Abstract
Human immunodeficiency virus type 1 (HIV-1) resistance development was evaluated in vitro by using combinations of the drugs tenofovir and emtricitabine or abacavir and lamivudine, as well as by using the compounds individually. Emtricitabine- and lamivudine-resistant HIV-1 isolates with the M184I or M184V mutation in reverse transcriptase were readily selected in the cultures with emtricitabine alone, lamivudine alone, and the two drug combinations and conferred high-level resistance to emtricitabine and lamivudine. Tenofovir-resistant HIV-1 isolates with the K65R mutation occurred in both the culture with tenofovir alone and the culture with the combination of emtricitabine and tenofovir. The S68N and S68K mutations were also observed in the tenofovir cultures, with no detectable impact on resistance, suggesting a possible compensatory role in viral fitness. At low concentrations of emtricitabine and tenofovir, the M184I mutation appeared first, followed by the K65R mutation, in a subset of viruses. At intermediate concentrations of emtricitabine and tenofovir, viruses harboring the K65R mutation or a novel K65N and K70R double mutation grew before they gave rise to mutants with K65R and M184V/I double mutations at higher emtricitabine concentrations. Abacavir resistance was characterized by the accumulation of the M184V, Y115F, and K65R mutations in the abacavir culture, while the M184V and L74V mutations were selected in combination with lamivudine. In the presence of the abacavir resistance mutations, viral growth was strong even in the presence of high concentrations of abacavir. In contrast, viral growth was markedly impaired in the cultures with high tenofovir concentrations, even in the presence of K65R. In conclusion, these studies show that HIV-1 mutants with a K65R and M184V genotype are generated under maximum selection pressure from the combination of tenofovir and emtricitabine.
As a result of antiretroviral drug therapy, human immunodeficiency virus type 1 (HIV-1) may develop resistance mutations which render the virus less susceptible to antiretroviral drugs. In vitro resistance selection experiments can be used to predict which genetic form of resistance may occur in vivo. For tenofovir (TFV), previous resistance selection experiments have shown the development of the K65R mutation in HIV-1 reverse transcriptase (RT), which resulted in an approximately threefold reduced susceptibility to TFV after extended passage in vitro (38). Isolates with the K65R mutation show reduced susceptibility to some nucleoside RT inhibitors (NRTIs), including an 8-fold reduced susceptibility to emtricitabine (FTC), a 12-fold reduced susceptibility to lamivudine (3TC), and a nearly 4-fold reduced susceptibility to abacavir (ABC), but maintain full susceptibility to zidovudine (ZDV) (6, 43). Development of the K65R mutation in vivo has been observed on an infrequent basis (in <3% of treated patients) after 8 to 80 weeks of combination therapy with tenofovir disoproxil fumarate (TDF), the Food and Drug Administration-approved prodrug formulation of TFV, and other agents (8). Development of the K65R mutation was not observed during a 28-day dose-escalation monotherapy study of TDF (1). No other patterns of resistance have been observed as a result of TDF therapy.
For FTC, previous in vitro resistance selection experiments have shown the development of the M184I or the M184V mutation in HIV-1 RT, which resulted in high-level resistance (>300-fold) to FTC as well as to 3TC (27, 32). HIV-1 isolates with the M184V/I mutation in RT remained fully susceptible to TFV, ZDV, and stavudine (d4T) but showed some cross-resistance to ABC and didanosine (ddI) (18, 21). In vivo development of the M184V/I mutation as a result of virologic failure of a combination regimen containing FTC has been observed on an infrequent basis (25). No other patterns of development of resistance to FTC or 3TC have been reported clinically.
Resistance to ABC has been studied both in vitro and in vivo; and investigators have shown that various combinations of the K65R, L74V, Y115F, and M184V mutations in RT were primarily responsible for resistance to ABC (29, 31, 39).
Fixed-dose combination pills with FTC and TDF and with ABC and 3TC were recently approved by the Food and Drug Administration under the brand names Truvada and Epzicom, respectively. The resistance profiles of these fixed-dose combination pills were analyzed in the present study by using in vitro resistance selection experiments. Implications for the treatment of HIV-1-infected patients are discussed.
MATERIALS AND METHODS
Reagents.
MT-2 cells were obtained through the NIH AIDS Research and Reference Reagent Program. Sup-T1 cells, used for the production of site-directed recombinant viruses, were purchased from ATCC (catalog no. CRL-1942). The first series of drug selection experiments was conducted with commercially available wild-type HIV-1 strain IIIb (Applied Biosystems, Foster City, CA), which was used to infect MT-2 cells at a multiplicity of infection of approximately 0.01. In a second series of experiments, wild-type HIV-1 strain LAI was used under the same conditions of infection as described above. Wild-type HIV-1 LAI was generated at Gilead Sciences by lipid-based transfection of the proviral DNA clone pLAI-2 into MT-2 cells. In both series of experiments, 2 × 106 MT-2 cells were infected and plated in six-well plates in a 5-ml volume for each selection. TFV and FTC were synthesized by Gilead Sciences. 3TC was obtained from Moravek Biochemicals (Brea, CA), and ABC was obtained from GlaxoSmithKline (Research Triangle Park, NC).
Selection conditions.
The first series of experiments was conducted with HIV-1 IIIb by using TFV and FTC either singly or in combination. These experiments were repeated with HIV-1 LAI in the second series of experiments, and the panel of drugs was extended to include 3TC and ABC either singly or in combination. The single-drug experiments were started at the effective concentration required to inhibit viral replication by 50% (EC50) of each drug (3.5 μM, 0.8 μM, 4 μM, and 0.4 μM for TFV, FTC, 3TC, and ABC, respectively). A total of four dual-combination drug selection experiments were initiated. Three of them were conducted with a combination of FTC and TFV (namely, cultures FT1, FT2, and FT3), and the other was conducted with a combination of 3TC and ABC (culture 3A). FT1 and FT2 were duplicate experiments set up with HIV-1 IIIb and HIV-1 LAI, respectively, while FT3 and 3A were set up with HIV-1 LAI. The selecting drugs were started at one-half the EC50 (1.75 μM and 0.4 μM for TFV and FTC, respectively; 2 μM and 0.2 μM for 3TC and ABC, respectively) for FT1, FT2, and 3A in order to provide an inhibitory power similar to that in the single-drug experiments. For FT3, the starting drug concentrations were based on the maximum concentrations of the drugs in serum (Cmax) (1 μM and 7.2 μM for TFV and FTC, respectively) (10, 11) and were set at one-fourth the Cmax (0.25 μM and 1.8 μM for TFV and FTC, respectively). A no-drug control was used for each experiment to ensure virus growth in the absence of compound. Syncytium formation was checked periodically to monitor virus growth. Viral growth was characterized as strong when extensive syncytia were present in the culture after 3 to 4 days (over 90% of cells with syncytia), and it was qualified as weak when the cultures showed little or no evidence of a cytopathic effect after 7 days or more in culture. When a full or a nearly full cytopathic effect was observed in the wells containing a drug(s), the cells in the cultures were centrifuged and the supernatants were collected and frozen at −80°C for later genotypic and/or phenotypic analysis. Subsequent passages were set up by using a fraction of the harvested virus (500 μl of 5 ml) to infect 2 × 106 fresh MT-2 cells. The amount of drug used in subsequent passages of virus was increased 2-fold in most cases and was occasionally increased 1.5-fold when virus growth was suboptimal at a high drug concentration. No-drug controls were set up at each new passage, and in addition, control cultures in which the harvested virus was passaged with no increase in drug levels were also set up to ensure that the virus was growing reproducibly and continued to result in a cytopathic effect. In the absence of a cytopathic effect, cells were split 1:3 or 1:4 every 5 to 8 days and fresh drug and culture medium were replenished.
