-TCAGCCATATGTCGATGTCGATGTCCTACACATGGACAGGC-3′ |
| 5B-D216Xhis | 5′-TCACTCGAGGCGGGGTCGGGCACGAGACAGGC-3′ |
| L419M F | 5′-ATC ATG TAT GCG CCC ACC ATG TGG GCA AGG ATG ATC CTG -3′ |
| L419M R | 5′-CAG GAT CAT CCT TGC CCA CAT GGT GGG CGC ATA CAT GAT-3′ |
| A421V F | 5′-ATG CGC CCA CCT TGT GGG TAA GGA TGA TCC TGA TG-3′ |
| A421V R | 5′-CAT CAG GAT CAT CCT TAC CCA CAA GGT GGG CGC AT-3′ |
| M423V F | 5′-CTT TGT GGG CAA GGG TGA TTC TGA TGA CTC-3′ |
| M423V R | 5′-GAG TCA TCA GAA TCA CCC TTG CCC ACA AAG-3′ |
| M426T F | 5′-TGG GCA AGG ATG ATC CTG ACC ACT CAT TTC TTC TCC ATC-3′ |
| M426T R | 5′-GAT GGA GAA GAA ATG AGT GGT CAG GAT CAT CCT TGC CCA-3′ |
| M426V F | 5′-CAA GGA TGA TTC TGG TGA CTC ACT TCT TCT C-3′ |
| M426V R | 5′-GAG AAG AAG TGA GTC ACC AGA ATC ATC CTT G-3′ |
The nomenclature lists the wild-type residue first and its position in the NS5B protein, followed by the residue it was changed to. F and R, forward and reverse primers, respectively.
Mutations are underlined. Restriction sites are indicated in boldface.
To generate pBB-L419M, pBB7-A421V, pBB7-M423V, pBB7-M426T and pBB7-M426V, a unique 917-bp Bsu36I fragment within the NS5B region from the constructs pET-(BB7)5BΔ21CHis containing the NS5B mutations was exchanged for the equivalent fragment in pBB7. The plasmids encoding multiple mutations within NS5B were constructed by swapping the Bsu36I fragment from the PCR4Blunt TOPO clones generated from the pool of resistant replicon cells with the equivalent fragment in pBB7. In cases in which the Bsu36I sites were absent or mutations occurred outside this fragment, mutations were generated by site-directed mutagenesis.
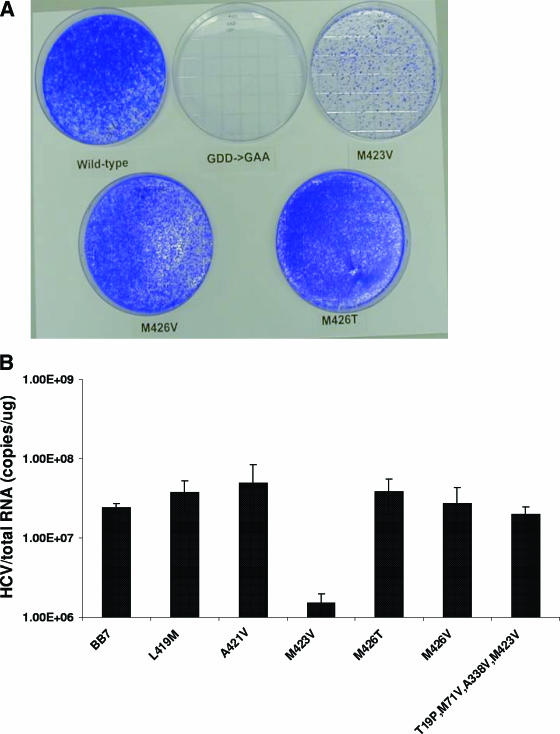
Replicon RNA for transfection was prepared by digesting the plasmid DNAs for all the replicon constructs with ScaI, followed by in vitro transcription and purification of RNA by using a T7 MegaScript kit (Ambion), according to the manufacturer's recommendations. The purified RNAs were quantified by determining the absorbance at 260 nm, and their integrities were determined by denaturing gel electrophoresis with 1% glyoxal-containing agarose gels (NorthernMax-Gly gel prep/running system; Ambion) with RNA size standards (0.24- to 9.5-kb RNA ladder; Invitrogen). Five micrograms of each transcribed RNA was transfected into Huh-7 cells with a BTX electroporator. The transfected cells were plated onto 150-mm dishes, and at 24 h after transfection, the medium was replaced with fresh medium containing 1.0 mg/ml G418 (Invitrogen) and colonies were selected. After 3 weeks, individual colonies were isolated with sterile cloning rings, trypsinized, and expanded to generate cell lines containing HCV replicons with the engineered changes in NS5B. Additionally, a separate plate was fixed with 5% glutaraldehyde for 1 h and then stained with crystal violet to visualize the number of colonies.
Inhibition kinetics.
