Abstract
The hepatitis C virus (HCV) RNA-dependent RNA polymerase NS5B is an important target for antiviral therapies. NS5B is able to initiate viral RNA synthesis de novo and then switch to a fast and processive RNA elongation synthesis mode. The nucleotide analogue 2′-C-methyl CTP (2′-C-Me-CTP) is the active metabolite of NM283, a drug currently in clinical phase II trials. The resistance mutation S282T can be selected in HCV replicon studies. Likewise, 2′-O-Me nucleotides are active both against the purified polymerase and in replicon studies. We have determined the molecular mechanism by which the S282T mutation confers resistance to 2′-modified nucleotide analogues. 2′-C-Me-CTP is no longer incorporated during the initiation step of RNA synthesis and is discriminated 21-fold during RNA elongation by the NS5B S282T mutant. Strikingly, 2′-O-methyl CTP sensitivity does not change during initiation, but the analogue is no longer incorporated during elongation. This mutually exclusive resistance mechanism suggests not only that “2′-conformer” analogues target distinct steps in RNA synthesis but also that these analogues have interesting potential in combination therapies. In addition, the presence of the S282T mutation induces a general cost in terms of polymerase efficiency that may translate to decreased viral fitness: natural nucleotides become 5- to 20-fold less efficiently incorporated into RNA by the NS5B S282T mutant. As in the case for human immunodeficiency virus, our results might provide a mechanistic basis for the rational combination of drugs for low-fitness viruses.
Hepatitis C virus (HCV) infection is the most common blood-borne infection and a major cause of chronic liver disease in developed countries. More than 170 million individuals in the world are infected with HCV, making these individuals at risk of developing liver cirrhosis and hepatocellular carcinoma. Persistent HCV infection can be controlled by antiviral therapy (30, 40) or even eradicated from infected patients (43).
Current antiviral therapies rely on the combination of pegylated alpha interferon and the nucleoside analogue ribavirin (16, 18, 29). Less than 50% of treated patients respond when they are infected with the most prevalent HCV genotype, i.e., genotype 1 (23). The HCV RNA-dependent RNA polymerase (RdRp), the NS5B protein, plays an essential role in the replication of the viral genome and is thus an attractive target for the development of new antiviral drugs (36). Intensive drug-screening programs led to the discovery of two classes of HCV NS5B inhibitors, namely, nonnucleoside inhibitors (NNIs) and nucleoside inhibitors (NIs). NNIs have been described to bind to one of the three allosteric sites present at the NS5B surface (for a review, see reference 11). All of the NNIs are noncompetitive inhibitors relative to nucleoside triphosphate (NTP) incorporation and target the alloenzyme free of substrate. They are inactive when the enzyme has entered into the processive elongation phase (3, 17, 31, 45, 46) of in vitro RdRp reactions or when NS5B is complexed with other nonstructural replicative proteins (28) in replicon-expressing cells. NIs, however, target the active site and are active on both the purified polymerase and the replicative complex (7, 28, 39). NM283 is such a compound. It is an oral prodrug of 2′-C-methyl cytidine (2′-C-Me-cytidine) which is currently under clinical evaluation. 2′-Modified nucleosides, modified with either 2′-C-Me or 2′-O-Me, inhibit RNA replication in the replicon system and in RdRp assays in vitro (7, 33, 39). Both types of 2′-modified nucleosides promote chain termination of the neosynthesized RNA when incorporated into nascent RNA by NS5B (7, 33).
Unfortunately, all NNI or NI compounds tested so far in the replicon system lose efficacy after selection of a single point mutation in the viral polymerase gene (22, 26, 33-35, 45, 46). Paralleling findings in the human immunodeficiency virus type 1 (HIV-1) antiretroviral field, this suggests that viral resistance to polymerase inhibitors may also occur in patients. To date, no resistant HCVs have been selected during the clinical testing of NM283 (2), although the increase of viral loads observed during monotherapy suggests the appearance of such resistant mutants. 2′-C-Me-purine nucleoside-resistant replicons can be isolated readily in vitro, and resistance is the result of the S282T substitution in the NS5B gene (S282T NS5B) (33). It has been shown that the S282T substitution increases both 50% inhibitory concentrations and Ki values for 2′-C-methyl-modified nucleotides as well as the ability to extend an analogue-terminated primer relative to that of the wild type (WT) (33). It was suggested that the S282T mutation induces resistance by increased discrimination of the 2′-C-methyl nucleotide analogue at the NS5B active site relative to the natural substrate NTP. However, the molecular details of S282T mutant-mediated resistance are still unclear. Discrimination can result from a selective decrease of either the binding of the analogue (reflected by an increased Km) or the catalytic step of analogue incorporation into RNA (reflected by the decrease of the maximum velocity value [Vmax]). Resistance can also be achieved by the excision of the incorporated drug (32). This particular mechanism is described in vitro for the bovine viral diarrhea virus RdRp, in the case of incorporation of 3′-dNMP (10). No reparation or excision of an NTP analogue has been reported yet for the HCV NS5B polymerase, and the mechanism leading to discrimination (increased Km or decreased Vmax) of the 2′-C-methyl analogue in the context of S282T mutant resistance is not known.