Genotypic analyses.
HIV-1 RNA was extracted from virus supernatants (viral RNA preparation kit; QIAGEN, Valencia, CA), and the PCR products obtained after reverse transcription (Titan RT-PCR; Roche Diagnostics, Indianapolis, IN) were analyzed by population-based sequencing with an ABI Prism BigDye Terminator v3.1 cycle sequencing kit and a 3100 ABI Prism genetic analyzer (Applied Biosystems, Foster City, CA). The RT sequence corresponding to amino acids 1 to 300 was assessed. The detection limit for characterization of minor-sequence species by this assay is approximately 25%. Sequences were compared to the sequence of the strain IIIb or strain LAI starting virus inoculum. In the earlier part of the experiments, genotypic analyses were conducted at each new virus harvest until genotypic resistance was detected. In the remainder of the experiments, genotypic analyses were done every 4 to 6 weeks and when the drug concentrations were increased. In addition, the final virus harvests were analyzed genotypically.
Single-genome sequencing (SGS).
On the basis of Poisson's distribution, it is estimated that a DNA template that gives rise to three positive PCR products in 10 reactions (∼30% positive) contains 1 copy of starting DNA template per reaction about 80% of the time (20). HIV-1 RNA was extracted from the virus supernatants (viral RNA preparation kit; QIAGEN), reverse transcribed into cDNA (Superscript II; Invitrogen, Carlsbad, CA), and amplified by PCR (rTth XL DNA polymerase; Applied Biosystems). In order to determine the optimal amount of starting material to be used, the cDNA was serially diluted and amplified in two separate rounds of PCR to determine the dilution that would yield an amplification approximately 30% positive. The resulting cDNA dilution was amplified in a 96-well plate in two separate rounds of PCR, and the products were analyzed on a 1% agarose gel. When no more than 30% of the PCRs were positive, the PCR products were sequenced as described above.
Phenotypic analyses and recombinant virus generation by site-directed mutagenesis.
The susceptibilities of the viruses of interest to TFV, FTC, ZDV, 3TC, d4T, ABC, and ddI were determined in MT-2 cells by using a 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay, as described earlier (13, 41). Briefly, 1.2 × 106 MT-2 cells were infected with either wild-type HIV-1 (HXB2D) or selected mutant virus and incubated in the assay plates for 5 days starting at an initial concentration of approximately 17,000 cells/well. The antiviral effects of the compounds tested were measured by determining the HIV-1 cytopathic effect by using the vital dye XTT (Sigma-Aldrich, St. Louis, MO). The EC50s were determined by using SigmaPlot (SPSS, Chicago, IL). Phenotypic resistance that measured between a 1.5- and a 3-fold increase in the EC50 above that for the wild-type control (HXB2D) was defined as low-level resistance. A greater than eightfold increase in the EC50 above that for the wild-type control was defined as high-level resistance. Recombinant viruses were generated by PCR site-directed mutagenesis by cotransfection of the HIV-1 proviral molecular clone pHXB2Δ2-261RT (plasmid DNA provided by C. Boucher, Utrecht Medical Center, Utrecht, The Netherlands) with a PCR product corresponding to the first 300 amino acids of HIV-1 RT on which the mutation(s) of interest (K65R, Y115F, M184V, or H221Y, alone or in combination) had been inserted. The DNA molecules were cotransfected in Sup-T1 cells, where replication-competent viruses were generated by homologous recombination and harvested after 10 to 20 days (2, 42). The nucleotide sequence corresponding to the first 300 amino acids of RT for the viruses obtained was confirmed by genotypic analysis with an ABI Prism 3100 genetic analyzer, as described above.
RESULTS
Single-drug resistance selections.
The starting drug concentrations for the four compounds tested in single-drug resistance selection experiments were their respective EC50s (3.5 μM, 0.8 μM, 4 μM, and 0.4 μM for TFV, FTC, 3TC, and ABC, respectively). The study was not intended to be a strict measure of time to resistance, as the detection of resistance could be delayed by the 3 to 4 days separating the inspection and/or harvesting of the cultures. The development of resistance to FTC and 3TC followed similar patterns (Table 1). For both drugs, resistance appeared early in the form of a mixture of the M184I mutation and the wild-type sequence at codon 184 with low drug concentrations, ranging from the EC50 to fourfold the EC50. For both FTC and 3TC, subsequent stepwise increases in the drug concentration could readily be achieved after the detection of initial genotypic resistance and led to highly resistant viruses harboring a full M184V or M184I mutation or a mixture of both resistance mutations. The resistant viruses showed strong viral growth at up to 288- and 128-fold the EC50 for FTC and 3TC, respectively, corresponding to >30-fold the Cmax.
TABLE 1.
Selection conditions and genotypic analyses
| Selecting drug(s) | Culture name | Starting conditions
|
First detection of genotypic resistance
|
Ending conditions
|
Comment | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Concn (μM [fold over EC50])a,b | Starting virus | Genotypec | Time (days) | Concn (μM [fold over EC50])a | Genotypec | Duration (days) | Concn (μM [fold over EC50])a | Genotypec | |||
| FTC | F1 | 0.8 (1) | IIIb | wt | 14 | 0.8 (1) | M184I/M/V | 48 | 230 (288) | M184V/I | Strong viral growth |
| FTC | F2 | 0.8 (1) | LAI | wt | 18 | 3.2 (4) | M184I/M | 58 | 230 (288) | M184V | Strong viral growth |
| 3TC | 3 | 4 (1) | LAI | wt | 11 | 4 (1) | M184I/M | 58 | 512 (128) | M184I | Strong viral growth |
| TFV | T1 | 3.5 (1) | IIIb | wt | 56 | 56 (16) | K65R | 99 | 112 (32) | K65R | Weak viral growth |
| TFV | T2 | 3.5 (1) | LAI | wt | 44 | 28 (8) | K65R/K | 328 | 168 (48) | K65R S68K H221Y | Weak viral growth |
| ABC | A | 0.4 (1) | LAI | wt | 44 | 6.4 (16) | M184I | 203 | 51.2 (128) | K65R Y115F M184V H221Y | Strong viral growth |
| FTC-TFV | FT1 | 0.4 (1/2), 1.75 (1/2) | IIIb | wt | 24 | 1.6 (2), 7 (2) | M184I/M | 133 | 12.8 (16), 56 (16) | K65R | Weak viral growth |
| FTC-TFV | FT2 | 0.4 (1/2), 1.75 (1/2) | LAI | wt | 40 | 1.6 (2), 7 (2) | M184I/M | 288 | 25.6 (32), 112 (32) | K65N K70Rg | Transition from intermediate K65R to K65N+K70R genotype (∼day 130) |
| FTC-TFV | FT3d | 1.8 (2.3), 0.25 (0.07) | LAI | wt | 18 | 1.8 (2.3), 0.25 (0.07) | M184I | 71 | 230 (288), 32 (9) | K65R M184V | Starting concentrations based on Cmax for each drug |
| FTC-TFV | FT1ae | 51.2 (64), 7 (2) | IIIb | M184I L214F | NAf | 42 | 51.2 (64), 28 (8) | K65R M184I L214F | [FTC] kept constant | ||
| FTC-TFV | FT1be | 25.6 (32), 28 (8) | IIIb | K65R | NA | 54 | 230 (288), 28 (8) | K65R M184V | [TFV] kept constant | ||
| 3TC-ABC | 3A | 2 (1/2), 0.2 (1/2) | LAI | wt | 18 | 2 (1/2), 0.2 (1/2) | M184I/M | 329 | 512 (128), 51.2 (128) | L74V Q161L M184V T200A H221Y | Strong viral growth |
Concentrations are shown in the same order as the selecting drugs. Expressed in μM.