HCV RdRp assays were performed under saturating nucleoside triphosphate (NTP) reaction conditions in which either the concentration of pOF RNA (20) or the concentration of a single NTP in the reaction mixture was varied. Reactions were carried out in either the absence or the presence of HCV-371 at 0, 0.05, 0.2, 1, 2, 4, and 8 μM. Inhibition data from these reactions were analyzed by double-reciprocal plots of the polymerase activity versus the concentration of the variable substrate for each HCV-371 concentration. Data from the double-reciprocal plots were fitted to equations for both linear competitive inhibition, v = VmaxA/[Km(1 + I/Ki) + A], and linear noncompetitive inhibition, v = VmaxA/{[Km + A](1 + I/Ki)}, where v is the initial velocity, Vmax is maximum velocity, Km is the Michaelis constant, A is the substrate concentration, I is the inhibitor concentration, and Ki is the inhibition constant, by using the programs of GraFit 5.0.3 (Erithacus Software, Surrey, United Kingdom). Determination of the best fit was made according to criteria established by Cleland (11).
De novo initiation.
One micromolar of RNA (5′-AAA AAA AAA AAA AAA AGU CC-3′) was incubated with 1 μM HCV NS5BdCT21-His (21) in a 50-μl reaction mixture containing 20 μM HEPES (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, 0.05 mg/ml bovine serum albumin, 0.4 U/ml RNase inhibitor (Promega), and an inhibitor dissolved in 15% DMSO (final DMSO concentration, 3% [vol/vol]). The reaction was initiated by adding 0.2 μM ATP, 0.016 μM CTP, 0.005 μM GTP, and 0.1 μM UTP spiked with 10 μCi [α-32P]NTP, as indicated. The reaction mixture was incubated at room temperature for 1 h, and the reaction was terminated by adding an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) (Gibco/BRL). The reaction products were extracted once with phenol-chloroform-isoamyl alcohol and precipitated with 3 volumes of ice-cold ethanol, 0.3 M sodium acetate (pH 5.5), and 20 μg of glycogen (Invitrogen). The precipitate was washed with 0.8 ml 95% ethanol, dried under vacuum for 5 min, and dissolved on ice with 25 μl TE buffer (10 mM Tris-HCl, pH 7.0, 1 mM EDTA). The reaction products were added to an equal volume of gel loading buffer (Ambion), and the mixture was heated at 85°C for 10 min before separation in a 25% urea-polyacrylamide gel. After electrophoresis, the gel was fixed in the presence of 10% glycerol, 40% ethanol, and 10% acetic acid and examined with a Fluor-S-MultiImager (Bio-Rad).
RESULTS
Selection of replicon variants with reduced susceptibilities to pyranoindoles.
Clone A cells were serially passaged in the presence (or the absence) of 30 μM (about four to five times the EC50) HCV-570 without G418 for 16 days, and aliquots of cells were collected from each passage to monitor the levels of HCV RNA. While the viral load of the control cells remained relatively stable throughout the treatment, the cells treated with HCV-570 showed a >15-fold reduction in HCV RNA levels within the first 3 days of treatment (data not shown). Continual treatment of the cells with the compound up to day 16 did not lead to a further reduction in the HCV RNA level, most likely due to the replication of resistant replicon variants.
To select for cells containing drug-associated resistant replicons, 0.5 mg/ml of G418 was applied to the cells treated with 30 μM of HCV-570 at the sixth passage. The concentration of G418 was subsequently increased to 1 mg/ml after four passages. During the course of G418 selection, cells that were “cured of” the HCV replicon or that contained low levels of HCV RNA did not sustain the antibiotic selection and were removed. Colonies of the drug-resistant cells (henceforth referred to as “570R cells”) began to appear after two to three passages in the presence of G418 and HCV-570. The pool of 570R cells was expanded and used for the subsequent experiments.
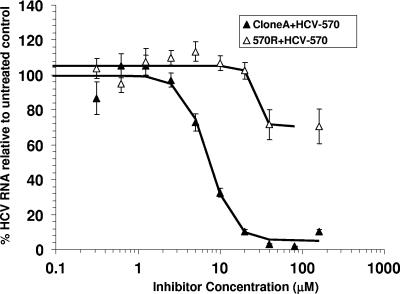
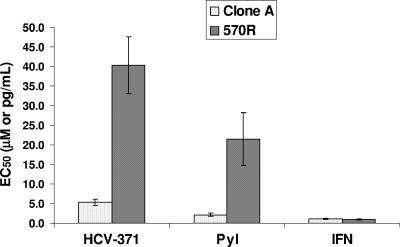
The susceptibilities of the 570R cells to pyranoindole inhibitors were evaluated and compared with those of the wild-type clone A cells. As shown in Fig. 1, a dose-dependent inhibition of the steady-state HCV RNA levels was observed in the control cells treated with HCV-570 (EC50, 7.2 ± 0.5 μM). In contrast, the 570R cells were more than 15-fold less susceptible to the compound inhibition (EC50s, >160 μM). As expected, 570R cells displayed a loss of susceptibility to HCV-371, the purified R enantiomer of HCV-570 (Fig. 2, HCV-371). A 10-fold reduced susceptibility was also observed in the 570R cells treated with another structurally related pyranoindole inhibitor (PyI, Fig. 2). The specificity of resistance was evaluated by treating the 570R cells with alpha interferon (IFN-α) (Fig. 2, IFN). No difference in susceptibility was observed between the wild-type clone A and the 570R cells (Fig. 2) for IFN-α.
FIG. 1.
Susceptibilities of 570R variants to HCV-570. Clone A and 570R cells were treated with increasing concentrations of HCV-570 for 3 days before total RNA isolation. The levels of HCV RNA were analyzed by a quantitative TaqMan RT-PCR. The y axis of the graph indicates the mean percentage of HCV RNA compared to that for untreated cells, with error bars indicating the standard deviations. Each datum point in the graph represents an average value obtained from four cell replicates; each was analyzed twice by the TaqMan RT-PCR.
