Abstract
The lfrA gene of Mycobacterium smegmatis encodes an efflux pump which mediates resistance to different fluoroquinolones, cationic dyes, and anthracyclines. The deletion of the lfrR gene, coding for a putative repressor and localized upstream of lfrA, increased the lfrA expression. In this study, reverse transcription-PCR experiments showed that the two genes are organized as an operon, and lacZ reporter fusions were used to identify the lfrRA promoter region. The lfrRA promoter assignment was verified by mapping the transcription start site by primer extension. Furthermore, we found that some substrates of the multidrug transporter LfrA, e.g., acriflavine, ethidium bromide, and rhodamine 123, enhance lfrA expression at a detectable level of transcription. LfrR protein was purified from Escherichia coli as a fusion protein with a hexahistidine tag and found to bind specifically to a fragment 143 bp upstream of lfrR by gel shift analysis. Furthermore, acriflavine was able to cause the dissociation of the LfrR from the promoter, thus suggesting that this molecule interacts directly with LfrR, inducing lfrA expression. These results suggest that the LfrR repressor is able to bind to different compounds, which allows induction of LfrA multidrug efflux pump expression in response to these ones. Together, all data suggest that the LfrA pump is tightly regulated and that the repression and induction can be switched about a critical substrate concentration which is toxic for the cell.
Multidrug resistance has emerged as a major clinical problem and can arise through a number of mechanisms, including the action of efflux transporters that pump out a wide variety of structurally and chemically dissimilar drugs, dyes, and other compounds (49). Multidrug efflux transporters are membrane proteins found in both prokaryotes and eukaryotes and are classified into five families. Two of these are large and ancient superfamilies known as the ATP-binding cassette (ABC) superfamily and the major facilitator superfamily (MFS). The other three are smaller families: the resistance-nodulation-cell division (RND) family, the small multidrug resistance (SMR) family, and the multidrug and toxic compound extrusion (MATE) family (25). The mechanism used to bind and export a broad range of substrates remains poorly understood, largely due to the difficulties posed by the structural analysis of integral membrane proteins. In the case of bacteria, an alternative approach has been the study of proteins that regulate the expression of specific multidrug transporters. The data available today show that multidrug transporters are often expressed under precise and elaborate control at the level of transcription (13). Examples of both repressors (12, 27, 28) and activators (1, 24) of transcription whose genes are adjacent to that for the transporter have been described. Many of these regulators are local repressors that directly interact with the promoter regions of multidrug resistance (MDR) efflux genes or operons. For example, repressors QacR (Staphylococcus aureus), MtrR (Neisseria gonorrhoeae), AcrR (Escherichia coli), and MexR (Pseudomonas aeruginosa) bind specifically to the promoter sequences of qacA, mtrCDE, acrAB, and mexAB efflux pump-encoding genes, respectively, thereby inhibiting transcription of these genes (9, 12, 15, 29).
Overexpression of multidrug resistance pumps, resulting in increased bacterial resistance, is usually due to mutations in these regulatory genes (9, 12, 16, 36). For these reasons, the study of the regulation of MDR efflux gene expression is an important issue in the field of antibiotic resistance. Furthermore, an increasing number of efflux pump genes has also been found to be controlled by global transcriptional activator proteins (13) or by two-component regulatory systems (25).
Mycobacterium tuberculosis is the infectious agent responsible for tuberculosis. Tuberculosis is often difficult to treat, because M. tuberculosis is intrinsically resistant to most common antibiotics, apparently because of its extremely low cell wall fluidity and permeability (3, 20, 46). The situation is made worse by the dramatic increase in multidrug-resistant strains. About 50 million people are presently infected with MDR M. tuberculosis strains, defined as resistant to both isoniazid and rifampin, the two first-line drugs used to treat tuberculosis (8). Along with cell wall permeability, active efflux systems also provide resistance by extruding the drugs that enter the cell. Therefore, it seems reasonable to characterize efflux pump-mediated multidrug resistance in mycobacteria by using Mycobacterium smegmatis as the model organism. Despite several mycobacterial efflux pumps having been characterized (5, 26, 48), their involvement in intrinsic and acquired drug resistance in mycobacteria remains unresolved (5), except for the study of the lfrA gene in M. smegmatis (40). LfrA is an MFS transporter that confers resistance to ethidium bromide, acriflavine, and some fluoroquinolones when overexpressed from a multicopy plasmid (43). Disruption of the lfrA gene rendered the mutant more susceptible to ethidium bromide, acriflavine, ciprofloxacin, doxorubicin, and rhodamine 123 (two- to eightfold decrease in MICs) (40). These results were also confirmed by Li et al. (26). The upstream region of lfrA contains a gene coding for a putative TetR family transcriptional repressor, named LfrR, hypothesized to be responsible for regulation of lfrA gene expression (6, 26). The deletion of lfrR increased the expression of lfrA and resulted in higher resistance to several drugs (26).
In this study, we demonstrate that lfrR and lfrA genes are cotranscribed by a common promoter. LfrR represses the transcription of the lfrRA operon by directly binding to the promoter region of lfrR-lfrA. We expressed and purified the LfrR protein from Escherichia coli cells, and we demonstrated that it binds to a 143-bp region upstream of the lfrR gene. Furthermore, we identified both the lfrRA promoter region and the transcriptional start site. To investigate the regulation of the lfrA gene, we analyzed its expression by reverse transcription-PCR (RT-PCR) in the presence of compounds hypothesized to be transported by the LfrA pump. Our results indicate that the LfrA pump is tightly regulated.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All cloning steps were performed in Escherichia coli DH5α grown in Luria-Bertani (LB) broth or on LB agar (38). M. smegmatis mc2155 wild-type and mc211 mutant strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) oleic acid/albumin/dextrose/catalase (OADC enrichment) and 0.05% Tween 80 or on Middlebrook 7H11 medium (Difco) supplemented with 10% (vol/vol) OADC. When necessary, antibiotics (Sigma) were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 34 μg/ml; carbenicillin, 50 μg/ml; and kanamycin, 25 μg/ml (for mycobacteria) and 50 μg/ml (for E. coli). All strains were grown aerobically at 37°C with shaking at 200 rpm.
DNA techniques.