Selections from wild-type virus were started at EC50 for single selections and at one-half the EC50 for each drug in dual selections, with the exception of the FT3 culture.
Genotypic analyses of the selected viruses captured the first 300 amino acids of RT. wt, wild type.
In the FT3 culture, starting concentrations were one-fourth the Cmax of each drug: FTC Cmax = 7.2 μM; TFV Cmax = 1 μM.
Both FT1a and FT1b were started from viral harvests obtained from culture FT1 that had genotypic resistance.
NA, not applicable.
Further selection at higher FTC concentration (32-fold EC50 for TFV and 128-fold EC50 for FTC) gave rise to a virus population with A62V + K65R + M184V.
Genotypic resistance to TFV and ABC was detected only at higher drug concentrations and after a longer time in culture than the concentrations and times for FTC and 3TC (Table 1). For TFV, genotypic resistance was detected at codon 65 (K65R) after 6 to 8 weeks in culture at a TFV concentration of 8 to 16 times the EC50. At completion, one selection experiment (experiment T1) generated a virus with the single K65R mutation which grew weakly at 32 times the EC50 over 14 weeks in culture. The other selection experiment (experiment T2) yielded a more complex virus with the K65R, S68K, and H221Y mutations after 46 weeks in culture in up to 48 times the EC50. In contrast to the highly resistant M184V- or M184I-containing virus obtained at the end of the FTC and 3TC cultures, the K65R-containing mutants selected in the presence of TFV grew weakly as the TFV concentration was increased stepwise. After 46 weeks in culture, viral growth was severely restricted, as evidenced by a lack of a cytopathic effect at TFV concentrations above 48-fold the EC50, whereas a strong cytopathic effect was observed in the no-drug controls that were cultured in parallel. Cell growth was unaffected at these TFV concentrations.
Genotypic resistance to ABC was detected after 6 weeks in culture, where viral growth had proceeded with an ABC concentration up to 16 times the EC50 (Table 1). Genotypic resistance to ABC was characterized by the stepwise accumulation of resistance mutations starting with M184I at week 6 (16-fold EC50), followed by addition of Y115F and the transition to M184V at week 16 (64-fold the EC50) and ending with the addition of K65R at week 23 (128-fold the EC50). An H221Y mutation also came up in the ABC selection at week 19 (128-fold the EC50). The final virus population contained the K65R, Y115F, M184V, and H221Y mutations and showed strong growth in the presence of ABC at 128-fold the EC50 (51.2 μM).
Dual drug resistance selections.
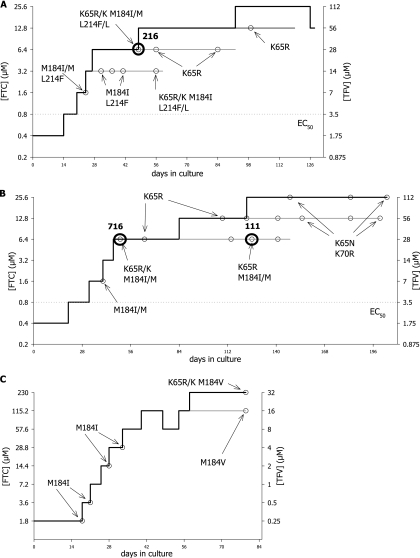
The starting drug concentrations used in the dual drug resistance selection experiments were one-half the EC50 of each drug (1.75 μM, 0.4 μM, 2 μM, and 0.2 μM for TFV, FTC, 3TC, and ABC, respectively) in all cultures except the FT3 culture, for which TFV and FTC were started at one-fourth the Cmax (Table 1). The first series of experiments with TFV and FTC were conducted with HIV IIIb (Table 1, cultures FT1, FT1a, and FT1b). The growth curve for the FT1 culture is shown in Fig. 1A. Genotypic resistance to FTC came up first as an M184I/M mixture after 24 days in culture, with the drug concentrations corresponding to twofold the EC50s of both drugs. An L214F polymorphism was also detected along with M184I/M. As the culture progressed, the M184I/M mixture became a K65R/K plus M184I/M double-mutation mixture. A full M184I mutant was also observed in a separate culture before the detection of a K65R/K plus M184I/M double-mutation mixture (data not shown). After 7 weeks in culture at 16-fold the EC50 of each drug, the K65R mutation persisted while the M184I mutation became undetectable by population sequencing (Fig. 1A). These findings suggest that at these drug concentrations the K65R mutation exhibited enough resistance to FTC and TFV to become the principal mutation for resistance to the two drugs. By switching from M184I to K65R, the virus became less resistant to FTC (∼10-fold resistance to FTC with K65R compared to the >200-fold resistance to FTC with M184I) but gained ∼3-fold resistance to TFV (Table 2, viruses J and 9). In addition, in vitro phenotypic data suggest that the presence of M184V or M184I increases susceptibility to TFV in the presence or absence of K65R (Table 2, TFV fold change in EC50 for virus K compared to those for virus J, virus 9, and virus 10). Altogether, a switch from M184I to K65R allowed the virus to survive better under the selective pressures from both drugs at these concentrations in vitro.
FIG. 1.
Resistance selection growth curves for cultures selected with FTC and TFV. The concentrations of both drugs (μM) are plotted against the time in culture (days). Bold lines, lead cultures where drug concentrations were increased whenever possible; thin lines, parallel cultures in which the drug concentrations were maintained at intermediate concentrations; dotted lines, EC50 for each drug; small open circles, virus harvests and the corresponding genotypes; bold open circles and numbers, viral harvests that were analyzed by single-genome sequencing. (A) Culture FT1; (B) culture FT2; (C) culture FT3.
TABLE 2.