FIG. 2.
Specificity of pyranoindole-associated resistance. 570R cells harboring replicons resistant to HCV-570 were treated with HCV-371 (a purified R enantiomer of HCV-570), a pyranoindole inhibitor (PyI), and IFN-α (IFN). After 3 days of treatment, total cellular RNA was extracted, and the levels of HCV RNA and total cellular RNA were determined by a quantitative duplex TaqMan RT-PCR. The y axis represents the mean compound concentrations (μM for HCV-371 and the pyranoindole inhibitor, pg/ml for IFN-α) required to inhibit 50% of the HCV RNA copies/μg total cellular RNA (EC50) in the replicon-containing cells. Each bar represents an average of at least three independent determinations, and the error bars are the associated standard deviations.
Mapping of amino acid changes in NS5B pyranoindole-resistant variants.
Since the pyranoindole class of inhibitors specifically targets the HCV NS5B RdRp (17, 20), the resistance observed in the 570R cells is likely due to the presence of mutations within the NS5B. To map the amino acid changes within the NS5B enzyme, total RNA was extracted from the 570R cells and the NS5B gene was amplified by RT-PCR. The NS5B gene was cloned into plasmids and transformed into E. coli, and bacterial colonies were isolated to generate a library of the NS5B gene variants. As a control, the HCV NS5B gene from the control clone A cells was prepared in parallel.
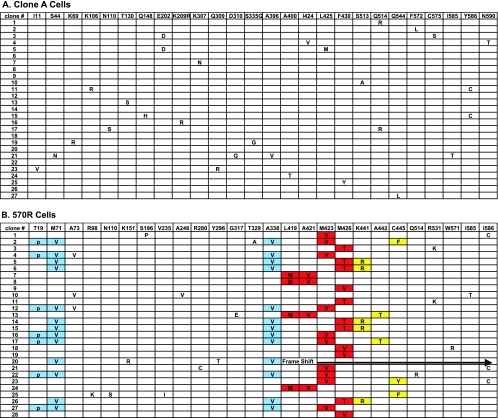
As expected from RNA viruses, sporadic mutations were observed within the NS5B gene derived from the control clone A cells (Fig. 3A). Among the 27 clones isolated, 9 clones contained the wild-type sequence, 9 clones had one mutation, 8 clones had two mutations, and 1 clone had four mutations. A total of 25 mutations among the 27 clones, with an average of about 1 mutation/clone, were observed within the NS5B gene derived from the control clone A cells. All mutations appeared to be random in control replicons due to the lack of a proofreading mechanism in the HCV RdRp, and no particular amino acid change provided a replication advantage relative to the original input replicon sequence.
FIG. 3.
Summary of amino acid changes with the HCV NS5B gene isolated from clone A (A) and 570R cells (B).
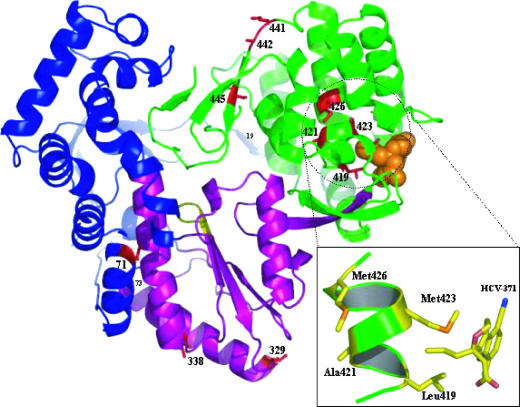
In contrast, specific patterns of mutations were detected in clones derived from the 570R cells. Among the 28 clones examined, 27 clones had a complete open reading frame for NS5B; 1 (clone 20) had a frameshift mutation that led to a premature termination of the NS5B open reading frame (Fig. 3B). Two clusters of amino acid changes were observed within the thumb subdomain of NS5B (Fig. 4). A detailed analysis of these clusters in the context of the crystal structure will be presented elsewhere (Chopra et al., unpublished). Briefly, the first cluster contains mutations in the vicinity of the compound's binding site, including changes at amino acids 419 (Leu to Met), 421(Ala to Val), 423 (Met to Val), and 426 (Met to Thr/Val) (Fig. 3B and 4). The mutations of L419M and A421V always coexist within the same replicon variants. In addition, the binding pocket within the same NS5B gene in the replicon variants appears to allow only one of the three patterns of mutations, namely, L419M linked with A421V, M423V, or M426T/V. The second cluster of mutations, which includes amino acids 441 (Lys to Arg), 442 (Ala to Thr), and 445 (Cys to Phe/Tyr), was located on β-strand 7 (Fig. 4). This structural element forms a part of the β-hairpin loop, unique to NS5B, which may play a role in positioning the 3′ end of the RNA template for proper initiation of RNA synthesis, as well as interfering with double-stranded RNA binding (19, 50).
FIG. 4.