DNA manipulations were performed by standard procedures as described by Sambrook and Russell (38). DNA restriction and modifying enzymes were used as recommended by the manufacturer (Amersham Biosciences). DNA fragments and PCR products were purified from agarose gels with the Qiaex kit (QIAGEN), unless otherwise specified, according to the manufacturer's instructions. Isolation of plasmid DNA was performed using the Plasmid Mini kit (QIAGEN) according to the manufacturer's instructions. Plasmid DNA was sequenced with the SP6 promoter primer by using an automatic DNA sequencer (ABI-PRISM 3100) (Applied Biosystems). Electroporation of E. coli cells was done as described previously (38). Electrocompetent mycobacterial cells were prepared and electroporated by using a Bio-Rad Gene Pulser, as described by Parish and Stoker (35). Following electroporation, M. smegmatis was plated onto Middlebrook 7H11 medium supplemented with 10% (vol/vol) OADC and 25 μg/ml of kanamycin.
PCR amplification.
All primers used for PCR are listed in Table 1. PCR amplifications were performed in a volume of 40 μl containing 200 μM of each deoxynucleoside triphosphate, 500 nM of each primer, 2.5 mM MgCl2, 2% dimethyl sulfoxide, 100 ng of mycobacterial DNA, and 5 U of Taq DNA polymerase (Sigma). The protocol used for amplification was as follows: denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing for 1 min at a temperature dependent on the primer pair used, and elongation at 72°C for a time dependent on the expected sizes of the products, with a final elongation at 72°C for 10 min.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Comment(s) |
|---|---|---|
| RG389 | TGCTGGTCCTGGCCGATCAC | Sense primer, amplifying a 672-bp fragment of the lfrRA operon |
| prexA | GCGATGAGCAGCACCGGAAGT | Antisense primer, amplifying a 672-bp fragment of the lfrRA operon |
| RG400 | TTGGATCCATCGGCGTCACTC (BamHI) | Sense primer for lfrR-lfrA intergenic region-lacZ fusion, amplifying a 75-bp DNA fragment |
| RG401 | TTGGATCCACTCGCGGCCTAC (BamHI) | Sense primer for lfrR-lfrA intergenic region-lacZ fusion, amplifying a 297-bp DNA fragment |
| RG402 | TAGGTACCTCATGAGAAGAGCT (KpnI) | Antisense primer for lfrR-lfrA intergenic region-lacZ fusion, used with RG400 and RG401 |
| RG404 | TAGGATCCATCCTCGGCGAG (BamHI) | Sense primer for upstream lfrR-lacZ fusion, amplifying a 351-bp DNA fragment of the lfrR upstream region |
| RG405 | TAGGATCCCGCCTCCTCCGC (BamHI) | Sense primer for upstream lfrR-lacZ fusion, amplifying a 227-bp DNA fragment of the lfrR upstream region |
| RG406 | TAGGATCCTGCCCGCCTATTC (BamHI) | Sense primer for upstream lfrR-lacZ fusion, amplifying a 143-bp DNA fragment of the lfrR upstream region |
| RG407 | TAGGATCCAGGTGAGTGGGA (BamHI) | Sense primer for upstream lfrR-lacZ fusion, amplifying a 62-bp or a 125-bp DNA fragment of the lfrR upstream region with primers RG409 and RG410, respectively |
| RG409 | TAGGTACCTGGTCATGTATCAA (KpnI) | Antisense primer for upstream lfrR-lacZ fusion, used with RG404, RG405, RG406 and RG407, respectively |
| RG408 | TAGGATCCACCAGCCCGAGCA (BamHI) | Sense primer for upstream lfrR-lacZ fusion, amplifying a 67-bp DNA fragment of the lfrR gene |
| RG410 | TAGGTACCGCATGGCGGCATCAA (KpnI) | Antisense primer for upstream lfrR-lacZ fusion, amplifying a 67-bp DNA fragment of the lfrR gene with RG408 |
| prexR | GAGTGCGGCGGTGGGGTGAT | Primer for primer extension |
| RG391 | CGCCCCGAGCACCGAGTT | Sense primer, amplifying a 547-bp DNA fragment of the lfrA gene |
| RG392 | GATGATCGACAGGAAGTTC | Antisense primer, amplifying a 547-bp DNA fragment of the lfrA gene |
| RG168 | TTGGATCCGATGACCAGCCCGAGCAT (BamHI) | Sense primer for lfrR expression into pET-15b, amplifying a 570-bp fragment of the lfrR gene |
| RG169 | TTGGATCCTCAGGTGCGCGGCAGG (BamHI) | Antisense primer for lfrR expression into pET-15b, amplifying a 570-bp fragment of the lfrR gene |
| A19 | GCGGCCTTTATCTATGTCAC | Sense primer, amplifying a 369-bp DNA fragment of the Rv0191 gene |
| A20 | CAGACTGGTTCCGATGTAGA | Antisense primer, amplifying a 369-bp DNA fragment of the Rv0191 gene |
| RG106 | CAGCTACATCGACTACGCC | Sense primer, amplifying a 317-bp DNA fragment of the gyrA gene |
| RG107 | GCGCTTCGGTGTAACGCAT | Antisense primer, amplifying a 317-bp DNA fragment of the gyrA gene |
Restriction sites are underlined in the sequence, and the corresponding enzymes are listed in parentheses following the sequences.
Construction of lfrR::lacZ and lfrA::lacZ fusions.
Various portions of the upstream lfrR region and of the lfrR-lfrA intergenic region were PCR amplified from M. smegmatis mc2155 genomic template by using different pairs of primers as follows: RG407-RG409, RG406-RG409, RG405-RG409, RG404-RG409, RG408-RG410, RG407-RG410, RG400-RG402, and RG401-RG402. The PCR fragments were cloned into pGEM-T Easy (Promega) to yield pECR1, pECR2, pECR3, pECR4, pECD1, pECD2, pECA1, and pECA2, respectively. All plasmids were sequenced to make sure that no mutations were introduced in the corresponding regions. Next, the various fragments were fused to lacZ in the 9.3-kb promoter probe vector pJEM12 (45). To do this, the pEC plasmid series was digested with BamHI and KpnI; the DNA fragments were purified from an agarose gel and then ligated to pJEM12 digested with the same enzymes to produce plfrR1, plfrR2, plfrR3, plfrR4, plfrD1, plfrD2, plfrA1, and plfrA2, respectively. The fusion plasmids were then transformed into M. smegmatis mc2155 by electroporation as described above for DNA techniques.
β-Galactosidase assay.