Phenotypic analyses of selected viruses and mutant viruses generated by site-directed mutagenesis
| Virus name | Culturea | Selecting drug(s) | Genotype | Avg fold change in EC50 from that of wild-type controlb
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TFV | FTC | ZDV | 3TC | d4T | ABC | ddI | ||||
| 1 | T1 | TFV | K65R | 1.9** | 5.9** | 1.1 | 8.6** | 1.2 | 3.1** | 3.6** |
| 2 | T2 | TFV | K65R S68N | 3* | 4.8 | 1.6 | ≫ | 1.2 | ≫ | 1.8 |
| 3 | T2 | TFV | K65R S68K H221Y | 4.1** | 7** | 1.2 | ≫ | 2.1** | 2.1 | 2.1 |
| 4 | FT1 | FTC-TFV | M184I L214F | 0.7 | 38.2** | 0.6 | ≫ | 0.6 | 1.9* | 2.7** |
| 5 | FT1 | FTC-TFV | K65R | 2.9** | 8.4** | 0.9 | 11.9** | 1 | 2.8** | 3.7** |
| 6 | FT1 | FTC-TFV | K65R M184I L214F | 1.6* | ≫ | 0.5 | ≫ | 0.7 | ≫ | 4.7** |
| 7 | FT2 | FTC-TFV | K65N K70R | 3.6** | 39* | 0.3* | 17* | 1 | 2.1 | 1.2 |
| 8 | FT2 | FTC-TFV | A62V K65R M184V | 2.6* | ≫ | 0.7 | ≫ | 1.5 | ≫ | 3 |
| 9 | F2 | FTC | M184I | 0.5** | ≫ | 0.5 | ≫ | 0.6 | 2.3** | 2.8** |
| 10 | F2 | FTC | M184V | 0.5** | ≫ | 0.6 | ≫ | 0.8 | 3.5** | 3.1** |
| 11 | A | ABC | Y115F M184V H221Y | 1.8 | ≫ | 0.8 | ≫ | 0.8 | ≫ | 2.2* |
| 12 | A | ABC | K65R Y115F M184V H221Y | 2.6* | ≫ | 0.8 | ≫ | 1.1 | ≫ | 3* |
| 13 | 3A | 3TC-ABC | L74V Q161L M184V T200A H221Y | 0.8 | ≫ | 0.3 | ≫ | 0.8 | ≫ | 2.6 |
| J | SDR | K65R | 3.3** | 11** | 1.3 | 10** | 2.2** | 2.9** | 3** | |
| K | SDR | K65R M184V | 2.1** | ≫ | 0.4 | ≫ | 1.4* | ≫ | 7.5** | |
| L | SDR | K65R H221Y | 3.6* | 13.6* | 1.3 | 11.2 | 2.3 | 5* | 3.3* | |
| M | SDR | K65R Y115F M184V | 4.5** | ≫ | 1.4 | ≫ | 1.3 | ≫ | 5.6** | |
| N | SDR | K65R Y115F | 5.9** | 25.5** | 1.4 | ≫ | 1.8* | ≫ | 2.4** | |
Culture details are presented in Table 1. SDR, recombinant viruses generated by site-directed mutagenesis, as described in Materials and Methods.
HXB2D wild-type control EC50s were 3.6, 0.77, 0.14, 3.8, 6.7, 0.43, and 4.4 μM for TFV, FTC, ZDV, 3TC, d4T, ABC, and ddI, respectively. Statistical significance is indicated as follows: **, P < 0.01 compared to the value for the wild-type control; *, P < 0.05 compared to the value for the wild-type control. ≫, at least one value was >43-fold, >210-fold, or >8-fold above the value for the wild type for 3TC, FTC, and ABC, respectively. Data are averages from at least three independent experiments.
Attempts to increase the drug concentrations to 32-fold the EC50s of TFV and FTC were unsuccessful, as viral growth was completely inhibited at these concentrations, while viral growth was strong in the no-drug control culture. In order to promote the selection process further and to elucidate the interplay between the K65R and the M184I/V mutations as the TFV and FTC drug levels increased, the drug concentrations were independently modified so that viral replication could occur. To test that approach, culture FT1a was set up starting from an M184I virus at an FTC concentration kept constant during the whole experiment at 64-fold above the EC50 and then increasing the concentration of TFV stepwise starting from 2-fold the EC50. Conversely, culture FT1b was set up starting from a K65R virus at a TFV concentration kept constant during the whole experiment at 8-fold the EC50 and then increasing the FTC concentration stepwise starting at 32-fold the EC50. Under these conditions, both cultures gave rise to K65R plus M184V/I double mutations, as evidenced by the absence of the wild-type peaks at positions 65 and 184 on the electropherogram after population sequencing. Culture FT1a went from an M184I virus to a K65R-M184I virus, while culture FT1b evolved from a K65R virus to a virus with a K65R plus M184V double mutation under selective pressure from both TFV and FTC.
The results for culture FT1a were confirmed in culture FT3, where the starting drug concentrations were based on the Cmaxs of the drugs found in plasma. One-quarter of the Cmaxs (corresponding to 1.8 μM FTC and 0.25 μM TFV) were chosen in order to provide strong selective pressure while allowing viral replication to occur. Lower drug levels would not provide adequate selective pressure in this experiment. By using the Cmaxs, the starting FTC concentration was higher than the TFV concentration relative to the EC50s, and the M184I mutation arose early in the experiment before giving rise to a K65R/K plus M184V double mutation at the end of the experiment (Fig. 1C). Thus, the earlier observation that dual selection with TFV and FTC leads to a resistant mutant carrying the single K65R mutation was most likely a consequence of the specific drug concentrations used in those experiments. With higher FTC concentrations that better mimic the situation in vivo, dual selection with TFV and FTC gave rise first to virus with M184V/I and then to a resistant virus harboring both the K65R and the M184V/I mutations.
In a second series of drug selection experiments with TFV and FTC with HIV-1 LAI as the starting virus (Table 1, culture FT2), results similar to those obtained with HIVIIIb were obtained initially—the early development of M184I/M, followed by the development of K65R/K plus M184I/M double mutations and then K65R alone—but a new mutant was observed in the later part of the experiment (Fig. 1B). At drug concentrations corresponding to 16-fold the EC50s, the K65R mutation was replaced by a K65N and K70R double mutation, and mutants with this double mutation were able to grow after the subsequent increase of the concentration of drugs to 32-fold their EC50s. Further drug increases to 64-fold the EC50s were not successful, as viral growth was fully inhibited at these concentrations for both cultures.
Conditions under which the FTC concentration was increased independently of the TFV concentration were also tested. In that experiment, FTC concentrations corresponding to 32-, 64-, and 128-fold the EC50 were used and the TFV concentration varied from 16- to 32-fold the EC50. The starting virus harboring the three mutations A62V, K65R, and M184I evolved into a virus harboring A62V, K65N, and K70R when it was grown in the presence of 32-fold the EC50 of FTC, while it evolved into a virus harboring A62V, K65R, and M184V when it was grown in the presence of 64- and 128-fold the EC50 of FTC (data not shown). Therefore, the emergence of the K65N-K70R mutation resistance pathway at an intermediate FTC concentration appears to represent a temporary step in the development of resistance to the FTC-TFV combination, where viruses carrying both K65R and M184V are the most likely outcomes in these in vitro models.
Resistance selection with 3TC and ABC led to a mutant virus containing an M184I/M mixture after 18 days in culture at the lowest concentrations of the compounds used (2 μM and 0.2 μM for 3TC and ABC, respectively). As the experiment progressed, the M184I/M mixture became a full mutant and the virus acquired the H221Y mutation, followed by the L74V mutation. After more than 46 weeks in culture, the final viral population had the L74V, Q161L, M184V, T200A, and H221Y mutations and showed strong growth in the presence of 3TC and ABC concentrations corresponding to 128-fold the EC50s (512 μM and 51.2 μM for 3TC and ABC, respectively).