Crystal structure showing pyranoindole resistance mutations as a ribbon diagram of HCV NS5B in complex with a pyranoindole inhibitor, HCV-371. The protein is represented as an idealized ribbon and is colored according to the domains: the thumb domain is green, the palm domain is purple, and the fingers domain is blue. Sites of drug resistance mutations are red and are labeled. The inset depicts a simplified view of the pyranoindole-binding site indicating the relative positions of Leu419, Ala421, Met423, and Met426 on α-helix N.
In addition to the amino acid changes proximal to the HCV-371-binding pocket, three mutations, at residues 19 (Thr to Pro), 71 (Met to Val), and 338 (Ala to Val), were found in the finger and palm subdomains of the polymerase (Fig. 3B and 4). M71V and A338V appear to have the same frequency of mutation and coexist within the same gene, without exception.
Characterization of drug-associated resistant replicon variants in Huh-7 cells.
To assess the contribution of the amino acid changes associated with drug resistance, replicon variants containing single amino acid mutations in NS5B in the background of the genotype 1b BB7 adaptive replicon were generated (4). Transfection of the replicon RNAs into Huh-7 cells resulted in colony formation in the presence of G418 within 20 days after transfection. No colonies were obtained from Huh-7 cells transfected with the RNAs containing a GAA mutation within NS5B (Fig. 5A) or mock transfected (results not shown). Replicon RNA containing the M423V mutation consistently produced less than 10% of the colonies generated by the wild-type replicon or other variants (Fig. 5A). The colony formation efficiency for the wild-type replicon and the L419M, A421V, or M426V/T variants ranged from 1,470 to 3,000 CFU/μg in in vitro-transcribed RNA, whereas the colony formation efficiency was 108 CFU/μg RNA for the M423V replicon. Colonies derived from the M423V replicon also grew more slowly. Following G418 selection, all replicon-containing cells with the exception of the M423V variant had similar steady-state levels of HCV RNA (approximately 107 to 108 copies of HCV RNA/μg total cellular RNA); the M423V variant had a 10-fold lower HCV RNA level (Fig. 5B). The lower colony formation efficiency, the slower growth rate, and the reduced steady-state viral RNA level suggested that the M423V replicon variant replicates suboptimally in these cells.
FIG. 5.
Characterization of cells transfected with HCV replicon RNAs. (A) Colony formation efficiencies of the replicon variants. HCV replicon RNAs molecularly engineered with mutations in the NS5B gene were transfected into Huh-7 cells by electroporation. After 3 weeks, the cells were fixed with 5% glutaraldehyde and stained with crystal violet to visualize the number of colonies. (B) Steady-state HCV RNA levels in replicon variants. Replicon-containing cells were seeded under subconfluent conditions in 96-well plates. The cells were allowed to grow for 3 days, until the cell density was confluent. Total cellular RNA was extracted, and the levels of HCV and total cellular RNAs were determined by a quantitative duplex TaqMan RT-PCR. The HCV RNA levels of each replicon variant were determined from at least three independent cell clones; each was measured in four replicates. The steady-state level of HCV RNA is expressed as the mean number of HCV copies per microgram of total cellular RNA, with the error bars indicating the standard errors of the means.
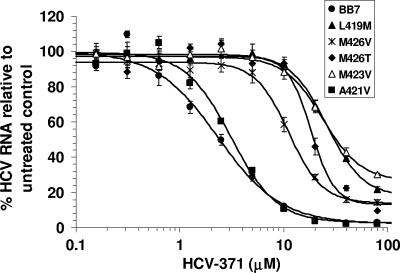
Susceptibility to HCV-371 in the replicon variant cell lines was evaluated in a 3-day assay. As shown in Fig. 6, HCV replication in the wild-type BB7 replicon was inhibited in a dose-dependent manner, with an EC50 of 3.3 ± 0.4 μM (Fig. 6 and Table 2). The replicon variant containing the A421V mutation exhibited susceptibility to HCV-371 similar to that of the wild-type BB7 replicon (Fig. 6 and Table 2). A mutation at amino acid 426 (M426V/T) resulted in a moderate decrease (three- to fourfold) in susceptibility to the compound (Fig. 6 and Table 2). The replicon variants containing L419M and M423V mutations displayed 8- to 10-fold reduced susceptibilities to HCV-371 (Fig. 6 and Table 2). L419 and M423 have direct contacts with the inhibitor (Fig. 4), which may account for their significant impact on resistance. The resistance of the replicon variant containing double mutations of L419M and A421V was not increased (compare L419M with L419M and A421V; Table 2). It should be noted that despite the increase in EC50s, no variants were completely resistant to HCV-371; inhibition of viral RNA was observed as the compound concentrations were increased (Fig. 6).
FIG. 6.
Effect of HCV-371 on cells containing drug-associated replicon variants. Cells harboring the wild type or variants of genotype 1b BB7 NS5B were treated with increasing concentrations of HCV-371 in medium containing 2% FCS and 0.5% DMSO without G418 for 3 days. Total cellular RNA was extracted, and the levels of HCV and total RNAs were determined by a quantitative duplex TaqMan RT-PCR. The amounts of HCV RNA were expressed as the number of HCV RNA copies per μg total cellular RNA. The y axis indicates the mean percentage HCV RNA level relative to that for untreated control cells, and the error bars indicate the associated standard deviations. At least three cell clones were evaluated for each variant. The figure shows representative results of a typical experiment obtained from one of the experiments. Each point represents an average of eight determinations. IC50s were determined by the MDL LSW data analysis.
TABLE 2.