β-Galactosidase activity was measured as described by Miller (32). Activities were determined in M. smegmatis mc2155 strains containing the plasmid pJEM12 and all promoter fusion constructs. The assay was carried out at 28°C using ortho-nitrophenyl β-d-galactopyranoside (ONPG) as substrate, and the enzyme activity, expressed in terms of Miller units, was detected by measuring the optical density at 420 nm (OD420). Briefly, transformed mycobacterial cells were grown in 7H9 medium supplemented with 10% (vol/vol) OADC, 0.05% Tween 80, and kanamycin (25 μg/ml) to an optical density at 600 nm of 0.5. No antibiotic or sublethal concentrations (0.4× MIC) of ethidium bromide (MIC, 8 μg/ml) or acriflavin (MIC, 12.5 μg/ml) were added, and the cultures were reincubated for 2 h. The cells were then harvested by centrifugation (4,000 × g, 15 min, 4°C), and the pellets were washed and resuspended in 1 ml of phosphate-buffered saline (20 mM phosphate buffer, 150 mM NaCl, pH 7.4). After adding 20 μl of chloroform and 10 μl of 10% sodium dodecyl sulfate (SDS), aliquots of the lysates were incubated with ONPG at 28°C. The enzymatic reaction was followed spectrophotometrically at A420. The β-galactosidase activity was determined as follows: A = (OD420 min−1)/(OD600 × milliliters of culture). Triplicate samples were measured for each bacterial clone.
RNA extraction and RT-PCR.
M. smegmatis was grown to an OD600 of 0.6 in 7H9 medium supplemented with 10% (vol/vol) OADC and 0.05% Tween 80. Ten milliliters of culture was harvested at 5,000 × g for 15 min at 4°C; the cells were resuspended in 200 μl of Tris-EDTA buffer (Tris-HCl 10 mM, EDTA 1 mM, pH 8.0) containing 4 mg/ml of lysozyme and sonicated twice for 10 s at 40 W. Total RNA was then extracted using the RNeasy Mini kit (QIAGEN) according to the manufacturer's protocol and treated with DNase I-RNase free (10 U/μg of RNA) (Roche) for 30 min at room temperature; the DNase was inactivated at 70°C for 10 min. The RT reactions were carried out by using 2 μg of RNA template in the presence of M-MLV Reverse Transcriptase (Promega). Reverse transcription was carried out as follows. Two micrograms of RNA and 100 pmol of gene-specific primers (prexA and RG392, respectively; see Table 1) were incubated at 70°C for 5 min and then cooled on ice. Five microliters of 5× reaction buffer, 5 μl of 10 mM deoxynucleoside triphosphates, and 200 U of M-MLV Reverse Transcriptase enzyme were added, and the reaction was carried out at 37°C for 1 h. The enzyme was inactivated at 95°C for 5 min, and the reaction was ethanol precipitated. cDNA was dissolved in 20 μl of deionized water, and 4 μl was used for subsequent PCRs. cDNAs were amplified using primer pairs prexA-RG389 and RG391-RG392 (Table 1), as described above, with an annealing temperature of 64°C and 54°C, respectively, and an elongation time of 1 min. Samples were analyzed by electrophoresis on 1.5% agarose gels containing 0.5 μg/ml of ethidium bromide and visualized under UV light. The same reactions were carried out for each sample without M-MLV Reverse Transcriptase to ensure that amplification was a result of cDNA and not of contaminating DNA molecules.
Primer extension analysis.
The primer prexR (Table 1), which is complementary to the beginning of the coding sequence of the lfrR gene, was used to map the 5′ transcriptional start site. It was end labeled with 3,000 Ci mmol−1 [γ-32P]ATP (Amersham Biosciences) by using T4 polynucleotide kinase, as described in the primer extension kit (Promega). Briefly, the labeled primer (6 pmol) was incubated with 60 μg of RNA and 5 μl of 2× avian myeloblastosis virus primer extension buffer at 70°C for 10 min to melt secondary structures within the template and chilled on ice. The annealing was performed at 68°C for 20 min and at room temperature for 10 min. Extension was carried out with avian myeloblastosis virus reverse transcriptase (Promega), according to the manufacturer's instructions, at 42°C for 30 min; the reaction was stopped by the addition of 20 μl of loading dye (98% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). The reaction products were separated on a 6% polyacrylamide-8 M urea sequencing gel and were run alongside the sequencing products obtained with the prexR primer. Sequencing reactions were carried out with the Sequenase version 2.0 kit (USB) according to the manufacturer's instructions. The gels were dried and then exposed to X-ray film for 24 h at −80°C.
Induction experiments.
In order to detect the expression of the lfrA gene by RT-PCR, M. smegmatis mc2155 cells were grown in 7H9 medium supplemented with 10% (vol/vol) OADC and 0.05% Tween 80 to an OD600 of 0.5. The culture was then split, sublethal concentrations (0.05× to 0.4× MIC) of ethidium bromide (MIC, 8 μg/ml), acriflavine (MIC, 12.5 μg/ml), ciprofloxacin (MIC, 0.16 μg/ml), doxorubicin (MIC, 4 μg/ml), and rhodamine 123 (MIC, 5 μg/ml) were added, and the cultures were reincubated for 1, 2, and 4 h, respectively. Cells were harvested at 4,000 × g for 15 min and stored at −20°C until RNA extraction and RT-PCR experiments.
Overproduction and purification of LfrR protein.
To produce hexahistidine-tagged LfrR (His6-LfrR), the lfrR gene was amplified from M. smegmatis mc2155 genomic DNA by PCR with primers RG168 and RG169, which incorporate a BamHI restriction site (Table 1). PCR conditions were as described above, except that the annealing temperature was 54°C. The PCR fragment was gel purified and cloned into pGEM-T Easy cloning vector (Promega) to generate pGEM/lfrR. The 570-bp BamHI fragment from pGEM/lfrR was then cloned into BamHI-restricted pET15-b (Novagen) to produce pET/lfrR. A fresh colony of E. coli BL21(DE3)pLysS harboring plasmid pET/lfrR was grown overnight at 37°C in LB medium containing chloramphenicol and carbenicillin at a final concentration of 34 μg/ml and 50 μg/ml, respectively. The culture was diluted 1:100 into 200 ml of the same medium and incubated at 37°C until it reached an OD at 600 nm of 0.5. Isopropyl-β-thiogalactopyranoside (IPTG) was then added at a final concentration of 0.125 mM, and incubation was continued for 2 h. Bacterial cells were harvested, resuspended in sonication buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole [pH 8.0]), and disrupted by sonication. The lysate was centrifuged at 10,000 × g for 45 min at 4°C, and the His6-LfrR protein was recovered from the supernatant by Ni-nitrilotriacetic acid chromatography (QIAGEN) as recommended by the manufacturer. The LfrR protein was eluted with the sonication buffer containing 200 mM imidazole. The homogeneity of the eluted protein was estimated to be 90% by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. His6-LfrR protein concentration was determined by using the Bradford assay (Bio-Rad) with bovine serum albumin (BSA) (Sigma) as the protein standard. Total yield was about 20 mg of purified His6-LfrR from 200 ml of culture.
