Abstract
We investigated the enzymatic efficiency and inhibition by quinolones of Mycobacterium tuberculosis DNA gyrases carrying the previously described GyrA G88C mutation and the novel GyrA G88A mutation harbored by two multidrug-resistant clinical strains and reproduced by site-directed mutagenesis. Fluoroquinolone MICs and 50% inhibitory concentrations for both mutants were 2- to 43-fold higher than for the wild type, demonstrating that these mutations confer fluoroquinolone resistance in M. tuberculosis.
The incidence of multidrug-resistant tuberculosis (MDR-TB), which is associated with high rates of mortality (7), has become a concern for TB control in many countries (23). Fluoroquinolones are one of the three bactericidal drug groups recommended for the treatment of MDR-TB (3, 11, 23). Unfortunately, acquired resistance to quinolones in Mycobacterium tuberculosis (5, 8, 18, 22), described since the first use (6), is increasing (12).
DNA gyrase, a tetrameric A2B2 protein acting by a transient double-stranded DNA break and cooperating to facilitate DNA replication and other key DNA transactions, is the sole target for quinolones in M. tuberculosis (9, 10, 14). Mutations involved in M. tuberculosis quinolone resistance described so far generated substitutions in the A subunit at positions 90, 91, and 94 and more rarely in the B subunit at position 510, as described for other bacteria (2, 6, 8, 18, 20, 22). These mutations confer cross-resistance to all members of the quinolone family (20).
Because fluoroquinolone therapy is associated with an improved outcome in MDR-TB (7, 23), each MDR M. tuberculosis clinical strain submitted to our laboratory is tested for fluoroquinolone susceptibility. In this context, we identified a new mutation leading to G88A in GyrA in two MDR-M. tuberculosis strains isolated from patients with pulmonary tuberculosis who relapsed after being treated by a fluoroquinolone, either ofloxacin or moxifloxacin. Even for bacteria other than M. tuberculosis (4, 15, 19, 20), the effect of mutations at position 88 in GyrA on quinolone susceptibility is poorly understood, and no study clearly demonstrated the implication of these mutations in quinolone resistance. We aimed to examine the effects at the molecular level of amino acid substitutions at position 88 in GyrA by expressing recombinant DNA gyrases harboring mutations at this position. These mutations lead to fluoroquinolone resistance in M. tuberculosis, but the resistance level depends on the quinolone structure with regard to the substitutions at the residues R6 and R7. Our observations bring new insight to the role of position 88 in the model of quinolone binding.
MICs were determined by the 1% standard proportion method on 7H11 agar supplemented with 10% oleic acid-albumin-dextrose-catalase for the two M. tuberculosis clinical isolates bearing GyrA G88A mutations and the quinolone-susceptible strain M. tuberculosis H37Rv (13).
Plasmids expressing M. tuberculosis gyrA genes containing the new mutation G88A or the mutation G88C, previously described (17, 18, 22), were generated from pATB (1), which contained the wild-type (WT) gyrA gene of M. tuberculosis, by using the QuickChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. Oligonucleotides synthesized by MWG-Biotech (Ebersberg, Germany) mimicked nucleotides 247 to 282 of the M. tuberculosis gyrA sequence containing the appropriate base substitutions (in boldface characters): G88A (5′ AACTACCACCCGCACGCCGACGCGTCGATCTACGAC 3′) and G88C (5′ AACTACCACCCGCACTGCGACGCGTCGATCTACGAC 3′), together with complementary strands 5′ GTCGTAGATCGACGCGTCGGCGTGCGGGTGGTAGTT 3′ for G88A and 5′ GTCGTAGATCGACGCGTCGCAGTGCGGGTGGTAGTT 3′ for G88C.
WT and mutant GyrA subunits were purified as previously described, as was the WT GyrB subunit. DNA supercoiling experiments were carried out as described previously (1, 2).
The two M. tuberculosis clinical strains carrying the G88A mutation were resistant to fluoroquinolones, the MICs of which were 2- to 16-fold higher than those for the WT H37Rv reference strain (Table 1). These two strains harbored a level of fluoroquinolone resistance lower than those of strains described with the GyrA G88C mutation, i.e., a laboratory-selected mutant of M. tuberculosis H37Rv for which the ciprofloxacin MIC was over 3 μg/ml (22) and a clinical strain isolated from a patient treated with ofloxacin as a monotherapy for which the levofloxacin MIC was 16 μg/ml (17).
TABLE 1.