Activities of HCV-371 in replicon variants containing amino acid mutations in NS5B
| Cella | EC50 (μM [mean ± SD])b | Mean fold resistancec |
|---|---|---|
| Wild-type (1b, BB7) | 3.3 ± 0.4 (14) | |
| L419M | 34.3 ± 8.1 (3) | 10d |
| A421V | 4.0 ± 1.1 (12) | |
| M423V | 27.9 ± 5.9 (4) | 8d |
| M426T | 14.4 ± 3.3 (6) | 4d |
| M426V | 10.5 ± 5.3 (5) | 3 |
| L419M, A421V | 22.9 ± 2.6 (3) | 7d |
| T19P, M71V, A338V, M423V | 32.5 ± 1.9 (3) | 10d |
| T19P, M71V, A338V, M423V, A442T | 56.2 + 13.4 (3) | 17d,e |
| T19P, M71V, T329A, A338V, M423V, C445F | 37.1 + 5.3 (3) | 11d |
The BB7 replicon was derived from pBB7, which was licensed from Apath, LLC. Replicon variants were constructed in the genotype 1b BB7 NS5B gene. The nomenclature for the replicon variants is listed with the wild-type residue first and its position in the NS5B protein, followed by the residue it was changed to. The amino acid changes in addition to the T19P, M71V, A338V, M423V variant are underlined.
Inhibitory activities are expressed as the mean IC50 ± standard deviation. At least three cell clones were analyzed for each resistant variant. The number of determinations is provided in parentheses.
Fold resistance was determined by comparing the IC50s of the replicon variants with that of BB7.
With the exception of M426V, all variants had significant fold resistance compared to that of the wild-type (genotype 1b BB7) replicon, with P values of <0.05.
P was <0.05 in comparison with the results for replicon variants T19P, M71V, A338V, and M423V.
Because a majority of the NS5B clones derived from the resistant replicons contained mutations of T19P, M71V, A338V, and M423V (Fig. 3B), the combined effects of these amino acid changes on resistance were evaluated by swapping a fragment of NS5B containing these mutations with that in the wild-type BB7 replicon. The EC50 of HCV-371 for the replicon variant containing the T19P/M71V/A338V/M423V mutations was similar to that for the replicon variant containing only the M423V mutation (Table 2), suggesting that the mutations at the finger (T19P, M71V) and palm (A338V) subdomains might not contribute directly to the drug-associated resistance. The presence of the T19P/M71V/A338V mutations in this replicon variant correlated with the increase in the HCV RNA level so that it was similar to that in the wild type (Fig. 5B). These mutations may play a compensatory role in restoring the fitness and replication capacity of the M423V replicon in the cells.
By using the T19P/M71V/A338V/M423V replicon variant as the reference, the effects of mutations located at the β-hairpin loop were examined. The presence of the A442T change gave an additional sevenfold increase in resistance (Table 2). In contrast, the presence of the T329A and C445F mutations did not add further resistance (Table 2).
Competition with GTP.
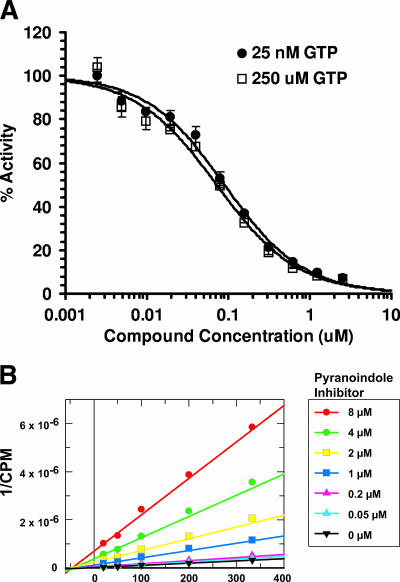
HCV NS5B binds to recombinant GTP (rGTP) at a secondary site 30 Å away from the catalytic pocket (6). This secondary rGTP-binding site is located at the molecular surface between the finger and thumb subdomains of the HCV polymerase. Although the pyranoindole inhibitor has no direct contact with the residues constituting this rGTP-binding pocket, the pyranoindole-binding pocket is 12Å from this secondary rGTP-binding site (Fig. 4). The inhibitor at the thumb cleft might affect GTP binding at this site (3, 28, 48). This low-affinity GTP-binding site has been proposed to play an allosteric regulatory role in initiating RNA synthesis and NS5B oligomerization (6, 38, 49).
The competitive nature of the pyranoindole inhibitors with GTP was evaluated in two independent experiments. In the first study, the inhibitory profile of HCV NS5B polymerase activity was determined in the presence of saturating and limiting GTP and a pyranoindole inhibitor. As shown in Fig. 7A, the dose-response curves generated under these two conditions were superimposed, indicating that the substantial increase in GTP concentration does not affect the ability of the pyranoindole compound to inhibit NS5B polymerase activity. In the second study, the kinetics of inhibition were examined by varying the concentrations of GTP in the absence or the presence of a pyranoindole inhibitor. The Lineweaver-Burk double-reciprocal plot of the polymerase activity versus GTP concentrations at various inhibitor concentrations is indicated in Fig. 7B. In this study, the Km for GTP remained unchanged, while the apparent Vmax of the enzyme decreased with increasing concentrations of the inhibitor. Similar results were obtained with CTP, ATP, UTP, and RNA templates (data not shown). The results of these kinetic studies suggest that the pyranoindole compound inhibits the HCV NS5B RdRp noncompetitively with respect to the substrate NTPs and RNA template, possibly through binding at an allosteric site in the HCV polymerase.