Phenotypic analyses.
In vitro drug susceptibility data for TFV and the most commonly used NRTIs are shown in Table 2 for a group of in vitro-selected viruses and a panel of mutants generated by site-directed mutagenesis. Three viruses selected in the presence of TFV harboring the K65R mutation were analyzed phenotypically and showed 1.9- to 4.1-fold increases in the EC50 of TFV compared to that of the wild-type control, while they showed various levels of cross-resistance to all other compounds except ZDV. Virus 1 was from an early passage of culture T1 and may have included some wild-type virus in the viral supernatant that could have been responsible for the low fold change in the EC50 observed for that virus (1.9-fold) compared to that for the reference recombinant virus with K65R generated by site-directed mutagenesis (virus J; 3.3-fold). The presence of S68K and H221Y in virus 3 appeared to increase the EC50 of TFV to 4.1-fold above that for the wild-type control, showing a trend similar to that observed for the mutant virus containing both K65R and H221Y, as generated by site-directed mutagenesis (virus L; 3.6-fold), while the presence of S68N in virus 2 did not appear to affect susceptibility to TFV compared to that with the presence of K65R alone (virus J).
Phenotypic analyses of viruses obtained under selection pressure from both TFV and FTC have confirmed the progression in the levels of resistance to the compounds. The early M184I virus (Table 2, virus 4) showed high-level resistance to FTC (>8-fold) and full susceptibility to TFV, followed by the K65R virus (Table 2, virus 5), which showed a 2.9-fold reduced susceptibility to TFV and a lower level of resistance to FTC (8.4-fold compared to >210-fold for viruses with the M184V and M184I mutations), and ending with the virus with the K65R and M184I mutations (Table 2, virus 6), which showed high-level resistance to FTC and lower-level resistance to TFV (1.6-fold). For both ABC and ddI, greater levels of cross-resistance were observed for viruses with both mutations together than for viruses with the mutations in isolation (>8-fold and 4.7-fold, respectively) (Table 2, virus 6). The novel mutant with the K65N plus K70R mutations (virus 7) showed notably increased resistance to FTC and 3TC compared to that for the K65R control (virus J), where resistance levels were 39- and 17-fold above those for the wild-type control for FTC and 3TC, respectively. In contrast to virus 7 (obtained at 32-fold EC50 for both FTC and TFV), which showed high but measurable FTC resistance, the virus from culture FT2 obtained at higher FTC concentration (128-fold EC50 for FTC and 32-fold EC50 for TFV) (Table 2, virus 8) showed complete resistance to FTC (>210-fold), emphasizing the primary role of M184V in high-level resistance to FTC, as opposed to the intermediary role of K65N-K70R (39-fold).
Resistance selection experiments conducted with FTC led to M184V/I mutants (viruses 9 and 10) that exhibited high-level resistance to FTC and 3TC and elevated but measurable resistance to ABC and ddI. The mutants also showed ∼0.5-fold hypersusceptibility to TFV, ZDV, and d4T.
Two viruses selected in the presence of ABC alone as well as one virus selected in the presence of both ABC and 3TC were tested phenotypically (viruses 11, 12, and 13). In all three cases, high-level resistance to FTC, 3TC, and ABC were observed due to the presence of M184V for FTC and 3TC and due to the presence of M184V plus either Y115F, K65R-Y115F, or L74V for ABC. For TFV, low-level resistance was observed in the presence of Y115F, M184V, and H221Y (1.8-fold above that for the wild-type control; virus 11), and the level of resistance was increased with the addition of K65R (2.6-fold above that for the wild-type control; virus 12). Of note, the mutant carrying K65R, Y115F, and M184V, as generated by site-directed mutagenesis (virus M), showed more pronounced resistance to TFV and ddI than ABC-selected virus 12. No phenotypic resistance to TFV was detected in the 3TC- and ABC-selected virus harboring L74V and M184V (virus 13).
Single-genome sequence analyses.
The observed replacement of the mutant with the M184I mutation by the mutant with the K65R mutation under selection pressure from TFV and FTC at critical drug concentrations between 8- and 16-fold their EC50s in the FT1 and FT2 series of cultures was of interest. As shown in Fig. 1A and B, a virus quasispecies harboring the M184I mutation was detected first. However, subsequently, complex mixtures of M184I, K65R, and wild-type HIV-1 viruses were observed by population sequencing. To determine whether K65R grew independently or with M184I, SGS experiments were conducted. Two of the viruses tested in the SGS analyses had mixtures of mutant and wild-type sequences at both positions, as determined by population sequencing. The viruses tested are circled in Fig. 1 (Fig. 1A, virus 216; Fig. 1B, viruses 716 and 111). The results for these analyses are shown in Table 3. The results for virus 216 showed the most evidence of the independence of the viral genomes, with 50% and nearly 40% of the clones exhibiting either the K65R or the M184I mutation, respectively. The nucleotide sequences of the 26 clones of virus 216 analyzed by SGS showed various clusters of association of the mutations of interest with a limited number of silent or coding nucleotide changes (data not shown). Analysis of these associations strongly suggested that one of the three clones with double mutations could have been generated by the recombination of mutants with single mutations. There was evidence that the second clone with a double mutation emerged after the addition of K65R to an M184I virus, while the third clone with a double mutation emerged after the addition of M184I to a K65R virus. In the last two cases, the mechanism by which the double mutants were generated—either de novo mutation or recombination—could not be inferred.
TABLE 3.
Single-genome sequencing analysis
| Major genotype by SGSa | No. (%) of the following viruses (culture no., day of harvest)b:
|
||
|---|---|---|---|
| 216 (FT1, day 48, n = 26) | 716 (FT2, day 50, n = 22) | 111 (FT2, day 126, n = 28) | |
| K65R alone | 13 (50.0) | 5 (22.7) | 15 (53.6) |
| M184I alone | 10 (38.5 ) | 11 (50.0) | |
| K65R/M184I | 3 (11.5) | 6 (27.3) | 11 (39.3) |
| Otherc | 2 (7.1) | ||
| Total K65R | 16 (61.5) | 11 (50.0) | 26 (92.9) |
| Total M184V/I | 13 (50.0) | 17 (77.3) | 11 (39.3) |
Other mutations found in two or more clones were D17N (n = 5, virus 216), L214F (n = 12, virus 216), R358K (n = 3, virus 716; n = 4, virus 111), and A62V (n = 5, virus 111).
The genotype of viruses 216 and 716 by population sequencing was K65R/K and M184I/M and that of virus 111 was K65R and M184I/M. n, total number of clones.
Virus 111 had two clones with K65N alone.
The dominant viral genome found in virus 716 had M184I alone (50% of clones), while the remaining clones were similarly divided within the double-mutant or the single-K65R-mutant categories. Virus 111 was tested as a follow-up to virus 716, after an additional 76 days in culture with concentrations of drugs eightfold the EC50s. The M184I mutation was still present, but only in combination with K65R. This suggests that with drug concentrations eightfold the EC50s, the fitnesses of viruses with K65R or K65R and M184I are comparable, but viruses with M184I alone are less fit in the presence of both TFV and FTC.