One milliliter of protein was dialyzed overnight at 4°C against 1 liter of dialysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol) to remove imidazole and used for gel mobility shift assay.
Electrophoretic mobility shift assay.
The DNA probes for gel shift experiments were obtained by BamHI-KpnI digestion of pECR1, pECR2, and pECA2 plasmids and isolated from agarose gel by using a Ultrafree-DA kit (Millipore) according to the manufacturer's instructions. The fragments were end labeled with [γ-32P]ATP (3,000 Ci mmol−1) (Amersham Biosciences) by using T4 polynucleotide kinase (Promega) according to the manufacturer's instructions. Approximately 3.5 nM of the labeled DNA fragments was incubated with 2 μM LfrR in reaction buffer (10 mM Tris HCl [pH 7.5], 0.5 mM dithiothreitol, 0.5 mM EDTA, 50 mM NaCl, 10 mM MgCl2, 50 ng of salmon sperm DNA, 1 mg/ml of BSA, 4% glycerol) at room temperature for 20 min. The samples were immediately loaded onto a 6% native polyacrylamide gel containing 0.5× TBE (0.05 M Tris base, 0.05 M boric acid, 1 mM EDTA-Na2 · 2H2O) and subjected to electrophoresis. The gels were then vacuum dried and exposed overnight to Biomax radiography film (Kodak). Electrophoretic mobility shift assay was also performed with acriflavine and ciprofloxacin (0.05× and 2× MIC) to test the LfrR modulatory effect upon binding to target DNA. Electrophoretic mobility shift assay competitions were performed with specific and nonspecific competitor DNA. The nonspecific DNA was amplified by PCR with primers A19 and A20 (Table 1) and 100 ng of M. tuberculosis DNA. The 369-bp PCR product, consisting of an internal portion of the Rv0191 gene coding for a probable conserved integral membrane protein (http://genolist.pasteur.fr/tuberculist), was isolated from agarose gel as described above.
Unlabeled specific and nonspecific competitor DNA (50- to 100-fold molar excess) were incubated with His6-LfrR for 10 min at room temperature, followed by the addition of the labeled probe and incubation for 20 min at room temperature. The resulting DNA-protein complexes were then subjected to electrophoresis and autoradiographed as described above.
RESULTS
Transcriptional analysis of the lfrRA operon using RT-PCR.
Previously, it has been reported that the upstream region of lfrA contained a gene coding for a putative TetR family transcriptional repressor, named LfrR, and that the lfrR deletion increased the lfrA expression (26). These data suggest that LfrR negatively regulates the production of LfrA. Moreover, lfrA disruption in M. smegmatis increased susceptibility to ethidium bromide (40). To demonstrate that lfrR and lfrA genes are part of the same transcriptional unit, RT-PCR experiments were performed. To this aim, total RNA from M. smegmatis strain mc2155, grown in the absence or in the presence of ethidium bromide at a final concentration of 3.2 μg/ml, was isolated and retrotranscribed with primer prexA that annealed to the lfrA gene (Table 1). The cDNA was amplified by PCR using primers internal to lfrR and lfrA sequences (RG389 and prexA, respectively), allowing the detection of cotranscription of the lfrR and lfrA genes. An amplification product of 672 bp according to the size predicted from the DNA sequence was obtained in the case of RNA isolated from cells grown in the presence of ethidium bromide (data not shown). The sequence analysis of the amplified product confirmed that this fragment corresponds to the region defined by RG389 and prexA primers. This result demonstrates clearly and for the first time the presumed operon structure of the lfrR and lfrA genes. In our conditions, no amplification was observed when the RNA was isolated from cells grown in the absence of ethidium bromide (data not shown).
Mapping of the lfrRA promoter.
Transcriptional lacZ gene fusions carrying various portions of the lfrR upstream regions as well as of the lfrR-lfrA intergenic region were constructed and used to map the lfrRA promoter region. The sizes of the fragments contained in the lacZ fusion plasmids are shown in Fig. 1A. The fusion plasmids were electroporated into M. smegmatis strain mc2155, and β-galactosidase activities were measured. Cells containing vector pJEM12 did not show any detectable β-galactosidase activity, indicating that the endogenous level of expression of the promoterless lacZ was low and negligible (Fig. 1B). Also, the cells harboring the plfrD1 plasmid, which contains the lfrR region from nucleotide positions +2 to +69 devoid of obvious promoter sequences, did not show any detectable activity (Fig. 1B). In contrast, plasmid plfrR3 (−220 to +7) showed the highest activity; however, expression of this activity was not higher in the case of plfrR4 (−351 to +7), which showed an activity comparable to that of plfrR3 plasmid, suggesting that the entire promoter region is located in the R3 fragment (Fig. 1B). This hypothesis was consistent with the fact that the plasmid plfrR2 (−136 to +7) directed expression of a reduced level of β-galactosidase activity (Fig. 1B), followed by plasmid plfrR1 (−55 to +7), which exhibited a low level of β-galactosidase activity.
FIG. 1.
Localization of the lfrRA promoter region using lacZ transcriptional fusions. A. Diagram showing the sizes and the positions of the various fragments (shown as black boxes) contained in the lacZ fusion plasmids. The white arrows indicate the lfrR and lfrA genes. B. β-Galactosidase activity of M. smegmatis cells containing the different constructs. Error bars indicate standard deviations from triplicate determinations of β-galactosidase activity.
The β-galactosidase activity of the different constructs has also been determined with cultures grown in the presence of ethidium bromide and acriflavine (data not shown). The promoter activity of each construct was similar to that observed in the case of cultures grown without inducers, indicating that these compounds did not affect the promoter activity. The high expression level of the reporter without inducer could be explained by the amplification effect of this region due to the cloning into pJEM12 vector.
In summary, these experiments localized the lfrRA promoter in a region extending 220 nucleotides upstream of lfrR.
