Abstract
VIM metallo-β-lactamase-producing serotype O11 or O12 Pseudomonas aeruginosa isolates infecting or colonizing 19 patients from seven hospitals in Hungary were characterized between October 2003 and November 2005. Macrorestriction analysis revealed the involvement of hospitals from three different towns in northwest Hungary in an outbreak caused by VIM-4-producing P. aeruginosa.
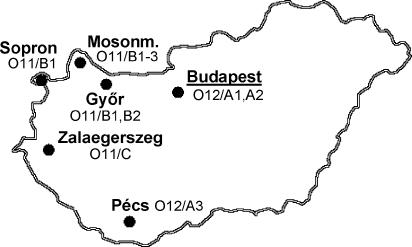
The worldwide spread of acquired metallo-β-lactamase (MBL)-producing gram-negative pathogens was observed in the past decade, with the blaVIM-type acquired MBL genes currently being the most prevalent in Europe (16, 23). The first VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary (isolates PA396 and PA450) were characterized at the National Center for Epidemiology in 2003 (7). We established routine screening of carbapenem-resistant Pseudomonas sp. isolates provided by collaborating regional laboratories for acquired MBL genes. In 2005, MBL-positive isolates from six towns in Hungary were detected (Fig. 1). Our aim was to characterize these isolates and to examine the clonal relationships between them and between the major European multiresistant serotype O12 P. aeruginosa clone, clone P12 (4, 8, 10).
FIG. 1.
Geographic distribution of the VIM-positive P. aeruginosa clinical isolates in Hungary characterized in the present study. Small black circles indicate the cities. Budapest (the capital) is underlined. Mosonm., Mosonmagyaróvár. The serotype and PFGE subtype of the VIM-positive P. aeruginosa isolates characterized are shown under or next to the names of the cities.
The VIM-positive clinical and environmental isolates tested in this study are listed in Table 1, together with the previously published control isolates PA396 and PA450 (7). Isolates P12-Q and P12-E were from French patients “Q” and “E,” respectively, who participated in a previous clone P12-related study (8). MICs were determined by the agar dilution method (1) for β-lactam antibiotics and by the Etest (AB Biodisk, Solna, Sweden) for other antibiotics. The MBL Etest and the imipenem-EDTA, ceftazidime-EDTA, and cefepime-EDTA double-disk methods were used for phenotypic screening (21, 22).
TABLE 1.
Characteristics of VIM-producing Pseudomonas sp. isolates from Hungarya
| Town | Hospitalb | Ward | Patientc | No. ofd isolates | Representative isolatee | Sample | Serotype | PFGE subtype | Integronf | Date of isolation (day.mo.yr) |
|---|---|---|---|---|---|---|---|---|---|---|
| Budapest | KHK | ICU | PA | 1 | PA396g | Urine | O12 | A1 | a | 26.08.02 |
| ICU | PB | 1 | PA450g | Urine | O12 | A1 | a | 24.09.02 | ||
| ICU | P1 | 4 | MB242 | Wound swab | O12 | A2 | a | 04.07.05 | ||
| ICU | — | 1 | MB248g | Spillway | O12 | A2 | a | 20.07.05 | ||
| ICU | — | 1 | PP524g | Sink | a | 10.12.03 | ||||
| Pécs | BMK | Infectious disease | P2 | 1 | PA555g | Urine | O12 | A3 | b | 22.10.03 |
| PTE-BK | Hematology | P3 | 1 | F69 | Blood | O12 | A3 | b | 03.08.04 | |
| PTE-BK | Hematology | — | 1 | MB143g | Shower | O12 | A3 | b | 20.04.05 | |
| PTE-BK | Hematology | P4 | 1 | MB397 | Urine | O12 | A3 | b | 19.08.05 | |
| Mosonmagyaróvár | KK | ICU | P5 | 3 | MB93g | Tracheal aspirate | O11 | B1 | d | 18.03.05 |
| ICU | P6 | 1 | MB94g | Tracheal aspirate | O11 | B2 | d | 18.03.05 | ||