DISCUSSION
The aim of the current studies was to characterize the in vitro drug resistance selection profile of the components of the anti-HIV dual-drug combination pill consisting of FTC and TDF. The first experiment with FTC and TFV gave rise to three possible viruses, depending on the drug concentrations used. At low concentrations of drugs, a mutant with a single M184I mutation was found, followed by a mutant with a single K65R mutation at intermediate concentrations and a mutant with K65R and M184V double mutations when each drug was used at the highest concentration that supported viral replication.
When the drug concentrations used in vitro were set to more closely mimic plasma drug levels in treated patients, an M184I mutation was observed first, followed directly by a double mutation, K65R and M184V. In the ongoing clinical trial GS-01-934, in which patients are receiving TDF, FTC and efavirenz, patients failing therapy were found to develop efavirenz resistance mutations and M184V, but no K65R mutation has been observed to date (7). In an analogous clinical trial—study GS-99-903—in which 3TC was used instead of FTC, patients failing therapy developed efavirenz resistance and either the M184V/I, M184V/I-K65R, or K65R mutation(s), in decreasing frequency of occurrence (8, 16). The results from these two clinical studies suggest that the hallmark TFV resistance mutation K65R develops at a much lower frequency than the M184V/I mutation in vivo. These clinical results are consistent with the later development of K65R observed in vitro.
These experiments have shown that the mutations obtained under selective pressure from TFV and FTC in vitro are strongly dependent on the drug concentrations used. The M184V/I and K65R mutations were observed on different genomes, with the development of the M184V/I mutation preceding the development of the K65R mutation. Similar results were obtained from a clonal analysis of the development of the M184V and K65R mutations in patients taking TDF, 3TC, and ABC (4). The earlier development of M184V/I in vitro also correlates with the higher prevalence of M184V/I compared to that of K65R observed in vivo. The slow development of the K65R mutation in the selections with both TFV and FTC and with TFV alone, as well as the viral growth impairment observed in these two cultures under selective pressure from TFV, suggests that the K65R virus may have a reduced replication capacity, as described previously (5, 40), and/or that TFV may have residual activity in the presence of K65R. The observation that the K65R mutation selected in the ABC selection experiment did not result in impaired viral growth suggests that the viral growth impairment seen in the TFV culture was more likely caused by the residual activity of TFV rather than reduced replication capacity of the K65R mutant virus in our in vitro system. The clinical implications of the potential residual activity of TFV against HIV-1 isolates with the K65R mutation are unknown; however, the continued antiviral benefit of TFV against a simian immunodeficiency virus with the K65R mutant mutation has been shown (34-36).
A novel resistance pathway, in the form of the K65N and K70R mutations with intermediate FTC concentrations, was observed in the present study. While the K70R mutation is one of the well-known thymidine analog-associated mutations and has been frequently reported (9, 15, 19), the K65N mutation is very rare and has been reported in only four patients, to our knowledge (14, 24, 25, 37). The antiviral treatments received by these patients at the time of detection of the K65N mutation varied: AZT-ddI (37), ABC-3TC-TDF (14), EFV-ddI-FTC (25), and EFV-ABC-3TC (24). The K65N mutation may play a role similar to that of the K65R mutation in NRTI resistance. Indeed, in the present study, the K65N and K70R pattern of mutations appeared to represent an alternate pathway to resistance to both TFV and FTC at intermediate drug concentrations. Analysis of the nucleotide sequence data for that virus indicates that the observed evolution from K65R to K65N seen in culture reflects the existence of two independent viral species, both of which emerged from a wild-type sequence. The K65R mutation was AAA to AGA, while the K65N mutation was AAA to AAT. This nucleotide sequence analysis suggests that both viruses were competing in that culture and that the K65N species had a growth advantage over the K65R species at the drug concentrations used (32-fold the EC50). The clinical importance of the K65N-K70R mutation pattern remains to be elucidated.
Interestingly, the presence of the K65N and K70R mutation pattern in the FT2 culture enabled the virus to grow in the presence of drug concentrations corresponding to 32-fold their EC50s, whereas the virus harboring K65R alone seen in the FT1 culture was unable to grow in the presence of drug concentrations greater than 16-fold the EC50. In addition, phenotypic analyses of the K65N-K70R virus (Table 2, virus 7) have shown nearly fourfold and nearly twofold increases in the levels of resistance to FTC and 3TC, respectively, compared to that of virus with K65R, while the level of resistance to TFV was only minimally increased. Conversely, the levels of resistance to ZDV, d4T, ABC, and ddI were reduced in the K65N-K70R virus compared to those in the K65R virus. Of note, the presence of the well-established thymidine analog-associated mutation K70R (3, 9, 15, 19) did not appear to affect resistance to ZDV and d4T when that mutation was combined with K65N. These observations suggest that the K65N and K70R mutations represent an alternate to the M184V/I resistance genotype and lead to intermediate resistance to FTC and 3TC. Under selective pressure from both FTC and TFV at 32-fold their EC50s, the K65N-K70R resistance genotype appears to be preferred to the K65R resistance genotype because of the greater resistance to FTC but similar resistance to TFV. At higher FTC concentrations, however, the preferred mutant had the K65R and M184V mutations.
The selection of resistance to the combination of 3TC and ABC or ABC alone led to the accumulation of well-characterized ABC resistance mutations at position 184, 65, 74, and 115 (27, 29, 31, 32). Of note, the M184V mutation also developed by selection with ABC alone. The development of the M184V mutation occurred readily in the 3TC-ABC selection experiment and may play a role in resistance to the combination of 3TC and ABC, since that mutation confers measurable resistance to both compounds. A recent report described virus with the M184V mutation as being present in 50% of patients failing 3TC-ABC therapy in clinical trial ESS30008 after 48 weeks (28). In addition, the T200A polymorphism was observed in the dual ABC-3TC selection experiment, as was the Q161L mutation. Q161L has not been reported in relation to ABC but has been described as a foscarnet resistance mutation in several studies (17, 30, 33). The role of that mutation in resistance to ABC, if any, is unknown. Importantly, the virus that developed Y115F and subsequently the K65R mutation in the ABC culture showed cross-resistance to TFV. Increased cross-resistance was also demonstrated with the mutant generated by site-directed mutagenesis containing both K65R and Y115F compared to the virus with K65R alone. This finding may have negative implications regarding the use of TDF and ABC together in the clinical setting, in which the risk of development of Y115F may be increased and the effect of antiviral therapy with both drugs may be compromised.
Additional mutations at positions 62, 68, and 221 were observed during the course of the selection experiments with TFV alone or in combination with FTC and with ABC alone or in combination with 3TC. A database search (http://hivdb.stanford.edu/) revealed that the H221Y mutation is strongly associated with antiretroviral treatment experience (12, 22), and it may represent a common secondary resistance mutation. Phenotypic data for mutants harboring that mutation in association with K65R showed that they had slightly higher levels of resistance to most NRTIs and TFV than mutants with K65R alone, when the levels of resistance in both viruses were compared with that of a wild-type virus (Table 2). However, the changes observed were not statistically significant compared to that for the K65R virus control. Interestingly, the H221Y mutation was recently found to be associated with non-NRTI resistance (26). The role of that mutation as a resistance or compensatory mutation remains to be determined.