FIG. 7.
Effect of GTP on enzyme activity. (A) Inhibitory activity of HCV-371 in the presence of high GTP concentrations. HCV NS5BdCT21-His was preincubated with 25 nM or 250 μM GTP, followed by incubation with increasing concentrations of HCV-371. The reaction was initiated by adding the RNA template and four NTPs spiked with [α-32P]UTP. The reaction products were quantified after 1 h at room temperature. The y axis represents the mean percentage of RdRp activity remaining for three replicates compared with that for the no-inhibitor control, with error bars indicating the associated standard deviations. IC50s for NS5B activity were determined by MDL LSW data analysis. (B) Lineweaver-Burk double-reciprocal plot of the polymerase activity versus GTP concentrations at various inhibitor concentrations. Enzyme activity in the absence or presence of increasing inhibitor concentrations was determined as described in Material and Methods. The y axis indicates the inverse of the amounts of radioactive nucleotide incorporated, and the x axis indicates the reciprocal of increasing GTP concentrations. Rates are means of duplicate experiments. Individual values were within 10% of the mean.
Therefore, the inhibitory activity of the pyranoindole inhibitors is not affected by GTP as a primary substrate during RNA synthesis or as a regulatory nucleotide.
Effects on primer initiation during RNA synthesis.
The pyranoindole-binding pocket lies adjacent to several structural elements, including the β loop; the C-terminal hydrophobic tail; and motif E, which is located at β strands 14 and 15 (Fig. 4) (2, 6, 24, 36) and which is implicated in de novo initiation and primer extension during RNA synthesis (1, 19, 39). The presence of an inhibitor at this binding site might perturb the interdomain contacts required for RNA synthesis.
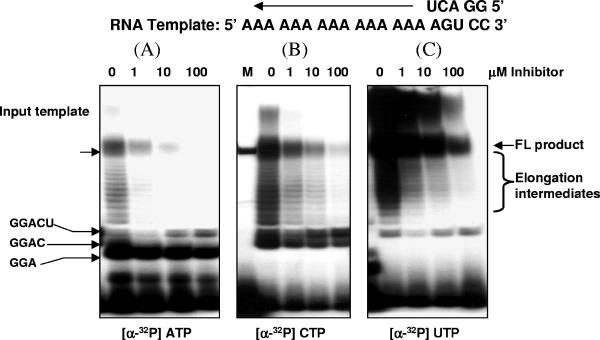
Purified recombinant HCV NS5B RdRp has been demonstrated to initiate in vitro RNA synthesis by three mechanisms, namely, de novo, copy-back, and template-primer initiations (29, 34). De novo initiation can be studied by using a single-stranded RNA template and high concentrations of GTP/GMP (22, 27, 41, 51). Under this experimental condition, the C-terminal truncated HCV NS5B, NS5BdCT21-His, synthesized a full-length product with a size similar to that of the input RNA template (data not shown). There was an accumulation of dinucleotides, possibly due to an abortive initiation. To monitor the effect of a pyranoindole inhibitor on the formation of a productive initiation complex, NS5BdCT21-His was incubated with a short single-stranded RNA template, 50 μM GMP, and the four NTPs spiked with radioactively labeled nucleotides. The RNA products were resolved in a denaturing polyacrylamide gel. As shown in Fig. 8, the presence of the inhibitor appeared to have no effect on the formation of the first 4 nucleotides (up to GGAC in Fig. 8B). However, a dose-dependent accumulation of a five-nucleotide oligomer (GGACU) with a concomitant decrease of the elongation intermediates and full-length product was evident in all samples examined (Fig. 8A to C). These results suggested that the initiation process might be interrupted after synthesis of the first 5 nucleotides. Although the initiation product containing 4 nucleotides was not detected in the samples labeled with radioactive ATP, a dose-dependent accumulation of the 5-nucleotide initiation product was detected in these samples. A similar result was observed when the 3′UTR derived from the minus-strand HCV RNA was used as the template RNA in the assay (data not shown).
FIG. 8.
De novo initiation of RNA. HCV NS5BdCT21-His was incubated with substrate RNA template and increasing concentrations of a pyranoindole inhibitor. The template and primer sequences are indicated at the top of the diagram. The reaction was initiated by the addition of four NTPs spiked with one radioactive NTP, as indicated. The reaction products were isolated from the reaction mixture, analyzed in a denaturing polyacryalmide gel, and examined with a Fluor-S MultiImager. FL, full-length.
DISCUSSION
HCV-371 has been shown to specifically target HCV RdRp and inhibit HCV replication in the replicon cell culture system (20). As a first step to understanding the molecular mechanism and viral resistance, HCV replicon-containing cells were used to select mutants with reduced susceptibilities to this class of inhibitors. A similar approach has been demonstrated with other HCV protease and polymerase inhibitors (25, 30, 33, 46). The replicon variants generated from the pyranoindole inhibitor remained susceptible to IFN (Fig. 2). Nucleotide sequence mapping revealed that the majority of mutations occurred around the inhibitor-binding pocket, with L419M and M423V contributing the greatest impact on resistance (Table 2 and Fig. 6). In addition, A442T, which is located at the β-hairpin loop, augmented the resistance in the presence of other mutations (Table 2). In the absence of an active infection in the replicon system, it is difficult to estimate the fitness of the replicon variants. Several amino acid changes in the replicon variants appear to affect viral replication. For example, M423V resulted in a lower colony formation efficiency and a reduced growth rate, whereas T19P, M71V, and A338V restored the steady-state replicon RNA level in the replicon variants.