Mycobacterial cells containing plfrA1, which carries the entire lfrR-lfrA intergenic region of 71 bp, showed a minimal activity, as did cells containing plfrA2 (−294 from lfrA to +3), indicating that no promoter activity was present in the intergenic region, thus confirming the RT-PCR data. The β-galactosidase activity of a plasmid containing the lfrR-lfrA intergenic region and the entire lfrR coding sequence was not evaluated.
Identification of the transcriptional start point of the lfrRA operon.
The transcription initiation point of the lfrRA operon was mapped in cells growing in the absence or presence of ethidium bromide. When total RNA, prepared from M. smegmatis mc2155 cells grown in the absence of ethidium bromide, was subjected to primer extension analysis, no signal could be identified, even using different oligonucleotides (data not shown). On the contrary, when total RNA was extracted from an M. smegmatis mc2155 culture grown in the presence of ethidium bromide (3.2 μg/ml) for 2 h, a single product was identified (Fig. 2A). The 5′ end of lfrRA is the A residue that is the first base of the translational initiation codon (Fig. 2A). No other signal could be identified further upstream with this oligonucleotide or with an oligonucleotide internal to the lfrA coding sequence (oligonucleotide prexA) (Table 1 and data not shown). It is noteworthy that the prexA oligonucleotide gave a product of the right size, in agreement with the prexR primer. The position of the 5′ end is consistent with the RT-PCR data, which indicated that the two genes are organized as an operon. It is likely that the primer extension experiment identified a bona fide transcriptional start site, because computer analysis of this region of the mRNA did not reveal any significant features, such as putative hairpin structures, known to prematurely arrest reverse transcription. When the sequence upstream of the lfrRA transcriptional start site was compared with sequences of other mycobacterial promoter structures, a putative −10 box (TATATT) could be identified (Fig. 2B). No homologous −35 regions with a canonical distance of 16 to 18 bp have been found; a −35 box (GGGACA) similar to the consensus sequence of E. coli was identified, but it was at a 28-bp distance from the −10 box (Fig. 2B).
FIG. 2.
A. Mapping of the transcriptional start point for the lfrRA operon. The position of the transcription start site was determined by primer extension with the oligonucleotide prexR (Table 1). Sequencing reactions, performed with the same oligonucleotide on a plasmid containing the entire lfrRA operon and the regions upstream and downstream of the genes, are reported in the last four lanes of the panel. The coordinates of the 5′ end are reported on the left of the panel. B. Nucleotide sequence of the lfrRA promoter region. The nucleotide sequences around the promoter region of the lfrRA genes are presented. The first 25 amino acids and the corresponding nucleotide sequence of the LfrR protein are also reported. The transcription start site is marked by an arrow. The putative −10 promoter element and the unusual −35 region at a distance of 28 bp are underlined. The “extended −10 promoter” is double underlined (see Discussion).
Some substrates of the LfrA transporter induce transcription of the lfrA gene.
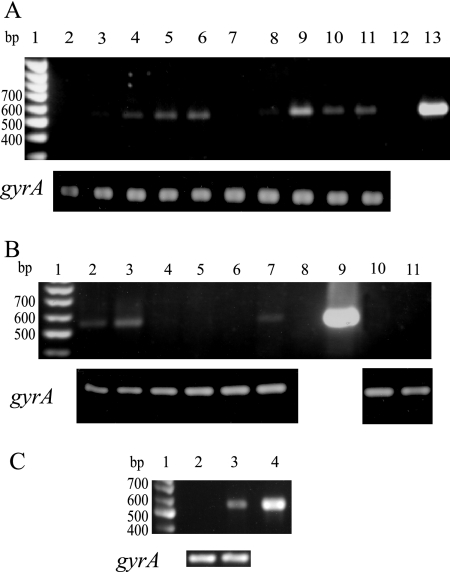
It was shown previously that the resistance of M. smegmatis to multiple structurally unrelated compounds, such as ethidium bromide, acriflavine, ciprofloxacin, doxorubicin, and rhodamine 123, can occur when LfrA is overexpressed by cloning the lfrA gene into a multicopy vector (40, 43). On the other hand, the disruption of the lfrA gene decreased resistance to ethidium bromide and acriflavine but resulted only in a twofold decrease in MICs for the other compounds (40). Since many of the known bacterial mechanisms of drug resistance are inducible by the corresponding drugs (23), it was interesting to determine if the expression of LfrA in M. smegmatis is inducible by LfrA substrates. To screen for factors influencing transcription of the lfrA gene, RT-PCR experiments were performed with primers RG391 and RG392 specific for lfrA (with an expected 547-bp product) (Table 1). The expression of the lfrA efflux pump gene was studied by analyzing the relative amount of lfrA mRNA expressed in M. smegmatis mc2155 grown in the absence and in the presence of ethidium bromide, acriflavine, ciprofloxacin, doxorubicin, and rhodamine 123, previously reported to be substrates of the LfrA efflux pump (40, 43). No detectable levels of the lfrA mRNA was observed when M. smegmatis was grown in the absence of any compounds (Fig. 3A, lanes 2 and 7). On the contrary, a detectable level of the lfrA mRNA was observed in response to different amounts of ethidium bromide (Fig. 3A, lanes 3 to 6 and 8 to 11). The lower concentration of ethidium bromide and the shorter time of the treatment were sufficient to induce the lfrA expression (Fig. 3A, lane 3). The same result was achieved after treatment of the M. smegmatis culture with 0.05× and 0.4× MIC of acriflavine (Fig. 3B, lanes 2 and 3) and with the highest concentration (0.4× MIC) of rhodamine 123 after 2 h of induction (Fig. 3B, lane 7). No lfrA mRNA was detected in the case of the RT-PCR amplification of RNA isolated from cultures treated either with doxorubicin or ciprofloxacin at concentrations corresponding to 0.05× and 0.4× MIC (Fig. 3B, lanes 4 and 5 as well as 10 and 11, respectively). In all experiments, the expression of the gyrA gene coding for the A subunit of the DNA gyrase was determined as an internal control to ensure that the differences observed were not due to variability in the RNA isolation and/or in the RT-PCR technique. The M. smegmatis gyrA expression is unaltered in the mid-logarithmic phase under different growth conditions (Fig. 3A and B), demonstrating that the differences detected in the amount of lfrA mRNA under different growth conditions are genuine.
FIG. 3.