| ICU | — | 2 | MB159 | Sink | O11 | B1 | d | 26.05.05 | ||
| ICU | P7 | 3 | MB197g | Blood | O11 | B1 | d | 06.06.05 | ||
| ICU | P8 | 6 | MB240g | Tracheal aspirate | O11 | B3 | d | 05.07.05 | ||
| ICU | P9 | 1 | MB200 | Tracheal aspirate | O11 | B1 | d | 08.06.05 | ||
| ICU | P10 | 1 | MB295g | Tracheal aspirate | O11 | B2 | c | 04.07.05 | ||
| ICU | P11 | 1 | MB296 | Tracheal aspirate | O11 | B1 | d | 28.07.05 | ||
| ICU | P12 | 1 | MB449 | Tracheal aspirate | O11 | B2 | c | 30.09.05 | ||
| Győr | PAK | Pediatrics | P13 | 1 | MB219 | Nasal swab | O11 | B1 | d | 17.06.05 |
| ICU | P14 | 1 | MB292 | Feces | O11 | B1 | d | 09.05.05 | ||
| ICU | P15 | 5 | MB329g | Wound swab | O11 | B2 | c | 13.08.05 | ||
| ICU | P16 | 1 | MB330 | Drain | O11 | B2 | c | 12.08.05 | ||
| ICU | P17 | 3 | MB447 | Tracheal aspirate | O11 | B2 | c | 14.10.05 | ||
| Zalaegerszeg | ZMK | ICU | P18 | 1 | MB346g | Urine | O11 | C | ND | 10.08.05 |
| Sopron | EK | ICU | P19 | 2 | MB387g | Urine | O11 | B1 | c | 05.09.05 |
All isolates are P. aeruginosa, with the exception of isolate PP524, which is a P. putida strain.
Hospitals are indicated by the abbreviation of their Hungarian name.
Patients are indicated by codes PA and PB for the control isolates and P1 to P19 for the isolates characterized in this study. —, environmental isolates.
The total number of characterized VIM-positive isolates from the patient. Except for P17, replicate isolates had the same serotype and the same PFGE type (A or B) and carried the same integron type (a, b, c, or d). P17 had two isolates carrying integron type c and one isolate carrying integron type d.
Mating-out assays were performed with the underlined isolates.
Integron codes correspond to those in Fig. 2. ND, not determined.
Sequencing of the blaVIM gene was performed, and the gene was identified as blaVIM-4 for these isolates, which represent the different PFGE subtypes and integron types.
blaVIM genes and class 1 integrons were detected by PCR (7). The variable regions of the integrons from isolates PA555 and MB197 were sequenced by using the following primers, together with those described previously (7): primer 197F (5′-AAT CGC TCA GTC GCC GAG-3′), primer 197R1 (5′-TAG TGC TTC TCC GTC GGG-3′), primer 197R2 (5′-AAT TCC GCA TTG CTG ATC G-3′), and primer 197R3 (5′-AGG TAT TGC TCC TGC ACT T-3′). Isolate PA555 was selected for full integron sequencing in 2003, as it was the first VIM-positive isolate from Pécs, Hungary, while isolate MB197 was selected as an invasive isolate from a cluster of clonally closely related VIM-positive isolates from northwest Hungary. For the other VIM-producing isolates, the integron structures were determined by PCR mapping and partial sequencing.
Pulsed-field gel electrophoresis (PFGE) was performed as described earlier (11), with modifications, and the patterns were interpreted by using Fingerprinting II Informatix software (Bio-Rad, Madrid, Spain). Pseudomonas aeruginosa antisera (Bio-Rad, Marnes-la-Coquette, France) were used for serotyping.
Conjugation experiments were carried out with strains Escherichia coli J5-3 Rifr and P. aeruginosa PAO4089Rp (6, 7) as the recipients. Transconjugants were selected on Mueller-Hinton agar plates containing 300 μg/ml rifampin and 32 μg/ml cefotaxime, 128 μg/ml piperacillin-tazobactam, or 128 μg/ml ticarcillin.