The presence of mutations at positions 62 and 68 has previously been observed in association with K65R by using the Monogram Biosciences (formerly ViroLogic) database, and these were in the form of the A62V and S68G mutations (D. J. McColl, N. T. Parkin, and M. D. Miller, Abstr. 7th Int. Congr. Drug Ther. HIV Infect., poster 113, 2004). These mutations are also found in treatment-naïve individuals but are enriched in antiretroviral treatment-experienced patients (http://hivdb.stanford.edu/) (12, 22). These mutations were found to occur along with K65R in three treatment-naïve patients from clinical trial GS-99-903 (16). Phenotypic analyses of mutant viruses with mutations generated by site-directed mutagenesis harboring the A62V or S68G mutation in association with K65R have revealed that the level of resistance of the mutants with double mutations was nearly identical to that of mutants with K65R alone (M. D. Miller, N. A. Margot, D. J. McColl, T. Wrin, D. F. Coakley, and A. K. Cheng, Abstr. XII Int. HIV Drug Resist. Workshop, abstr. 135, 2003). The S68N substitution has also been reported in association with K65R in one patient from study GS-99-903 (16) and was shown to be phenotypically similar to a mutant with K65R alone in the phenotypic analyses conducted for the present study. The S68K mutation has not previously been reported and may impart low-level increased TFV resistance or may be compensatory in nature, as hypothesized for the S68G and S68N mutations (16, 23).
In conclusion, the resistance selection studies reported here have shown that mutants with a K65R-M184V genotype are generated under maximum selection pressure from both TFV and FTC. Interestingly, mutants with only single M184V mutation, as opposed to the mutants with the predicted K65R and M184V double mutations, have been observed clinically in patients who have failed FTC and TDF therapy to date (7). Longer-term virologic failure, however, may result in the predicted K65R and M184V double mutation. To date, the novel K65N and K70R double mutation observed in these in vitro studies has not been observed in patients receiving FTC and TDF.
Acknowledgments
We thank Kirsten Stray for providing virus stocks. In addition, we thank Katyna Borroto-Esoda, Kirsten White, and Hans Reiser for critical review of the manuscript and Margaret Benton for technical assistance with the production of the manuscript.
The work was supported entirely by Gilead Sciences, Inc.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Barditch-Crovo, P., S. G. Deeks, A. Collier, S. Safrin, D. F. Coakley, M. Miller, B. P. Kearney, R. L. Coleman, P. D. Lamy, J. O. Kahn, I. McGowan, and P. S. Lietman. 2001. Phase I/II trial of the pharmacokinetics, safety, and antiretroviral activity of tenofovir disoproxil fumarate in human immunodeficiency virus type 1-infected adults. Antimicrob. Agents Chemother. 45:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher, C. A. B., W. Keulen, T. van Bommel, M. Nijhuis, D. de Jong, M. D. de Jong, P. Schipper, and N. K. T. Back. 1996. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 40:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jong, M. D., J. Veenstra, N. I. Stilianakis, R. Schuurman, J. M. A. Lange, R. J. de Boer, and C. A. B. Boucher. 1996. Host-parasite dynamics and outgrowth of virus containing a single K70R amino acid change in reverse transcriptase are responsible for the loss of human immunodeficiency virus type 1 RNA load suppression by zidovudine. Proc. Natl. Acad. Sci. USA 93:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaunay, C., F. Brun-Vezinet, R. Landman, G. Collin, G. Peytavin, A. Trylesinski, P. Flandre, M. Miller, and D. Descamps. 2005. Comparative selection of the K65R and M184V/I mutations in human immunodeficiency virus type 1-infected patients enrolled in a trial of first-line triple-nucleoside analog therapy (Tonus IMEA 021). J. Virol. 79:9572-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deval, J., K. L. White, M. D. Miller, N. T. Parkin, J. Courcambeck, P. Halfon, B. Selmi, J. Boretto, and B. Canard. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279:509-516. [DOI] [PubMed] [Google Scholar]
- 6.Feng, J. Y., F. T. Myrick, N. A. Margot, G. B. Mulamba, L. Rimsky, K. Borroto-Esoda, B. Selmi, and B. Canard. 2006. Virologic and enzymatic studies revealing the mechanism of K65R- and Q151M-associated HIV-1 drug resistance towards emtricitabine and lamivudine. Nucleosides Nucleotides Nucleic Acids 25:89-107. [DOI] [PubMed] [Google Scholar]
- 7.Gallant, J. E., E. DeJesus, J. R. Arribas, A. L. Pozniak, B. Gazzard, R. E. Campo, B. Lu, D. McColl, S. Chuck, J. Enejosa, J. J. Toole, and A. K. Cheng. 2006. Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N. Engl. J. Med. 354:251-260. [DOI] [PubMed] [Google Scholar]
- 8.Gallant, J. E., S. Staszewski, A. L. Pozniak, E. DeJesus, J. M. A. H. Suleiman, M. D. Miller, D. F. Coakley, B. Lu, J. J. Toole, A. K. Cheng, and 903 Study Group. 2004. Efficacy and safety of tenofovir DF vs stavudine in combination therapy in antiretroviral-naive patients: a 3-year randomized trial. JAMA 292:191-201. [DOI] [PubMed] [Google Scholar]
- 9.Gao, Q., Z. Gu, M. A. Parniak, X. Li, and M. A. Wainberg. 1992. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J. Virol. 66:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilead Sciences, Inc. 2004. EMTRIVA® (emtricitabine) capsules. U.S. prescribing information. Gilead Sciences, Inc., Foster City, Calif.
- 11.Gilead Sciences, Inc. 2005. VIREAD® (tenofovir disoproxil fumarate) tablets. U.S. prescribing information. Gilead Sciences, Inc., Foster City, Calif.