The Flaviviridae NS5b protein is able to initiate RNA synthesis without an RNA primer in a so-called de novo RNA synthesis process as well as to elongate an existing RNA primer (27, 38, 50). The RNA synthesis initiation phase is defined as the formation of the first phosphodiester bridge between two nucleotides to form a diribonucleotide primer. So far, primer-independent RNA synthesis is unique to viral RNA polymerases. Through evolution, these polymerases have selected unique structural features essential to the synthesis of their own short RNA primers (4, 6, 9). The crystal structure of the HCV polymerase and elegant enzymatic assays have identified some of these structural determinants (4, 19, 24). A peculiar β-strand-turn-β-strand (flap) subdomain might belong to the latter. The flap actually obstructs the polymerase active site when the site is compared to those of related primer-dependent polymerases. It has been proposed that the flap gives physical support to initiating nucleotides, up to the point where a conformational change occurs at the polymerase active site (4, 19, 49). Subsequently, the polymerase has to adopt an alternate conformation in which the primer is elongated in a processive fashion, giving rise to double-stranded RNA (dsRNA). Recently, the flap was shown to play a role in the repression of primer-directed RNA synthesis in favor of the initiation of RNA synthesis (41).
Using different template systems discriminating initiation from elongation, we have previously shown that these two steps of RNA synthesis are structurally and kinetically distinct and may thus have different nucleotide analogue susceptibilities (15). For example, 2′-O-methyl GTP is incorporated during the RNA elongation step only by WT NS5B. In view of optimal future HCV drug combination therapies, it is of prime importance to understand, at the molecular level, which viral RNA synthesis steps are targeted by a given drug as well as which resistance mechanisms are at play at the NS5B active site.
In this work, we have analyzed the use of 2′-C-methyl and 2′-O-methyl CTP (2′-C-Me-CTP and 2′-O-Me-CTP) and the natural CTP substrate by WT and S282T NS5B during initiation and elongation of RNA synthesis. We show that they have opposite resistance profiles during initiation and elongation of RNA synthesis and that the S282T mutant induces a general decrease in the efficiency of incorporation of natural nucleotides.
MATERIALS AND METHODS
HCV 1b polymerase, plasmid constructs, enzyme preparation, and reagents.
WT and mutant NS5B-D55 genes were tagged with six C-terminal histidines and expressed from the pDest 14 vector (Invitrogen) in Escherichia coli BL21(DE3) cells (Novagen). Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). All constructs were verified by DNA sequencing. WT and S282T NS5B proteins were purified as described previously (15). RNA molecular weight markers were synthesized using T7 RNA polymerase and the appropriate template RNAs as previously described (15). RNA oligonucleotides were obtained from MWG-Biotech. DNA oligonucleotides were obtained from Life Technologies. For RNA elongation experiments, RNA oligonucleotides were 5′ α-32P labeled, using T4 polynucleotide kinase (New England Biolabs). γ-32P-labeled ATP (3,000 Ci/mmol), α-32P-labeled GTP (3,000 Ci/mmol), and α-32P-labeled CTP (3,000 Ci/mmol) were purchased from Amersham. 2′-O-Me-CTP was purchased from Trilink, Inc. 2′-C-Me-CTP was kindly provided by Idenix, Inc.
Steady-state kinetics of RNA synthesis using homopolymeric templates.
Polymerase activity was assayed by monitoring the incorporation of radiolabeled guanosine into 15-mer cytidine RNA oligonucleotide templates, as described previously (15).
Determination of Vmax and Km.
An RNA oligonucleotide corresponding to the 3′ end of the negative strand of the HCV genome [RNA H (−) (5′-UCGGGGGCUGGC-3′)] or corresponding to a modified 3′ end of the positive strand of the HCV genome [RNA H (+2) (5′-CAGAUCAGGU-3′)] was used to analyze the synthesis of the first GC or AC phosphodiester bond. Reactions were performed as described previously (15), using 100 μM [α-32P]CTP (1 μCi) and GTP or ATP (1, 5, 10, 50, 100, and 500 μM). For Vmax and Km values for CTP or CTP analogue incorporation upon initiation, reactions were assayed with 100 μM [α-32P]GTP (1 μCi), using increasing concentrations of either CTP, 2′-C-Me-CTP, or 2′-O-Me-CTP (1, 5, 10, 50, 100, and 500 μM). RNA elongation was measured using a 5′ α-32P-labeled hairpin RNA template (HP4 [5′-UGACGGCCCGGAAAACCGGGCC-3′] for GTP incorporation, HP3 [5′-ACUGGGCCCGGAAAACCCGGGCC-3′] for CTP incorporation, and HP2 [5′-GACUGGCCCGGAAAACCGGGCC-3′] for ATP incorporation). Products were separated using sequencing gel electrophoresis and quantitated using photostimulated plates and a Fuji imager. Product formation during the initial reaction was assayed as described previously (15) and fitted to a linear curve as follows: product = Vit, where t is the time in seconds. The dependence of Vi on the NTP concentration is described by the hyperbolic equation Vi = Vmax(NTP)/(Km + NTP), where Vmax and Km are the maximal velocity and the apparent affinity constant of NTP incorporation by NS5B, respectively. Vmax and Km were determined by curve fitting using Kaleidagraph (Synergy Software).
Molecular modeling.
An initiation complex model was built from the superposition of NS5B structures in complex with UTP (Protein Data Bank no. 1GX6) (4) or ssRNA (Protein Data Bank no. 1NB7) (37) onto the ternary complex of HIV-1 reverse transcriptase (RT; Protein Data Bank no. 1RTD) (20). The final model consists of WT NS5B, ssRNA, GTP in the +1 position, and CTP in the +2 position. CTP was then replaced by 2′-C-Me-CTP or 2′-O-Me-CTP. The replacement of residue 282 by a threonine led to the mutant model of the initiation complex. All models were constructed with the TURBO program (42).