Effects of different LfrA substrates on lfrA expression. The expression of lfrA was analyzed by RT-PCR with primers RG391 and RG392 on RNA isolated from M. smegmatis mc2155 cultures. A. Expression of lfrA in the presence of increasing concentrations of ethidium bromide after 1 (lanes 3 to 6) and 4 h (lanes 8 to 11) of treatment. Lane 1, molecular size marker (Fermentas); also, the sizes of some bands in base pairs are provided on the left; lanes 2 and 7, no ethidium bromide; lanes 3 and 8, 0.4 μg/ml (0.05× MIC); lanes 4 and 9, 0.8 μg/ml (0.1× MIC); lanes 5 and 10, 1.6 μg/ml (0.2× MIC); lanes 6 and 11, 3.2 μg/ml (0.4× MIC); lane 12, negative control; lane 13, positive control, which is DNA amplification with primers RG391 and RG392 (expected size, 547 bp). The expression of the gyrA gene (317 bp) of M. smegmatis was determined as an internal control of all the RT-PCRs and is shown at the bottom of the panel. B. Expression of lfrA in the presence of acriflavine (lanes 2 and 3, 0.625 and 5 μg/ml [0.05× and 0.4× MIC]), doxorubicin (lanes 4 and 5, 0.2 and 1.6 μg/ml [0.05× and 0.4× MIC]), rhodamine 123 (lanes 6 and 7, 0.25 and 2 μg/ml [0.05× and 0.4× MIC]), and ciprofloxacin (lanes 10 and 11, 0.008 and 0.064 μg/ml [0.05× and 0.4× MIC]) after 2 h of treatment. Lanes 1, 8, and 9 are the same as lanes 1, 12, and 13 in panel A. The expression of the gyrA gene (317 bp) of M. smegmatis was determined as an internal control of all the RT-PCRs and is shown at the bottom of the panel. C. Expression of lfrA in M. smegmatis mc2155 wild-type (lane 2) and M. smegmatis mc211 mutant (lane 3) strains grown in the absence of inducers. Lanes 1 and 4 are the same as lanes 1 and 13 in panel A. The expression of the gyrA gene (317 bp) of M. smegmatis was determined as an internal control of all the RT-PCRs and is shown at the bottom of the panel.
Taken together, these results are in favor of the hypothesis that the LfrR protein could act as a repressor of the lfrA transcription. Previously in our laboratory we isolated the M. smegmatis mutant mc211 resistant to different fluoroquinolones, cationic dyes, and anthracyclines (6, 40). We demonstrated that the LfrA efflux pump was involved in the resistance profile (6, 40).
Further characterization pointed out that this mutant has an insertion of 18 bp within the lfrR coding region (E. De Rossi, personal communication). Consequently, we hypothesized that the mutated LfrR protein does not repress the lfrA transcription. To verify this hypothesis, RT-PCR experiments with primers RG391 and RG392 were performed using total RNA isolated from M. smegmatis mc211 cells grown in the absence of proper inducer. As shown in Fig. 3C (lane 3), a good level of lfrA expression was observed, thus confirming that LfrR acts as a repressor. On the contrary, no lfrA transcription was observed when RT-PCR experiments were performed with total RNA extracted from M. smegmatis mc2155 wild-type cells (Fig. 3C, lane 2). The M. smegmatis gyrA expression is unaltered (Fig. 3C), demonstrating that the differences detected in the amount of lfrA mRNA are genuine.
Expression and purification of the LfrR protein.
The M. smegmatis lfrR gene was amplified by PCR, cloned, overexpressed, and purified as a hexahistidine-tagged protein under control of the T7 promoter of the plasmid pET-15b (Novagen), creating a His-tagged protein. The His6-LfrR fusion protein was expressed in E. coli BL21(DE3)pLysS cultures, after induction with IPTG at a final concentration of 0.125 mM, and purified to near homogeneity by Ni2+ affinity chromatography. Only one band was visualized on SDS-polyacrylamide gel electrophoresis, which represents the purified protein; this band has an apparent molecular mass of about 21 kDa (data not shown), which is close to value calculated for the 189-amino-acid fusion protein monomer. The purified LfrR protein was used for all subsequent band-shift analyses.
Binding of LfrR to the region upstream of lfrR.
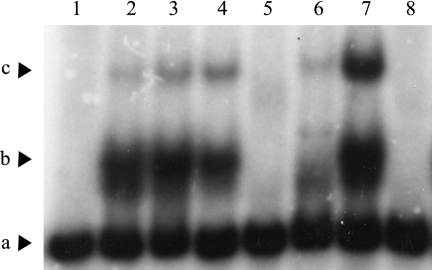
To determine if LfrR regulates the lfrRA operon via direct interaction with the promoter upstream of lfrR, gel mobility shift assays were performed. The purified His6-LfrR protein, when incubated with the end-labeled 143-bp lfrR promoter, isolated by BamHI-KpnI digestion of plasmid plfrR2 (Fig. 1A), showed a clear shift in the DNA-binding pattern, although two different LfrR-DNA complexes could be detected (Fig. 4, lanes 2 and 3); these retarded complexes are shown in Fig. 4 by the letters b and c, while the letter a indicates the free labeled 143-bp probe. This would suggest the labile formation of higher-molecular-weight complexes because of multimerization of LfrR on the DNA probe. Alternatively, this would indicate the presence of two potential binding sites in the upstream sequence of lfrR and that LfrR binds with different affinities to these two sites within the lfrR upstream sequence. The observed shift was not complete, as demonstrated by the presence of free labeled probe, probably due to incomplete labeling of the promoter fragment leaving unlabeled “competitor” DNA. However, similar results were achieved when the 227-bp fragment from plfrR3 was used. His6-LfrR binding to the 143-bp fragment was specific, since the band shift was completely and partially inhibited in the presence of a 100- and 50-fold molar excess of unlabeled fragment, respectively (Fig. 4, lanes 5 and 6). The specificity of binding was further investigated in a competition assay with a 100-fold molar excess of an unrelated labeled fragment (a 369-bp fragment amplified from the Rv0191 gene of M. tuberculosis) (Fig. 4, lane 4); the specific binding activity of LfrR to the 143-bp fragment is retained, indicating that LfrR binding to the lfrRA promoter region is sequence specific. Gel shift assays were also performed with His6-LfrR and the end-labeled 62-bp or 297-bp fragment (Fig. 1A), isolated by BamHI-KpnI digestion of plasmids plfrR1 and plfrA2, respectively. These two fragments did not cause any band shift when incubated with the His6-LfrR protein (data not shown).
FIG. 4.