There are several possibilities, not all mutually exclusive, that can explain the mechanism of a thumb domain NNRPI. These include (i) displacement of the GTP binding at the secondary site (6), (ii) disruption of the NS5B dimerization (38, 49), (iii) interference of polymerase-RNA interactions, and (iv) allosteric hindrance of the polymerase conformations during RNA synthesis. Although the proposed secondary GTP-binding site offers an attractive explanation because of its proximity to the inhibitor-binding pocket, the results from the mechanistic studies indicated that the inhibitory effect of the compound was not affected in the presence of high GTP concentrations (Fig. 7A) and that the compound inhibited the polymerase activity noncompetitively with respect to GTP (Fig. 7B). Sucrose gradient and ultracentrifugation analyses showed that the NS5BdCT21-His used for the in vitro studies was in a homogeneous monomeric form (data not shown). Therefore, it is unlikely that the mechanism of inhibition is due to blocking of the dimerization of NS5B. Consistent with the results of inhibition kinetics, in which the pyranoindole inhibitor acted noncompetitively with respect to the substrate NTPs and RNA, the results of the studies that used mass spectrometry-gel filtration and filter binding assays showed that the pyranoindole class of inhibitors did not bind to or compete with RNA for their interaction with the polymerase (unpublished results). On the basis of the results from the mechanistic studies, we favor allosteric hindrance as the most probable mechanism of the inhibitor's mode of action. Footprints of amino acid mutations at the structural elements essential for RNA initiation, such as the thumb domain, the unique β-hairpin loop, and the fingers domain, were detected in the pyranoindole-resistant replicons (Fig. 3B and Fig. 4). The results from the de novo initiation (Fig. 8) and the lack of inhibition in the truncated forms of NS5B lacking the β-hairpin loop or C-terminal tail (unpublished results) unequivocally suggested that this thumb domain NNRPI mediates long-range inhibition of polymerase activity. It is hypothesized that binding of the pyranoindole inhibitor at the lower thumb domain near the interface of the C-terminal tail of NS5B (Fig. 4) might allosterically interfere with the structural conformational change that is required for the transition from initiation to elongation during RNA synthesis (also see comments below).
The inhibition mediated by the pyranoindole inhibitors reveals an intriguing mechanism of RNA synthesis in HCV reminiscent of that described in the bacteriophage T7 RNA polymerase (T7 RNAP). In the case of T7 RNAP, the initiation of RNA synthesis involves binding of the N-terminal subdomain of the T7 RNAP to the specific DNA promoter sequence, melting of the duplexed DNA (transcription bubble), and the initiation of RNA synthesis by use of the coding strand of the DNA as a template and a single ribonucleotide. During the early stage of transcription, an abortive cycling and synthesis of a short 2- to 3-nucleotide transcript occurs (23, 43). Transition from an unstable binary initiation complex to a competent elongation complex is a multistep process which results in an 8-base-paired RNA-DNA hybrid during the intermediate step and the emergence of a 10- to 14-nucleotide RNA from the surface of the enzyme at the final stage (10, 44). The crystal structures of the T7 RNAP initiation and elongation complexes show that the RNAP undergoes remarkable structural reorganization during this transition. Formation of a competent elongation complex leads to processive RNA synthesis and the generation of a full-length product. In the case of HIV reverse transcriptase, conformational changes have also been demonstrated in thumb subdomains upon binding of a nucleotide substrate (31). Although a detailed analysis of RNA initiation and elongation in HCV has not been fully completed, it is tempting to postulate that HCV RdRp might also undergo a transformation during the initiation steps preceding productive polymerization. Abortive dinucleotide formation was observed during the de novo RNA synthesis in HCV (unpublished results). The blockade of RNA synthesis at the fifth nucleotide in the presence of the pyranoindole inhibitor (Fig. 8) correlates with the observation showing that oligonucleotide primers longer than 5 nucleotides are detrimental to de novo initiation (39, 50), suggesting that a structural conformation might be required in order to proceed with RNA synthesis. Interestingly, the first clash of the RNA-DNA hybrid with the core domain of the T7 RNAP is reported to occur at about the fourth to the fifth nucleotides (21, 32). A comparison of two distinct apo crystal forms (3) of genotype 2a HCV NS5B revealed a rigid-body rotation of the thumb domain, as well as movements of the unique β-hairpin loop and the Δ1 loop extending from the fingers domain to the thumb domain. More significantly, the spatial rearrangement of these structural motifs has been linked to the presence of an inhibitor bound to the pyranoindole-binding site. Taken together, these results suggest that a structural transformation occurs after the formation of an initiation complex, i.e., after the bound RNA exceeds 5 nucleotides; and it is likely that this step is inhibited by the thumb domain pyranoindole NNRPI.