RESULTS
2′-C-Me-CTP and 2′-O-Me-CTP give opposite S282T mutant-mediated resistance profiles during initiation.
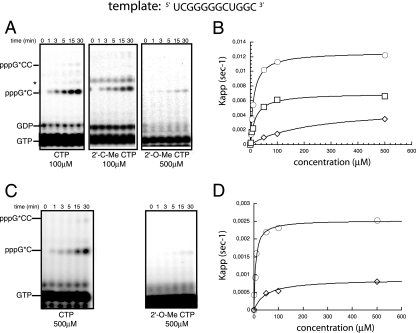
Figure 1 shows RNA synthesis using the HCV (−) 3′-end RNA as a template. Table 1 shows the corresponding enzymatic constants determined after product quantitation as described previously (15). WT NS5B incorporates 2′-C-Me-CMP and CMP to the same extent. Comparatively, although still incorporated, 2′-O-Me-CTP yields a pppGC2′-O-Me product with a much lower efficiency (Table 1 and Fig. 1B). Indeed, the Km and Vmax values are similar for 2′-C-Me-CTP and CTP, whereas there is a 14-fold increase in Km for 2′-O-Me-CTP (Km 2′-O-methyl-CTP = 189 μM and Km CTP = 13 μM). As previously reported, 2′-C-Me-cytidine and 2′-O-Me-cytidine are effective chain terminators when incorporated into nascent RNA. No elongation of the pppGC product was observed when products were terminated with these analogues, whereas the pppGCC product was formed in the presence of the natural CTP (Fig. 1A).
FIG. 1.
Incorporation of CTP, 2′-O-Me-CTP, and 2′-C-Me-CTP into RNA during initiation. (A) Time course of CTP, 2′-O-Me-CTP, and 2′-C-Me-CTP incorporation into RNA during initiation by WT NS5B polymerase, analyzed by 20% denaturing gel electrophoresis. The RNA template (shown at the top) allows the synthesis of pppGC and ppGCC products as described in Materials and Methods. The migration positions of radiolabeled GTP, pppGC, and pppGCC are shown on the left. (B) The experiment shown in panel A was repeated for various CTP and CTP analogue concentrations, the pppGC product was quantitated, and the initial velocity was calculated for each CTP or CTP analogue concentration. Results are represented as the initial velocity for each CTP (○), 2′-C-Me-CTP (□), or 2′-O-Me-CTP (⋄) concentration as a function of the NTP concentration. Hyperbolic fitting of the data was used to determine values of the enzymatic constants Vmax and Km. (C) The experiment shown in panel A was performed using the S282T NS5B mutant under the same conditions. (D) Product formation by the S282T NS5B mutant obtained with different CTP and CTP analogue concentrations was quantitated, and results were fitted to a hyperbolic equation to determine Vmax and Km values.
TABLE 1.
Steady-state constants of wild-type and S282T NS5B polymerases for RNA templates during initiationa
| NS5B polymerase | Nucleotide | Km (μM) | Vmax (s−1) | Vmax/Km (s−1/μM) | Discrimination (fold) | Resistance (fold) |
|---|---|---|---|---|---|---|
| WT | CTP | 13 ± 0.4 | 12 × 10−3 ± 0.7 × 10−3 | 9.2 × 10−4 | ||
| 2′-O-Me-CTP | 189 ± 24 | 4.7 × 10−3 ± 0.1 × 10−3 | 2.5 × 10−5 | 36 | ||
| 2′-C-Me-CTP | 20 ± 6 | 7 × 10−3 ± 0.3 × 10−3 | 3.5 × 10−4 | 2.5 | ||
| S282T mutant | CTP | 7 ± 1 | 2.5 × 10−3 ± 0.07 × 10−3 | 3.5 × 10−4 | ||
| 2′-O-Me-CTP | 48 ± 10 | 0.9 × 10−3 ± 0.05 × 10−3 | 1.6 × 10−5 | 21 | 0.7 | |
| 2′-C-Me-CTP | ND | ND | 0 |
Discrimination was determined as the ratio (Vmax/Km)CTP/(Vmax/Km)analogue. Resistance was determined as the ratio (Vmax/Km)WT/(Vmax/Km)mutant. Km and Vmax were calculated as described in Materials and Methods. Vmax is expressed in pmol s−1 · pmol−1 enzyme. ND, not detectable.
S282T NS5B is able to incorporate the natural CTP substrate leading to the formation of the pppGC and pppGCC products (Fig. 1C). However, there is a threefold decrease in the catalytic efficiency in the incorporation of CTP (Vmax/ Km CTP = 0.35 × 10−3) relative to that of the WT (Vmax/ Km CTP = 0.92 × 10−3) (Table 1) due to a fivefold decrease of Vmax. We then measured the incorporation of CTP analogues but could not detect any use of 2′-C-Me-CTP, reflecting a drastic effect of the S282T mutation. However, 2′-O-Me-CTP is still a substrate for the S282T mutant (Fig. 1C), but there is a dramatic effect on Vmax, which is sixfold lower (Vmax = 0.8 × 10−3 s−1) than that of the WT enzyme (Vmax = 4.7 × 10−3 s−1) (Table 1 and Fig. 1D). This Vmax decrease is nevertheless balanced by a fourfold increase in affinity (Km 2′-O-Me-CTP = 48 μM for the S282T mutant and 189 μM for the WT) (Table 1 and Fig. 1D). This leads to a catalytic efficiency similar to that of the WT enzyme and, therefore, to no resistance (Table 1).