LfrR binds specifically to the DNA sequence upstream of the lfrRA operon. Lane 1, free labeled 143-bp probe; lanes 2 and 3, probe with 20 and 40 ng of LfrR, respectively; lane 4, probe with 20 ng of LfrR and a 100-fold molar excess of nonspecific Rv0191 fragment (noncompetitive probe); lanes 5 and 6, probe with 20 ng of LfrR and 100- and 50-fold molar excesses of 143-bp fragment (competitive probe), respectively; lane 7, probe with 20 ng of LfrR and 0.32 μg/ml of ciprofloxacin (2× MIC); lane 8, probe with 20 ng of LfrR and 25 μg/ml of acriflavine (2× MIC). The letter a indicates the free labeled 143-bp probe, while letters b and c indicate two different retarded complexes (see Results).
To confirm the hypothesis that LfrR acts as a repressor of the lfrRA operon, dissociating from the upstream region upon drug binding, the effects of ciprofloxacin and acriflavine were also tested for their ability to dissociate the LfrR-DNA complex. Addition of an excess amount of acriflavine (25 μg/ml; 2× MIC) abolished the LfrR binding to the 143-bp labeled fragment (Fig. 4, lane 8), suggesting that this drug acts as an inducer that antagonizes the interaction between LfrR and DNA. These data are consistent with RT-PCR experiments, which showed a detectable level of lfrA mRNA only in the presence of acriflavine as well as ethidium bromide, both substrates of the LfrA efflux pump. This indicates that the presence of this compound causes the dissociation of the protein from the DNA, thus allowing the transcription to proceed. On the contrary, the addition of ciprofloxacin (0.32 μg/ml; 2× MIC) did not have any effect on LfrR binding to the 143-bp labeled fragment (Fig. 4, lane 7), suggesting that this drug does not dissociate the LfrR-DNA complex. These data are broadly consistent with the substrate specificity of the LfrA pump and strongly confirm RT-PCR results.
DISCUSSION
The first efflux pump described in mycobacteria, the protein LfrA, was identified in M. smegmatis. LfrA confers resistance to ciprofloxacin, other fluoroquinolones, ethidium bromide, and acriflavine (43), but also to anthracyclines and rhodamine 123 when cloned into a multicopy plasmid (40). More recently, a gene coding for a putative TetR family transcriptional repressor, named LfrR, was identified upstream of the lfrA gene (26).
In this work we show, by RT-PCR experiments, that lfrR and lfrA genes are organized as an operon. The use of lacZ fusions narrowed the lfrRA promoter to a region located within 220 nucleotides upstream of lfrR. Primer extension analysis confirmed that the transcriptional start site of lfrA is indeed upstream of the lfrR gene, and it was found that the A residue is the first base of the translational initiation codon of the lfrR gene. The leaderless mRNA is consistent with the lack of a Shine-Dalgarno sequence upstream of the lfrR gene (11, 19). Alignment of the region centered around position −10 (TATATT) consensus sequences from the transcriptional start site of the lfrRA genes showed significant sequence similarities to E. coli consensus promoters according to the study of Bashyam et al. (2). Sequence similarities were found also in respect to other mycobacterial promoters, in particular to the acetamidase gene (100% identity) (30) and to the S16 promoter (83% identity) (2) of M. smegmatis. One putative −35 region (GGGACA) was found positioned at an unusual spacing, 28 bp from the −10 region, showing 66% identity with the E. coli consensus promoter and with the −35 region (TTGACA) of the M. smegmatis rrnAP3 and rrnAPCL1 promoters (10). No other homologous −35 regions with a canonical 16- to 18-bp distance have been found; it is noteworthy that in mycobacteria there is a greater heterogeneity at the −35 region, reflecting a higher GC content, and in many cases there is no identifiable −35 element (42, 47). This could also explain why plfrR1 does not express LacZ: maybe the plfrR1 construct does not contain the lfr promoter. Moreover, the upstream region is also important for regulation and promoter activity, as demonstrated with plfrR3 and plfrR4 constructs. However, promoters that lack a canonical −35 sequence but are still functional have been reported in mycobacteria (2). For mycobacterial promoters, where apparent conservation in −35 region is absent, many of them possess TGN nucleotides immediately upstream of the −10 region, and thus they are termed “extended −10 promoters” (42). This “extended −10 promoter” is also present as TGC upstream of the −10 region of the lfrR gene (Fig. 2B).
It was not surprising that lfrA expression was inducible by ethidium bromide and acriflavine and, in part, by rhodamine 123, as this was reported for other drug efflux pumps of different bacteria, such as Bmr of Bacillus subtilis (1) and TtgABC (44) and SrpABC (24) of Pseudomonas putida. It is interesting to note that the two compounds for which the MICs strongly decrease in an M. smegmatis lfrA-deleted strain (40) are the ones that can induce the expression of the pump itself. In this way we demonstrate what was postulated in the previous work (40): ethidium bromide is a better inducer of the expression of lfrA than ciprofloxacin. It is noteworthy that the lfrA gene is highly expressed in the M. smegmatis mc211 mutant characterized by a mutated LfrR protein.
The critical role of LfrR on the regulation of LfrA in M. smegmatis and its binding site were defined by electrophoretic mobility shift assay: LfrR represses the transcription of lfrA. This feature resembles the control of TetA by TetR, in which the basal level of expression of tetA is minimal in the absence of tetracycline (13). This is probably due to the fact that both LfrA and the TetA pumps are specific for specific substrates and constitutive expression of lfrA, and tetA is not required in the absence of these compounds. On the other hand, the overproduction of efflux pumps in the absence of selection pressure or substrates has been demonstrated to be deleterious to some organisms (7, 33, 39). Therefore, there is a need for regulatory systems to modulate the expression of MDR efflux pumps in bacteria. In this respect, LfrR acts as a moderator to maintain balanced production of LfrA to meet the physiological needs and facilitate the adaptation of M. smegmatis to environmental changes, including antibiotic treatments.
It was previously reported that the expression of MDR efflux pumps can be conditionally induced by structurally diverse substrates of these pumps (1, 4, 12, 21, 22, 29, 31, 37). This induction is due to the direct interaction of the substrates with repressor molecules, which interferes with the binding of repressors to operator DNA and which results in increased levels of expression of MDR pump genes. Here we show that the inducible expression of LfrA following treatment with ethidium bromide, acriflavine, or rhodamine 123 is mediated by LfrR. Indeed, acriflavine is able to abolish the binding of LfrR to the 143-bp promoter region (Fig. 4), thus allowing the transcription to proceed. This finding is consistent with the in vivo induction of lfrA expression by acriflavine and strongly indicates that acriflavine-mediated inhibition of LfrR binding to lfrRA promoter is responsible for the enhanced transcription of lfrA.