A total of 758 carbapenem-resistant Pseudomonas sp. isolates collected on a voluntary basis from 85 epidemiological settings in 42 towns in Hungary were screened for MBL production between October 2003 and November 2005. The settings are distributed in every geographical region of Hungary. Fifty P. aeruginosa isolates and one P. putida isolate (isolate PP524) proved to be positive by the phenotypic tests. Seven of these positive isolates from a Budapest hospital were not included in this study. PCRs with VIM- and integron-specific primers (7) gave positive results for all these isolates. The blaVIM genes of the isolates indicated by a superscript g in Table 1 were sequenced and were identified as blaVIM-4 in every case. P12-Q and P12-E were negative by the MBL phenotypic tests.
Antibiotic susceptibility values for representative VIM-positive isolates and isolate P12-Q are shown in Table 2. All isolates were multidrug resistant, and some of them were only sensitive to aztreonam. Isolate MB397 was panresistant to all antimicrobials tested.
TABLE 2.
Antibiotic MICs determined for VIM-producing Pseudomonas sp. isolates and isolate P12-Q
| Isolate | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ATM | CAZ | FEP | TZP | GEN | AMK | CIP | PO | |
| PA396b | 64 | >32 | 32 | 256 | 256 | >256 | 8 | 32 | >32 | 2 |
| PA450b | 256 | >32 | 16 | 256 | 256 | >256 | 8 | 32 | >32 | 2 |
| MB242 | >256 | >32 | 32 | 128 | 256 | >256 | 32 | 64 | >32 | 2 |
| MB248 | >256 | >32 | 16 | 128 | 256 | >256 | >256 | 32 | >32 | 2 |
| PP524 | 256 | >32 | 64 | 256 | 256 | >256 | 8 | 32 | 1 | 2 |
| PA555 | >256 | >32 | 8 | 256 | 256 | >256 | >256 | >256 | >32 | 4 |
| F69 | 256 | >32 | 4 | >256 | >256 | >256 | 4 | 8 | >32 | 4 |
| MB143 | >256 | >32 | 16 | >256 | >256 | >256 | >256 | >256 | >32 | 2 |
| MB397 | >256 | >32 | 16 | 256 | >256 | >256 | >256 | >256 | >32 | 4 |
| MB93 | >256 | >32 | 32 | 256 | >256 | 256 | 64 | >256 | >32 | 2 |
| MB94 | >256 | >32 | 32 | 256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB159 | >256 | >32 | 8 | >256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB197 | >256 | >32 | 8 | 128 | >256 | >256 | 64 | >256 | >32 | 4 |
| MB240 | >256 | >32 | 32 | 256 | >256 | >256 | 16 | 256 | >32 | 1 |
| MB200 | 256 | >32 | 8 | 128 | 256 | >256 | 64 | >256 | >32 | 2 |
| MB295 | >256 | >32 | 8 | >256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB296 | >256 | >32 | 16 | 128 | 256 | >256 | 64 | >256 | >32 | 2 |
| MB449 | >256 | >32 | 8 | >256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB219 | >256 | >32 | 8 | 32 | 64 | 128 | 64 | >256 | >32 | 2 |
| MB292 | >256 | >32 | 16 | 128 | 256 | >256 | 32 | >256 | >32 | 2 |
| MB329 | 256 | >32 | 8 | >256 | >256 | >256 | 128 | >256 | >32 | 2 |
| MB330 | 256 | >32 | 16 | >256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB447 | >256 | >32 | 8 | >256 | >256 | >256 | 64 | >256 | >32 | 2 |
| MB346 | >256 | >32 | 8 | 32 | 64 | >256 | 16 | >256 | >32 | 4 |
| MB387 | >256 | >32 | 4 | 64 | 128 | >256 | 32 | >256 | >32 | 2 |
| P12-Q | 4 | 2 | 8 | 32 | 32 | >256 | 128 | 16 | 32 | 4 |
Abbreviations: IPM, imipenem; MEM, meropenem; ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; TZP, piperacillin-tazobactam; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin; PO, polymyxin B.
Antibiotic MICs determined previously for control isolates PA396 and PA450 (7) are also shown for comparison.