Modeling of CTP and CTP analogues in the NS5B initiation complex.
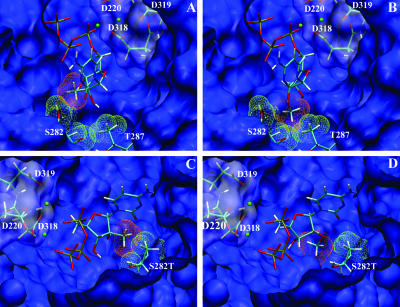
In order to gain further insight into the mechanism leading to the discrimination of 2′-O-Me-CTP, we generated a model based on the NS5B crystal structure (5, 24) and various structural polymerase-substrate complexes (4, 37). Available structural data are believed to be representative of the initiation complex. Thus, our model should only be relevant to results obtained from the analysis of the initiation steps of RNA synthesis. As previously described (33), 2′-C-Me-CTP is found in a position similar to that observed for the natural CTP (not shown), and no steric hindrance is observed (Fig. 2A). In contrast, the addition of the methyl group at the 2′-O position of the sugar changes the natural 3′-endo conformation of the sugar to a 2′-endo conformation (44). Therefore, the 2′-O-methyl group is found in close proximity to threonine 287 (Fig. 2B). This leads to an apparent steric hindrance between the methyl group of the modified CTP and both the methyl group of threonine 287 and the backbone of serine 282.
FIG. 2.
Modeling of 2′-C-Me-CTP and 2′-O-Me-CTP in the active site of the WT NS5B and S282T mutant polymerases. The GTP and ssRNA of initiation complex models are not shown. The active site of WT (A and B) or S282T (C and D) NS5B appears as a blue surface, with catalytic residues represented by sticks with a transparent surface and Mg2+ ions represented as green spheres. Residues 282 and 287 appear as sticks, with part of the surface shown as a green grid. Nucleotide analogues (2′-C-Me-CTP [A and C] and 2′-O-Me-CTP [B and D]) appear as sticks, with the surface of the methyl group shown as an orange grid. The figure was created with VMD (21).
To understand the effect of the resistance mutation S282T, the relative position of the 2′-modified nucleotide was also analyzed in the context of this substitution (Fig. 2C and D). For the sake of clarity, the view in Fig. 2C and D is rotated 180° relative to that in Fig. 2A and B. Strikingly, in the S282T NS5B active site, the 2′-C-methyl group is found in a position leading to a drastic steric clash with both the methyl group and the hydroxyl group of the substituted threonine at position 282 (Fig. 2C). This steric clash might not be compatible with binding of this analogue by the S282T NS5B and is consistent with the lack of incorporation observed in vitro at the initiation step of RNA synthesis (Fig. 1 and Table 1). In contrast, the accommodation of the 2′-O-Me-CTP is similar to that for the WT. The steric hindrance involving T287 is still present, and no additional hindrance is induced by the mutation of serine 282 to threonine (Fig. 2D). This observation is consistent with the 21-fold discrimination of this analogue by S282T NS5B, as measured in polymerase assays (Fig. 1 and Table 1).
Both 2′-C-Me-CTP and 2′-O-Me-CTP are discriminated by S282T NS5B during elongation.
We made use of a hairpin RNA primer template mimicking the elongation step of RNA synthesis (7, 33) (Fig. 3). In contrast to the case for initiation, both 2′-O-Me-CTP and 2′-C-Me-CTP are incorporated into the hairpin RNA to a similar extent as that for CTP (threefold discrimination) (Table 2 and Fig. 3B). This result reflects a marked relaxation of 2′-O-methyl group discrimination at the polymerase active site, leading to a 10-fold increase in inhibitor sensitivity once the enzyme has entered the elongation phase. Surprisingly, in the case of S282T NS5B, this scenario is exactly the opposite of what is observed during the initiation step: no incorporation of 2′-O-Me-CTP can be detected, while 2′-C-Me-CTP is incorporated into the hairpin RNA (Fig. 3C and D). However, the S282T substitution promotes a decrease in the apparent affinity for 2′-C-Me-CTP, which is reflected by an increased Km (170 μM) (Table 2). The 2′-C-Me-CTP analogue is discriminated 21-fold by the S282T enzyme relative to CTP, resulting in a ninefold resistance to 2′-C-Me-CTP (Table 2). In the case of 2′-O-Me-CTP, no incorporation can be detected, leading to complete resistance.
FIG. 3.
Incorporation of CTP, 2′-C-Me-CTP, and 2′-O-Me-CTP into RNA during elongation. (A) Time course of incorporation of CTP, 2′-C-Me-CTP, and 2′-O-Me-CTP into RNA hairpin template (1 mM, shown on the top) by WT NS5B, analyzed by 14% acrylamide denaturing gel electrophoresis. Migration positions of the hairpin and hairpin +1 template are shown on the left. (B) The experiment shown in panel A was repeated for various CTP and CTP analogue concentrations, the +1 product was quantitated, and the initial velocity was calculated for each CTP or CTP analogue concentration. Results are represented as the initial velocity for each CTP (○), 2′-C-Me-CTP (□), or 2′-O-Me-CTP (⋄) concentration as a function of the NTP concentration. Hyperbolic fitting of the data was used to determine values of the enzymatic constants Vmax and Km. (C) The experiment shown in panel A was performed using the S282T NS5B mutant under the same conditions. (D) Product formation by the S282T NS5B mutant with different CTP and CTP analogue concentrations was quantitated, and the results were fitted to a hyperbolic equation to determine Vmax and Km values.