Transcriptional regulators of the TetR family are characterized by a conserved helix-turn-helix-containing DNA-binding domain at the N-terminal region and a divergent C-terminal sequence that is involved in binding to various inducing ligands (14, 17, 18). Binding by an inducing compound to the C-terminal region triggers conformational changes in the N-terminal DNA-binding domain, reducing the affinity of a regulator to its target promoter DNA (13). Such structural changes in a repressor induced by the binding of a ligand have been confirmed for several regulator proteins in the TetR family, such as TetR and QacR (34, 41). Similar to other TetR family regulators, LfrR has a typical N-terminal DNA-binding helix-turn-helix motif and a potential ligand-binding region in the C-terminal portion (26). Therefore, it is likely that acriflavine interacts with the C-terminal region of LfrR and induces conformational changes in the repressor, resulting in a great reduction in its DNA-binding affinity.
The absence of an interaction among ciprofloxacin and LfrR was surprising given earlier observations of reproducible ninefold increases in the MIC of this compound when lfrA was overexpressed, establishing that it is a LfrA substrate (40). The explanation for this apparent paradox simply may be that not all LfrA substrates must be inducers, and LfrA may be more promiscuous than LfrR. A relevant example of such a behavior is given by the QacA efflux pump, which extrudes compounds that do not induce its expression or bind to the QacR repressor (12).
The ability of LfrR to interact functionally with dissimilar molecules can seemingly have two explanations. The first hypothesis suggests that the function of the LfrR-LfrA system is to protect bacteria from diverse environmental toxins. The alternative hypothesis suggests that LfrA has evolved to efflux-specific compounds while the ability of this protein to bind and efflux diverse drugs is merely a fortuitous side effect of its normal function.
There is no known homolog of the lfrA gene in the M. tuberculosis genome. This indicates that M. smegmatis (for which a genome size of 7.5 Mb has been estimated, versus the 4.4-Mb size of M. tuberculosis) will contain many other MFS transporters different from those identified in M. tuberculosis.
In conclusion, we demonstrate definitively that LfrR is the lfrA transcriptional repressor. Furthermore, the expression of the LfrA efflux pump is regulated by some of its substrates, and this regulation is mediated by its repressor LfrR.
Acknowledgments
We thank J. Timm for supplying plasmid pJEM12, required for this study.
This work was supported by Fondo d'Ateneo per la Ricerca 2004 and 2005 (E.D.R.), Istituto Superiore Sanità, Ricerca Finalizzata 2005 (G.R.), and Integrated Research Actions Italy-Spain (E.D.R.).
Footnotes
Published ahead of print on 16 October 2006.
REFERENCES
- 1.Ahmed, M., M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Bashyam, M. D., D. Kaushal, S. K. Das Gupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. H., T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 5.De Rossi, E., J. A. Ainsa, and G. Riccardi. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36-52. [DOI] [PubMed] [Google Scholar]
- 6.De Rossi, E., M. C. Blokpoel, R. Cantoni, M. Branzoni, G. Riccardi, D. B. Young, K. A. De Smet, and O. Ciferri. 1998. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 42:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert, B., and C. F. Beck. 1989. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol. 171:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Bikin, C. Dye, R. Williams, M. C. Raviglione, et al. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 9.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexABoprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667-674. [DOI] [PubMed] [Google Scholar]
- 11.Grill, S., C. O. Gualerzi, P. Londei, and U. Blasi. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 19:4101-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 13.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic, S., K. M. Hardie, M. H. Brown, and R. A. Skurray. 2003. Interactions of the QacR multidrug-binding protein with structurally diverse ligands: implications for the evolution of the binding pocket. Biochemistry 42:15226-15236. [DOI] [PubMed] [Google Scholar]
- 15.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 17.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 18.Hinrichs, W., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 19.Janssen, G. R. 1993. Eubacterial, archaebacterial, and eukaryotic genes that encode leaderless mRNA, p. 59-67. In R. H. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. ASM Press, Washington, D.C.
- 20.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 21.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, S., J. Nishimura, Y. Yufu, H. Ideguchi, T. Umemura, and H. Nawata. 1992. Modulation of expression of multidrug resistance gene (mdr-1) by adriamycin. FEBS Lett. 308:175-178. [DOI] [PubMed] [Google Scholar]
- 23.Kieboom, J., J. J. Dennis, G. J. Zylstra, and J. A. M. De Bont. 1998. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180:6769-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köhler, T., S. F. Epp, L. K. Curty, and J.-C. Pechère. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 26.Li, X. Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 30.Mahenthiralingam, E., P. Draper, E. O. Davis, and M. J. Colston. 1993. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J. Gen. Microbiol. 139:575-583. [DOI] [PubMed] [Google Scholar]
- 31.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 34.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 35.Parish, T., and N. G. Stoker. 1998. Electroporation of mycobacteria, p. 129-144. In T. Parish, and N. G. Stoker (ed.), Methods in molecular biology, vol. 101: mycobacteria protocols. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 36.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 40.Sander, P., E. De Rossi, B. Boddinghaus, R. Cantoni, M. Branzoni, E. C. Bottger, H. Takiff, R. Rodriquez, G. Lopez, and G. Riccardi. 2000. Contribution of the multidrug efflux pump LfrA to innate mycobacterial drug resistance. FEMS Microbiol. Lett. 193:19-23. [DOI] [PubMed] [Google Scholar]
- 41.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 42.Smith, I., W. R. Bishai, and V. Nagaraja. 2005. Control of mycobacterial transcription, p. 219-231. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, D.C.
- 43.Takiff, H. E., M. Cimino, M. C. Musso, T. Weisbrod, R. Martinez, M. B. Delgado, L. Salazar, B. R. Bloom, and W. R. Jacobs. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 93:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teràn, W., A. Felipe, A. Segura, A. Rojas, J. L. Ramos, and M. T. Gallegos. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trias, J., and R. Benz. 1994. Permeability of the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 14:283-290. [DOI] [PubMed] [Google Scholar]
- 47.Unniraman S., M. Chatterji, and V. Nagaraja. 2002. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 184:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viveiros, M., C. Leandro, and L. Amaral. 2003. Mycobacterial efflux pumps and chemotherapeutic implications. Int. J. Antimicrob. Agents. 22:274-278. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, C. 2003. Antibiotics: actions, origins, resistance, p. 89-155. ASM Press, Washington, D.C.