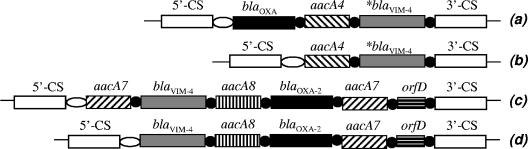
Two major groups could be identified among the VIM-positive P. aeruginosa isolates by serotyping and PFGE. Their geographical distribution is shown in Fig. 1. The first group comprised serotype O12 isolates from Budapest and Pécs (Table 1). These isolates were possibly related to each other, as determined by PFGE, and also to representative isolates of the major European P12 clone when a cutoff value of ≥80% was used (17-19). The O12 isolates from Budapest and Pécs carry an integron-borne blaVIM-4 gene with an identical 170-bp duplication in the last position, preceded by an aacA4 gene (Fig. 2, integrons a and b). An additional blaOXA cassette is present in the first position of integron a from Budapest (Table 1) (7).
FIG. 2.
Comparison of the schematic structures of the blaVIM-4-carrying class 1 integrons from Hungary. Open ellipses, attI1 site; black circles, 59-base elements; asterisks, blaVIM-4 cassettes with the 170-bp duplication; 5′-CS and 3′-CS, 5′ and 3′ conserved sequences, respectively. GenBank accession numbers and isolates harboring the integrons are as follows: (a) AY509609 and isolate PA396, (b) AY702100 and isolate PA555, (c) isolate MB387, (d) DQ357197 and isolate MB197.
The second group comprised all isolates from the Győr, Sopron, and Mosonmagyaróvár hospitals. These serotype O11 isolates were identical or closely related to each other by PFGE by use of a Dice coefficient of ≥95% (Table 1) and carried a blaVIM-4 gene without the 170-bp duplication on an integron that also harbored a blaOXA-2 cassette. Two variants of this integron were identified (Fig. 2, integrons c and d), with the only difference being that an additional aacA7 cassette was present in the first position of integron c. We identified a carrier patient (patient P14) who was transferred between the intensive care units (ICUs) of the Győr and Mosonmagyaróvár hospitals, providing an epidemiological link between them. Another patient (patient P13) with no related clinical history was identified as a carrier on admission to the hospital, suggesting the presence of VIM-positive strains in the community. These results demonstrate the involvement of institutions from three towns in an outbreak of VIM-4-producing P. aeruginosa. Our observations and the available literature (2, 20, 23) underscore the role of human carriers and the hospital environment as potential reservoirs for MBL-producing P. aeruginosa strains.
Mating-out assays were performed with the isolates underlined in Table 1. These experiments did not result in transconjugants under the experimental conditions applied, suggesting that the integrons are located either on the chromosome or on nonconjugative plasmids (12, 15). While integrons a, c, and d have so far been reported only from Hungary, class 1 integrons carrying the same gene cassettes in their variable regions as integron b were also identified in Poland and Belgium (3, 7, 9; P. Bogaerts, H. Rodriguez, C. Bauraing, A. Deplano, Y. Glupczynski, and M. J. Struelens, Abstr. 15th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P929, 2006).
The rates of imipenem resistance among P. aeruginosa clinical isolates in Hungary rose from 5.4% in 1996 to 13% in 2005 (5). The prevalence of VIM-positive P. aeruginosa isolates was estimated in the epidemiological settings from which the isolates were collected. On the basis of our experimental results and data from the National Bacteriological Surveillance database, we estimate that in 2004, VIM producers constituted about 0.4% and 0.05% of the imipenem-resistant isolates and all P. aeruginosa clinical isolates, respectively. In 2005, these values rose to about 6.5% and 0.8%, respectively. The observed increase could mainly be attributed to the clonal spread of the serotype O11 VIM-4-positive P. aeruginosa isolates in northwest Hungary.
This is the first report of an outbreak caused by acquired MBL-producing pathogens in Hungary. In hospitals in Greece, Italy, Korea, and Colombia, the dissemination of integrons carrying the blaVIM-1, blaVIM-2, or blaVIM-8 gene mostly occurred in serotype O11 and O12 clones of P. aeruginosa (2, 6, 14, 20). These studies, as well as ours, indicate that serotype O11 and O12 multiresistant clones of P. aeruginosa (13) play an important role in the dissemination of blaVIM through clonal spread but that other mechanisms, such as horizontal transfer, are also involved (3, 16, 23).
Nucleotide sequence accession numbers.
The sequences of blaVIM harboring integrons were deposited in GenBank under accession numbers AY702100 and DQ357197.