TABLE 2.
Steady-state constants of wild-type and S282T NS5B polymerases for RNA templates during elongationa
| NS5B polymerase | Nucleotide | Km (μM) | Vmax (s−1) | Vmax/Km (s−1/μM) | Discrimination (fold) | Resistance (fold) |
|---|---|---|---|---|---|---|
| WT | CTP | 12 ± 4 | 68 × 10−3 ± 3 × 10−3 | 5.6 × 10−3 | ||
| 2′-O-Me-CTP | 16 ± 3 | 34 × 10−3 ± 1 × 10−3 | 2 × 10−3 | 3 | ||
| 2′-C-Me-CTP | 20 ± 6 | 45 × 10−3 ± 3 × 10−3 | 2.3 × 10−3 | 2.5 | ||
| S282T mutant | CTP | 23 ± 9 | 25 × 10−3 ± 3 × 10−3 | 1 × 10−3 | ||
| 2′-O-Me-CTP | ND | ND | ND | |||
| 2′-C-Me-CTP | 170 ± 80 | 8 × 10−3 ± 1 × 10−3 | 4.7 × 10−5 | 21 | 9 |
Discrimination was determined as the ratio (Vmax/Km)CTP/(Vmax/Km)analogue. Resistance was determined as the ratio (Vmax/Km)WT/(Vmax/Km)mutant. Km and Vmax were calculated as described in Materials and Methods. Vmax is expressed in pmol s−1 · pmol−1 enzyme. ND, not detectable.
General loss of efficiency toward natural nucleotides by S282T NS5B.
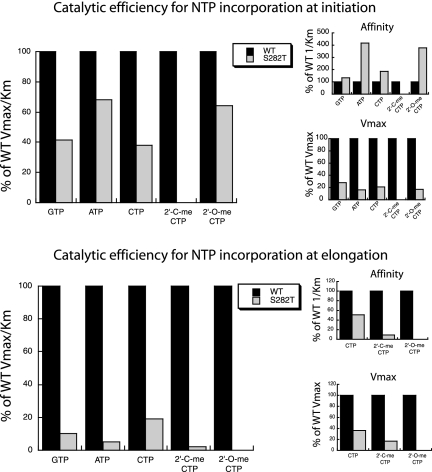
We noticed that the CTP incorporation efficiency is decreased threefold for S282T NS5B relative to that of the WT. If such a loss of efficiency is true for other NTPs, then this single NS5B substitution might be one of the factors responsible for a loss of fitness at the viral level. In order to gain insight into the molecular mechanism of this reduced substrate efficiency, we measured the efficiencies of incorporation of natural nucleotides into RNA. We first looked at the kinetics of incorporation of GTP, using an oligo(C)15 RNA template (Fig. 4). The synthesis of the G2 product, reflecting the initiation step of RNA synthesis (15), is barely affected by the S282T mutation, nor are the G3 and G4 products (Fig. 4). However, the amounts of G5 and longer products clearly decrease (30% of the amount of product synthesized by the WT enzyme) (Fig. 4B), although they are still detectable, suggesting a more drastic effect of the S282T mutation on the elongation step of RNA synthesis. It is also apparent from Tables 1 and 2 that the S282T mutation impedes incorporation of the natural CTP substrate (see above). GTP or ATP and CTP are the nucleotides incorporated during initiation in vivo at the +1 and +2 positions, respectively. Thus, since UTP is never used during this step, it was not investigated. For both initiation and elongation steps, GTP and ATP incorporation was affected by the S282T substitution, with a general decrease in catalytic efficiency (Fig. 5 and Tables 3 and 4). However, a more pronounced effect was observed during elongation, where the catalytic efficiencies for all substrates, either natural or 2′ modified, were below 20% of that for WT NS5B (Fig. 5 and Tables 2 and 4). UTP incorporation by WT NS5B was barely detected and was undetectable when assayed with S282T NS5B, confirming the same trend (not shown). The reduced catalytic efficiency for the incorporation of CTP or CTP analogues (Tables 1 and 2) and ATP or GTP (Tables 3 and 4) is the consequence of a general decrease of Vmax during both initiation and elongation (Fig. 5, insets). However, during initiation only, this Vmax decrease is balanced by a two- to fourfold increase of the apparent affinity for CTP or ATP (Fig. 5, insets). In summary, the S282T NS5B catalytic efficiency during initiation is 2-fold lower than that of the WT enzyme (Table 3), while it is reduced 5- to 20-fold during elongation (Fig. 5 and Table 4).
FIG. 4.
Kinetics of RNA synthesis by WT and S282T NS5B polymerases, using oligo(C) as a template. (A) RNA products of reactions by either WT or S282T NS5B were resolved in a 14% polyacrylamide-7 M urea gel. RNA markers were synthesized using T7 RNA polymerase as described in Materials and Methods. Each product band is indicated on the right. (B) Comparative quantitation of reaction products from the experiment shown in panel A. Each individual Pi product band from WT or S282T NS5B was quantitated, and the percentage of the ratio of S282T(Pi)/WT(Pi) products was plotted for each i-mer product.