Acknowledgments
We thank Anne Deatly and Janis Upeslacis for their critical review and helpful discussion of the manuscript. Special thanks are extended to Xiaoqun Jiang, Steve Swanberg, and the Wyeth Core Sequencing Facility for their technical assistance. We are also grateful to Susan Nastasee and Frances Del Vecchio for editorial support with manuscript preparation.
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Brechot, C. 1996. Hepatitis C virus: molecular biology and genetic variability. Dig. Dis. Sci. 41:6S-21S. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabot, B., J. I. Esteban, M. Martell, J. Genesca, V. Vargas, R. Esteban, J. Guardia, and J. Gomez. 1997. Structure of replicating hepatitis C virus (HCV) quasispecies in the liver may not be reflected by analysis of circulating HCV virions. J. Virol. 71:1732-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabot, B., M. Martell, J. I. Esteban, S. Sauleda, T. Otero, R. Esteban, J. Guardia, and J. Gomez. 2000. Nucleotide and amino acid complexity of hepatitis C virus quasispecies in serum and liver. J. Virol. 74:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham, G. M., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117-123. [DOI] [PubMed] [Google Scholar]
- 11.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 12.Colgrove, R., and A. Japour. 1999. A combinatorial ledge: reverse transcriptase fidelity, total body viral burden, and the implications of multiple-drug HIV therapy for the evolution of antiviral resistance. Antivir. Res. 41:45-56. [DOI] [PubMed] [Google Scholar]
- 13.Davis, G. L. 1999. Hepatitis C virus genotypes and quasispecies. Am. J. Med. 107:21S-26S. [DOI] [PubMed] [Google Scholar]
- 14.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 15.Drake, J. W. 1999. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann. N. Y. Acad. Sci. 870:100-107. [DOI] [PubMed] [Google Scholar]
- 16.Farci, P., and R. H. Purcell. 2000. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 20:103-126. [PubMed] [Google Scholar]
- 17.Gopalsamy, A., K. Lim, G. Ciszewski, K. Park, J. W. Ellingboe, J. Bloom, S. Insaf, J. Upeslacis, T. S. Mansour, G. Krishnamurthy, M. Damarla, Y. Pyatski, D. Ho, A. Y. Howe, M. Orlowski, B. Feld, and J. O'Connell. 2004. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 47:6603-6608. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, C. P., and P. A. Keller. 2005. Control of hepatitis C: a medicinal chemistry perspective. J. Med. Chem. 48:1-20. [DOI] [PubMed] [Google Scholar]
- 19.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 20.Howe, A. Y., J. Bloom, C. J. Baldick, C. A. Benetatos, H. Cheng, J. S. Christensen, S. K. Chunduru, G. A. Coburn, B. Feld, A. Gopalsamy, W. P. Gorczyca, S. Herrmann, S. Johann, X. Jiang, M. L. Kimberland, G. Krisnamurthy, M. Olson, M. Orlowski, S. Swanberg, I. Thompson, M. Thorn, A. Del Vecchio, D. C. Young, M. van Zeijl, J. W. Ellingboe, J. Upeslacis, M. Collett, T. S. Mansour, and J. F. O'Connell. 2004. Novel nonnucleoside inhibitor of hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 48:4813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imburgio, D., M. Rong, K. Ma, and W. T. McAllister. 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry (Moscow) 39:10419-10430. [DOI] [PubMed] [Google Scholar]
- 22.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzmine, I., and C. T. Martin. 2001. Pre-steady-state kinetics of initiation of transcription by T7 RNA polymerase: a new kinetic model. J. Mol. Biol. 305:559-566. [DOI] [PubMed] [Google Scholar]
- 24.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 28.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. (Erratum, 77:10176.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 31.Morris, M. C., C. Berducou, J. Mery, F. Heitz, and G. Divita. 1999. The thumb domain of the P51-subunit is essential for activation of HIV reverse transcriptase. Biochemistry (Moscow) 38:15097-15103. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee, S., L. G. Brieba, and R. Sousa. 2003. Discontinuous movement and conformational change during pausing and termination by T7 RNA polymerase. EMBO J. 22:6483-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel, P. H., and B. D. Preston. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. USA 91:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 38.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 39.Ranjith-Kumar, C. T., L. Gutshall, M. J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76:12526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shim, J., G. Larson, V. Lai, S. Naim, and J. Z. Wu. 2003. Canonical 3′-deoxyribonucleotides as a chain terminator for HCV NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:243-251. [DOI] [PubMed] [Google Scholar]
- 41.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Summa, V., A. Petrocchi, P. Pace, V. G. Matassa, R. De Francesco, S. Altamura, L. Tomei, U. Koch, and P. Neuner. 2004. Discovery of alpha,gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 47:14-17. [DOI] [PubMed] [Google Scholar]
- 43.Tahirov, T. H., D. Temiakov, M. Anikin, V. Patlan, W. T. McAllister, D. G. Vassylyev, and S. Yokoyama. 2002. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature 420:43-50. [DOI] [PubMed] [Google Scholar]
- 44.Temiakov, D., P. E. Mentesana, K. Ma, A. Mustaev, S. Borukhov, and W. T. McAllister. 2000. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 97:14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, M. P., N. Yao, and Z. Hong. 2003. Promising candidates for the treatment of chronic hepatitis C. Expert Opin. Investig. Drugs 12:1269-1280. [DOI] [PubMed] [Google Scholar]
- 48.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]