Acknowledgments
This work was supported by grant T-08/186/2001 of the Hungarian Scientific Council for Health.
We thank L. Keresztes, L. Kereszturi, A. Kovács, A. Kvarda, K. Pintér, and J. Szentandrássy for their help.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 2.Crespo, M. P., N. Woodford, A. Sinclair, M. E. Kaufmann, J. Turton, J. Glover, J. D. Velez, C. R. Castaneda, M. Recalde, and D. M. Livermore. 2004. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali, Colombia. J. Clin. Microbiol. 42:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiett, J., A. Baraniak, A. Mrowka, M. Fleischer, Z. Drulis-Kawa, L. Naumiuk, A. Samet, W. Hryniewicz, and M. Gniadkowski. 2006. Molecular epidemiology of acquired-metallo-β-lactamase-producing bacteria in Poland. Antimicrob. Agents Chemother. 50:880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grattard, F., O. G. Gaudin, B. Pozzeto, A. Ris, and A. D. Mbida. 1993. Genotypic homogeneity of nosocomial Pseudomonas aeruginosa O12 strains demonstrated by analysis of protein profiles, DNA fingerprints and rRNA gene restriction patterns. Eur. J. Clin. Microbiol. Infect. Dis. 12:57-61. [DOI] [PubMed] [Google Scholar]
- 5.Johan Béla National Public Health Institute. 1997. Antibiotic susceptibility of the most important gram-negative bacteria, p. 159-173. In The 1996 annual report of the Johan Béla National Public Health Institute. Johan Béla National Public Health Institute, Budapest, Hungary. (In Hungarian.)
- 6.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libisch, B., M. Gacs, K. Csiszár, M. Muzslay, L. Rókusz, and M. Füzi. 2004. Isolation of an integron-borne blaVIM-4 type metallo-β-lactamase gene from a carbapenem-resistant Pseudomonas aeruginosa clinical isolate in Hungary. Antimicrob. Agents Chemother. 48:3576-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mifsud, J. A., J. Watine, B. Picard, J. C. Charet, C. Solignac-Bourrel, and T. L. Pitt. 1997. Epidemiologically related and unrelated strains of Pseudomonas aeruginosa serotype O12 cannot be distinguished by phenotypic and genotypic typing. J. Hosp. Infect. 36:105-116. [DOI] [PubMed] [Google Scholar]
- 9.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 10.Pitt, T. L., D. M. Livermore, D. Pitcher, A. C. Vatopoulos, and N. J. Legakis. 1989. Multiresistant serotype O12 Pseudomonas aeruginosa: evidence for a common strain in Europe. Epidemiol. Infect. 103:565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poh, C. L., C. C. Yeo, and L. Tay. 1992. Genome fingerprinting by pulsed-field gel electrophoresis and ribotyping to differentiate Pseudomonas aeruginosa serotype O11 strains. Eur. J. Clin. Microbiol. Infect. Dis. 11:817-822. [DOI] [PubMed] [Google Scholar]
- 12.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Réseau Épidémiologique des Utilisateurs du Système SIR. 2001. Le portage prolongé et la diffusion clonale interhospitaliére des Pseudomonas aeruginosa multirésistants de sérotype O12 sont-ils liés? Étude multicentrique. Pathol. Biol. 49:620-623. [DOI] [PubMed] [Google Scholar]
- 14.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossolini, G. M. 2005. Acquired metallo-β-lactamases: an increasing clinical threat. Clin. Infect. Dis. 41:1557-1558. [DOI] [PubMed] [Google Scholar]
- 17.Scott, F. W., and T. L. Pitt. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609-615. [DOI] [PubMed] [Google Scholar]
- 18.Speijer, H., P. H. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struelens, M. J., V. Schwam, A. Deplano, and D. Baran. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan, J. J., J. J. Wu, S. H. Tsai, and C. L. Chuang. 2004. Comparison of the double-disk, combined disk, and Etest methods for detecting metallo-β-lactamases in gram-negative bacilli. Diagn. Microbiol. Infect. Dis. 49:5-11. [DOI] [PubMed] [Google Scholar]
- 22.Yong, D., K. Lee, J. H. Yum, H. B. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]