FIG. 5.
Comparative incorporation efficiencies for nucleotide utilization by WT and S282T mutant NS5B polymerases during initiation and elongation. Calculations were based on the values taken from Tables 1 and 3 for comparative incorporation during initiation and from Tables 2 and 4 for comparative incorporation during elongation. The catalytic efficiency (Vmax/Km) for each nucleotide is expressed as a percentage of the catalytic efficiency determined for WT NS5B polymerase. The percent binding affinity (1/Km) and maximal velocity (Vmax) were calculated in the same manner and are shown in the insets.
TABLE 3.
Steady-state constants of wild-type and S282T NS5B polymerases for RNA templates during initiationa
| NS5B polymerase | Nucleotide | Km (μM) | Vmax (s−1) | Vmax/Km (s−1/μM) | Efficiency decrease (fold) |
|---|---|---|---|---|---|
| WT | CTP | 13 ± 4 | 12 × 10−3 ± 7 × 10−3 | 0.92 × 10−3 | |
| GTP | 9.97 ± 0.9 | 11 × 10−3 ± 0.3 × 10−3 | 1 × 10−3 | ||
| ATP | 46 ± 7 | 38 × 10−3 ± 1.6 × 10−3 | 0.82 × 10−3 | ||
| S282T mutant | CTP | 7 ± 1 | 2.5 × 10−3 ± 0.07 × 10−3 | 0.35 × 10−3 | 2.6 |
| GTP | 7.46 ± 3.5 | 3.1 × 10−3 ± 0.3 × 10−3 | 0.415 × 10−3 | 2.4 | |
| ATP | 11 ± 3 | 6.2 × 10−3 ± 0.3 × 10−3 | 0.56 × 10−3 | 1.46 |
The efficiency decrease was determined as the ratio (Vmax/Km)WT/(Vmax/Km)mutant. Km and Vmax were calculated as described in Materials and Methods. Vmax is expressed in pmol s−1 · pmol−1 enzyme.
TABLE 4.
Steady-state constants of wild-type and S282T NS5B polymerases for RNA templates during elongationa
| NS5B polymerase | Nucleotide | Km (μM) | Vmax (s−1) | Vmax/Km (s−1/μM) | Efficiency decrease (fold) |
|---|---|---|---|---|---|
| WT | CTP | 12 ± 4 | 68 × 10−3 ± 3 × 10−3 | 5.2 × 10−3 | |
| GTP | 63 ± 19 | 6.2 × 10−3 ± 0.7 × 10−3 | 0.1 × 10−3 | ||
| ATP | 41 ± 7 | 7.5 × 10−3 ± 0.4 × 10−3 | 0.2 × 10−3 | ||
| S282T mutant | CTP | 23 ± 9 | 25 × 10−3 ± 3 × 10−3 | 1 × 10−3 | 5.2 |
| GTP | ND | ND | 0.01 × 10−3 | 10 | |
| ATP | ND | ND | 0.01 × 10−3 | 20 |
The efficiency decrease was determined as the ratio (Vmax/Km)WT/(Vmax/Km)mutant. Km and Vmax were calculated as described in Materials and Methods. Vmax is expressed in pmol s−1 · pmol−1 enzyme. ND, not detectable.
We conclude that the molecular determinants of S282T NS5B-mediated 2′-C-Me analogue resistance are completely discriminated during initiation and discriminated 21-fold during the elongation step of RNA synthesis. Strikingly, for 2′-O-Me-CTP incorporation, the picture is opposite, leading to a similar incorporation during initiation but to apparently complete discrimination during elongation. This S282T mutation-mediated resistance, however, drastically reduces the polymerization efficiency of natural nucleotides, raising interesting perspectives about viral fitness and its use in optimized anti-HCV therapeutic protocols.
DISCUSSION
In this study, we have characterized the molecular mode of action of triphosphate derivatives of 2′-O-Me- and 2′-C-Me-cytidine, with the latter currently being evaluated in clinical trials. 2′-C-Me-CTP is incorporated as efficiently as CTP by the WT enzyme during either initiation or elongation, consistent with the antiviral activities observed in patients after 2 weeks of treatment (1). 2′-O-Me-CTP has also been described as an HCV polymerase inhibitor which is active as a chain terminator (7), with a 50% inhibitory concentration measured by RdRp assay similar to that of 2′-C-Me-ATP. In contrast to 2′-C-Me-CTP, we show that 2′-O-Me-CTP is preferentially incorporated during elongation, as is 2′-O-Me-GTP (15). The 36-fold discrimination of 2′-O-Me-CTP at initiation is probably due to steric hindrance by the methyl group of threonine 287 during this step. During elongation, no discrimination is observed, and the 2′-O-Me-CTP is incorporated equally as well as the natural CTP or 2′-C-Me-CTP. This suggests that important modifications of the active site occur during the switch from initiation to elongation, leading to the displacement of threonine 287 and the relaxation of the steric hindrance induced by this residue during initiation. These results are consistent with our previous observation that the conformation of the active site during elongation is more permissive to sugar modifications than that during initiation (15).