- 12.Gonzales, M. J., T. D. Wu, J. Taylor, I. Belitskaya, R. Kantor, D. Israelski, S. Chou, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS 17:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, Z., H. Salomon, J. M. Cherrington, A. S. Mulato, M. S. Chen, R. Yarchoan, A. Foli, K. M. Sogocio, and M. A. Wainberg. 1995. K65R mutation of human immunodeficiency virus type 1 reverse transcriptase encodes cross-resistance to 9-(2-phosphonylmethoxyethyl)adenine. Antimicrob. Agents Chemother. 39:1888-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoogewerf, M., R. M. Regez, W. E. M. Schouten, H. M. Weigel, P. H. J. Frissen, and K. Brinkman. 2003. Change to abacavir-lamivudine-tenofovir combination treatment in patients with HIV-1 who had complete virological suppression. Lancet 362:1979-1980. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. Schapiro, A. Telenti, and D. Richman. 2005. Update of the drug resistance mutations in HIV-1: 2005. Top. HIV Med. 13:51-57. [PubMed] [Google Scholar]
- 16.Margot, N. A., B. Lu, A. Cheng, and M. D. Miller. 2006. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in study 903. HIV Med. 7:442-450. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, P. R., S. E. Matsuura, D. Zonarich, R. R. Chopra, E. Pendarvis, H. Z. Bazmi, J. W. Mellors, and W. A. Scott. 2003. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 77:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 19.Odriozola, L., C. Cruchaga, M. Andreola, V. Dolle, C. H. Nguyen, L. Tarrago-Litvak, A. Perez-Mediavilla, and J. J. Martinez-Irujo. 2003. Non-nucleoside inhibitors of HIV-1 reverse transcriptase inhibit phosphorolysis and resensitize the 3′-azido-3′-deoxythymidine (AZT)-resistant polymerase to AZT-5′-triphosphate. J. Biol. Chem. 278:42710-42716. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer, S., N. Margot, H. Gilbert, N. Shaw, R. Buckheit, Jr., and M. Miller. 2001. Tenofovir, adefovir, and zidovudine susceptibilities of primary human immunodeficiency virus type 1 isolates with non-B subtypes or nucleoside resistance. AIDS Res. Hum. Retrovir. 17:1167-1173. [DOI] [PubMed] [Google Scholar]
- 22.Rhee, S.-Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roge, B. T., T. L. Katzenstein, N. Obel, H. Nielsen, O. Kirk, C. Pedersen, L. Mathiesen, J. Lundgren, and J. Gerstoft. 2003. K65R with and without S68: a new resistance profile in vivo detected in most patients failing abacavir, didanosine and stavudine. Antivir. Ther. 8:173-182. [PubMed] [Google Scholar]
- 24.Ross, L. L., R. Dretler, P. Gerondelis, E. G. Rouse, M. L. Lim, and E. R. Lanier. 2006. A rare HIV reverse transcriptase mutation, K65N, confers reduced susceptibility to tenofovir, lamivudine and didanosine. AIDS 20:787-789. [DOI] [PubMed] [Google Scholar]
- 25.Saag, M. S., P. Cahn, F. Raffi, M. Wolff, D. Pearce, J.-M. Molina, W. Powderly, A. L. Shaw, E. Mondou, J. Hinkle, K. Borroto-Esoda, J. B. Quinn, D. W. Barry, F. Rousseau, and FTC-301A Study Team. 2004. Efficacy and safety of emtricitabine vs stavudine in combination therapy in antiretroviral-naive patients: a randomized trial. JAMA 292:180-190. [DOI] [PubMed] [Google Scholar]
- 26.Saracino, A., L. Monno, L. Scudeller, D. C. Cibelli, A. Tartaglia, G. Punzi, C. Torti, S. Lo Caputo, F. Mazzotta, G. Scotto, G. Carosi, and G. Angarano. 2006. Impact of unreported HIV-1 reverse transcriptase mutations on phenotypic resistance to nucleoside and non-nucleoside inhibitors. J. Med. Virol. 78:9-17. [DOI] [PubMed] [Google Scholar]
- 27.Schinazi, R. F., R. M. Lloyd, Jr., M.-H. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sosa, N., C. Hill-Zabala, E. Dejesus, G. Herrera, A. Florance, M. Watson, C. Vavro, and M. Shaefer. 2005. Abacavir and lamivudine fixed-dose combination tablet once daily compared with abacavir and lamivudine twice daily in HIV-infected patients over 48 weeks (ESS30008, SEAL). J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 40:422-427. [DOI] [PubMed] [Google Scholar]
- 29.Stone, C., M. Ait-Khaled, C. Craig, P. Griffin, and M. Tisdale. 2004. Human immunodeficiency virus type 1 reverse transcriptase mutation selection during in vitro exposure to tenofovir alone or combined with abacavir or lamivudine. Antimicrob. Agents Chemother. 48:1413-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tachedjian, G., J. Mellors, H. Bazmi, C. Birch, and J. Mills. 1996. Zidovudine resistance is suppressed by mutations conferring resistance of human immunodeficiency virus type 1 to foscarnet. J. Virol. 70:7171-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tramontano, E., G. Piras, J. W. Mellors, M. Putzolu, H. Z. Bazmi, and P. La Colla. 1998. Biochemical characterization of HIV-1 reverse transcriptases encoding mutations at amino acid residues 161 and 208 involved in resistance to phosphonoformate. Biochem. Pharmacol. 56:1583-1589. [DOI] [PubMed] [Google Scholar]
- 34.Van Rompay, K. K., M. D. Miller, M. L. Marthas, N. A. Margot, P. Dailey, D. R. Canfield, R. P. Tarara, J. M. Cherrington, N. L. Aguirre, N. Bischofberger, and N. C. Pedersen. 2000. Prophylactic and therapeutic benefits of short-term 9-[2-(R)phosphonomethoxy)propyl]adenine (PMPA) administration to newborn macaques following oral inoculation with simian immunodeficiency virus with reduced susceptibility to PMPA. J. Virol. 74:1767-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Rompay, K. K. A., R. P. Singh, L. L. Brignolo, J. R. Lawson, K. A. Schmidt, B. Pahar, D. R. Canfield, R. P. Tarara, D. L. Sodora, N. Bischofberger, and M. L. Marthas. 2004. The clinical benefits of tenofovir for simian immunodeficiency virus-infected macaques are larger than predicted by its effects on standard viral and immunologic parameters. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 36:900-914. [DOI] [PubMed] [Google Scholar]
- 36.Van Rompay, K. K. A., R. P. Singh, B. Pahar, D. L. Sodora, C. Wingfield, J. R. Lawson, M. L. Marthas, and N. Bischofberger. 2004. CD8+-cell-mediated suppression of virulent simian immunodeficiency virus during tenofovir treatment. J. Virol. 78:5324-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venturi, G., L. Romano, M. Catucci, M. L. Riccio, A. De Milito, A. Gonnelli, M. Rubino, P. E. Valensin, and M. Zazzi. 1999. Genotypic resistance to zidovudine as a predictor of failure of subsequent therapy with human immunodeficiency virus type-1 nucleoside reverse-transcriptase inhibitors. Eur. J. Clin. Microbiol. Infect. Dis. 18:274-282. [DOI] [PubMed] [Google Scholar]
- 38.Wainberg, M. A., M. D. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]
- 39.Walter, H., B. Schmidt, M. Werwein, E. Schwingel, and K. Korn. 2002. Prediction of abacavir resistance from genotypic data: impact of zidovudine and lamivudine resistance in vitro and in vivo. Antimicrob. Agents Chemother. 46:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber, J., B. Chakraborty, J. Weberova, M. D. Miller, and M. E. Quinones-Mateu. 2005. Diminished replicative fitness of primary human immunodeficiency virus type-1 isolates harboring the K65R mutation. J. Clin. Microbiol. 43:1395-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weislow, O. S., R. Kiser, D. L. Fine, J. Bader, R. H. Shoemaker, and M. R. Boyd. 1989. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 81:577-586. [DOI] [PubMed] [Google Scholar]
- 42.White, K. L., J. M. Chen, N. A. Margot, T. Wrin, C. J. Petropoulos, L. K. Naeger, S. Swaminathan, and M. D. Miller. 2004. Molecular mechanisms of tenofovir resistance conferred by human immunodeficiency virus type 1 reverse transcriptase containing a diserine insertion after residue 69 and multiple thymidine analog-associated mutations. Antimicrob. Agents Chemother. 48:992-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, K. L., N. A. Margot, T. Wrin, C. J. Petropoulos, M. D. Miller, and L. K. Naeger. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46:3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]