Conformational changes of the active site are also illustrated by the analysis of the S282T mutant enzyme. This mutant was selected and isolated after treatment of replicon-expressing cells with a suboptimal concentration of 2′-C-Me adenosine or guanosine (33). It was reported that the S282T substitution confers resistance to both 2′-C-Me nucleosides but does not affect 2′-O-Me nucleoside susceptibility. The molecular basis of this differential resistance profile for these closely related 2′-modified analogues is interesting. We show in this study that the S282T mutation-mediated 2′-C-Me-CTP resistance is the result of a steric hindrance translated at the molecular level, with an increased Km and an unchanged Vmax during elongation. During initiation, this steric hindrance is drastic, and no incorporation is detected, probably due to a steric clash between the methyl group of the modified analogue and the methyl group of the substituting threonine (Fig. 2). It was recently reported that HCV and bovine viral diarrhea virus polymerase resistance to nucleotide analogues could also be the result of pyrophosphorolysis leading to the excision of the analogue after its incorporation into the nascent RNA (10, 14). No pyrophosphorolysis could be observed in our experimental system with either WT or S282T NS5B, probably because of specific template requirements for this particular mechanism (14). Nevertheless, it was shown that S282T NS5B is able to excise chain-terminator analogues to the same extent as WT NS5B (14). Therefore, S282T mutant-mediated resistance to 2′-C-Me-NTP is only due to Km-based discrimination (Table 2).
Surprisingly, we noticed that the S282T substitution also impairs the incorporation of 2′-O-Me-CTP (Tables 1 and 2). This mutation, however, does not lead to resistance toward this analogue in the replicon system (33). We have no explanation for this discrepancy. However, RNA synthesis in vivo occurs in a replication complex involving other nonstructural proteins, which might influence the three-dimensional conformation of NS5B. Thus, the structure of the NS5B active site might be slightly different in the replication complex than the one determined with the isolated enzyme. Alternatively, and perhaps more likely, 2′-O-Me-CTP may inhibit another target besides NS5B when RNA synthesis is measured either from a purified replicative complex or in the replicon system. Nevertheless, in our in vitro RdRp assay, S282T NS5B was fully resistant to 2′-O-Me-CTP during elongation (Table 2) but was as competent as WT NS5B for incorporation during initiation. Again, our results illustrate an important conformational change occurring during the switch from initiation to elongation.
Mutations conferring resistance to HCV polymerase or protease inhibitors are often associated with a decrease in replicative fitness (25, 26, 34), although the molecular mechanisms are not fully understood. The S282T mutation is associated with such replicative defects (26). Our study suggests that reduced fitness may be due to a decrease in the catalytic rate (Vmax) of incorporation for all natural nucleotides, with the affinity (Km) being unaffected. Surprisingly, the S282T mutation also affects the incorporation of GTP and ATP during initiation, although this mutation is located 10 Å away from the +1 position, where GTP or ATP is positioned during this step (4). This suggests that the S282T mutation may have a long-range effect, e.g., an effect transmitted through the stacking of nucleotides +2 and +1. We did not observe a burst of product formation under any circumstances, using either a preformed NS5B-template or NS5B-nucleotide complex with either WT or S282T NS5B, ruling out a low dissociation rate as a rate-limiting step (not shown). Thus, variation of the dissociation rate of S282T NS5B does not explain the differences in Vmax observed with different nucleotides.
For the poliovirus 3D polymerase, it has been suggested that the correct positioning of the NTP is dependent on its correct binding at both the sugar binding site and the triphosphate binding site (8). The HCV NS5B sugar binding site is made up of serine 282, asparagine 291, and glutamic acid 225 (4). As in the poliovirus model, the ribose in the NS5B active site is thought to be held firmly by an interaction between its 3′-OH and the backbone of D225, in addition to its 2′-OH and both N291 and S282. Indeed, any alteration in this hydrogen bond network, by either sugar modification or amino acid substitutions, would induce a reduced stabilization of the ribose moiety and a probable loosening of binding at both the sugar and triphosphate binding sites (8). Based on this selectivity model, the S282T substitution in the HCV polymerase is indeed able to alter the sugar binding site, and hence triphosphate positioning, after promoting local rearrangement up to the catalytic center (Vmax effect).
Our results might have clinical relevance. Although no resistant viruses have been reported yet for NM283 clinical trials, the increase in viral load during monotherapy suggests the appearance of such resistant mutants (1). If this mutation appears in the clinic and in the absence of compensatory mutations (34), the resulting mutant viruses will have an impaired replication capacity that may translate into better control by the immune system or by other drugs. It is important to draw from the HIV-1 experience. The nucleoside analogue lamivudine is an RT inhibitor eliciting resistance through the emergence of the M184V substitution in RT (48). The M184V mutation alters viral fitness through a decreased incorporation efficiency of natural nucleotides by RT (13). It has been suggested, however, that there is a clinical benefit to maintaining the presence of the M184V substitution by uninterrupted lamivudine treatment (47). Indeed, the loss of viral fitness potentiates the efficacy of additional HIV-1 inhibitors (12). If the S282T mutation arises in patients, then the S282T mutation and 2′-C-methyl cytosine may well be the first example of such a rational approach of drug-resistant HCV therapy through the control of the fitness/resistance balance.
Acknowledgments
We thank B. Selisko, K. Alvarez, J. C. Guillemot, E. Decroly, and D. Storer for critical readings of the manuscript and helpful discussions. We thank J. L. Romette for handling intellectual property rights.
This work was supported by the European Community (Flavitherapeutics European contract no. QLK3-CT-2001-00506), the Association pour la Recherche contre le Cancer (ARC), and the Centre National de Recherche Scientifique (CNRS). H.D. is an INSERM appointee.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Afdhal, N., E. Godofsky, J. Dienstag, V. Rustgi, L. Shick, D. Duncan, X. J. Zhou, G. Chao, C. Fang, B. Fielman, M. Myers, and N. Brown. 2004. Final phase I/II trial results for NM283, a new polymerase inhibitor for hepatitis C: antiviral efficacy and tolerance in patients with HCV-1 infection, including previous interferon failure. AASLD, Boston, Mass.
- 2.Afdhal, N., M. Rodriguez-Torres, F. Lawitz, E. Godofsky, G. Chao, B. Fielman, S. Knox, and N. Brown. 2005. Enhanced antiviral efficacy for valopicitabine (NM283) plus peg-interferon in hepatitis C patients with HCV genotype-1 infection: results of a phase IIa multicenter trial. EASL, Paris, France.
- 3.Beaulieu, P. L., M. Bos, Y. Bousquet, G. Fazal, J. Gauthier, J. Gillard, S. Goulet, S. LaPlante, M. A. Poupart, S. Lefebvre, G. McKercher, C. Pellerin, V. Austel, and G. Kukolj. 2004. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery and preliminary SAR of benzimidazole derivatives. Bioorg. Med. Chem. Lett. 14:119-124. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 8.Castro, C., J. J. Arnold, and C. E. Cameron. 2005. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 107:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K. H., J. M. Groarke, D. C. Young, R. J. Kuhn, J. L. Smith, D. C. Pevear, and M. G. Rossmann. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. USA 101:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Abramo, C. M., L. Cellai, and M. Gotte. 2004. Excision of incorporated nucleotide analogue chain-terminators can diminish their inhibitory effects on viral RNA-dependent RNA polymerases. J. Mol. Biol. 337:1-14. [DOI] [PubMed] [Google Scholar]
- 11.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 12.Deval, J., J. M. Navarro, B. Selmi, J. Courcambeck, J. Boretto, P. Halfon, S. Garrido-Urbani, J. Sire, and B. Canard. 2004. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J. Biol. Chem. 279:25489-25496. [DOI] [PubMed] [Google Scholar]
- 13.Deval, J., K. L. White, M. D. Miller, N. T. Parkin, J. Courcambeck, P. Halfon, B. Selmi, J. Boretto, and B. Canard. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279:509-516. [DOI] [PubMed] [Google Scholar]
- 14.Deval, J., C. M. D'Abramo, L. Celai, and M. Gotte. 2005. Selective excision of chain terminating nucleotides by hepatitis C virus NS5B polymerase. Presented at 12th Symposium on HCV, Montreal, Canada, 2 to 6 October 2005.
- 15.Dutartre, H., J. Boretto, J. C. Guillemot, and B. Canard. 2005. A relaxed discrimination of 2′-O-methyl-GTP relative to GTP between de novo and elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J. Biol. Chem. 280:6359-6368. [DOI] [PubMed] [Google Scholar]
- 16.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 17.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 18.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 19.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 20.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27-28, 33-38. [DOI] [PubMed] [Google Scholar]
- 22.Kukolj, G., G. A. McGibbon, G. McKercher, M. Marquis, S. Lefebvre, L. Thauvette, J. Gauthier, S. Goulet, M. A. Poupart, and P. L. Beaulieu. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280:39260-39267. [DOI] [PubMed] [Google Scholar]
- 23.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 24.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 26.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, H., V. Leveque, A. De Witte, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 29.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 30.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 31.McKercher, G., P. L. Beaulieu, D. Lamarre, S. LaPlante, S. Lefebvre, C. Pellerin, L. Thauvette, and G. Kukolj. 2004. Specific inhibitors of HCV polymerase identified using an NS5B with lower affinity for template/primer substrate. Nucleic Acids Res. 32:422-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 33.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 34.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni, Z. J., and A. S. Wagman. 2004. Progress and development of small molecule HCV antivirals. Curr. Opin. Drug Discov. Dev. 7:446-459. [PubMed] [Google Scholar]
- 37.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 38.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 41.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 42.Roussel, A., and C. Cambillau. 1991. TURBO-FRODO. Silicon Graph. Geometry Partners Directory 86:77-78. [Google Scholar]
- 43.Somsouk, M., G. M. Lauer, D. Casson, A. Terella, C. L. Day, B. D. Walker, and R. T. Chung. 2003. Spontaneous resolution of chronic hepatitis C virus disease after withdrawal of immunosuppression. Gastroenterology 124:1946-1949. [DOI] [PubMed] [Google Scholar]
- 44.Tomassini, J. E., K. Getty, M. W. Stahlhut, S. Shim, B. Bhat, A. B. Eldrup, T. P. Prakash, S. S. Carroll, O. Flores, M. MacCoss, D. R. McMasters, G. Migliaccio, and D. B. Olsen. 2005. Inhibitory effect of 2′-substituted nucleosides on hepatitis C virus replication correlates with metabolic properties in replicon cells. Antimicrob. Agents Chemother. 49:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner, D., B. G. Brenner, J. P. Routy, M. Petrella, and M. A. Wainberg. 2004. Rationale for maintenance of the M184V resistance mutation in human immunodeficiency virus type 1 reverse transcriptase in treatment experienced patients. New Microbiol. 27:31-39. [PubMed] [Google Scholar]
- 48.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti-Infect. Ther. 2:147-151. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]