Quinolone and coumarin activities against M. tuberculosis WT and mutated strains (MICs) and DNA gyrases (IC50s)
| Drug | MIC (μg/ml)
|
IC50 (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| WT | G88Aa | WT | G88A | G88C | D94H | |
| Quinolones | ||||||
| Sparfloxacin | 0.25 | 0.5; 1 | 2.5 | 10 | 32 | ND |
| Gemifloxacin | 4 | 32; 64 | 3.5 | 90 | 150 | ND |
| Gatifloxacin | 0.12 | 1 | 4 | 7 | >128 | 150b |
| Moxifloxacin | 0.25 | 2 | 4 | 10 | 35 | 90b |
| Levofloxacin | 0.5 | 1 | 5 | 30 | 100 | 320b |
| Ofloxacin | 0.5 | 2 | 10 | 40 | 50 | 800b |
| Temafloxacin | 4 | 8 | 30 | 70 | 250 | ND |
| Enoxacin | 4 | 64 | 100 | 320 | 320 | ND |
| Flumequine | 64 | 32; 128 | 700 | 650 | 600 | >2,000 |
| Oxolinic acid | 32 | 16; 32 | 800 | 800 | 800 | ND |
| Nalidixic acid | 128 | 64; 128 | 1,100 | 1,200 | 1,150 | >3,200 |
| Pipemidic acid | 256 | 256 | 2,200 | 2,000 | 1,500 | ND |
| Coumarin | ||||||
| Novobiocin | NDc | ND | 0.3 | 0.3 | 0.2 | ND |
When the MICs were different for the two clinical strains, both values are presented.
From Aubry et al. (2).
ND, not determined.
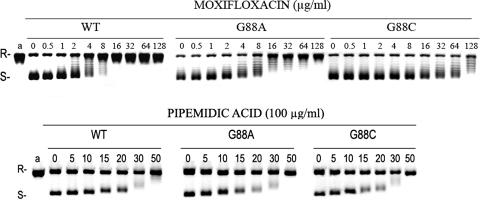
The recombinant gyrase complexes bearing GyrA G88A and G88C were 2- to 26- and 3- to 43-fold more resistant than WT gyrase to inhibition by fluoroquinolones, respectively (Table 1; Fig. 1). The GyrA G88C mutant showed fluoroquinolone 50% inhibitory concentrations (IC50s) similar to that observed for the classical GyrA A90V mutant well known to be implicated in fluoroquinolone resistance in M. tuberculosis (2). These results establish clearly that the GyrA G88C and G88A mutations confer resistance to fluoroquinolones in M. tuberculosis by decreasing gyrase inhibition. However, even if both mutations are implicated in fluoroquinolone resistance, increases in IC50s were higher for the G88C mutant than for the G88A mutant for sparfloxacin, gemifloxacin, gatifloxacin, moxifloxacin, levofloxacin, and temafloxacin and were similar for ofloxacin and enoxacin (Table 1).
FIG. 1.
Inhibitory activity of moxifloxacin and pipemidic acid on the supercoiling activity of M. tuberculosis DNA gyrase and wild-type (WT) and mutant proteins carrying GyrA mutations at position 88. Relaxed pBR322 DNA (0.4 μg) was incubated with gyrase (2 U) reconstituted from WT GyrA with WT GyrB and from WT GyrB with G88A or G88C GyrA. Incubation was carried out in the presence of 1 mM ATP and in the absence or presence of the indicated amounts (in micrograms/milliliter) of moxifloxacin or pipemidic acid. Reactions were stopped, and the DNA was examined by electrophoresis in 1% agarose. R and S denote relaxed and supercoiled DNA, respectively.
Glycine at position 88 in M. tuberculosis, which corresponds to the glycine at position 81 in Escherichia coli, is a small and flexible amino acid lying at the N-terminal end of the α4 helix, which may be part of the drug binding site (16). Since alanine is a hydrophobic amino acid whereas cysteine is polar and more bulky than alanine and glycine, it is consistent that the level of resistance to fluoroquinolone inhibition observed with the gyrase harboring GyrA G88C was higher than with the gyrase harboring GyrA G88A.
Intriguingly, for the four classical quinolones tested which do not have either an R7 ring (nalidixic acid and flumequine) or a fluor in R6 (oxolinic acid, pipemidic acid, and nalidixic acid), MICs were similar for the G88A mutants and the H37Rv strain, and IC50s of both G88C and G88A mutant gyrases were similar to the IC50s of the WT gyrase (Table 1; Fig. 1). These results are consistent with the observation of the mutation G81D in E. coli that conferred resistance to ciprofloxacin but susceptibility to nalidixic acid (4), whereas mutations at positions 83, 84, and 87 in E. coli conferred resistance to ciprofloxacin as well as to nalidixic acid (20). Since resistance to quinolones is usually crossed between all drugs of the family (20), we controlled these results by measuring IC50s of these four quinolones for the mutant gyrase with a GyrA D94G known to be implicated in fluoroquinolone resistance in M. tuberculosis (2). For the D94G mutant, IC50s of flumequine and nalidixic acid were at least threefold higher than for the WT and reached >2,000 and >3,200 μg/ml, respectively (Table 1).
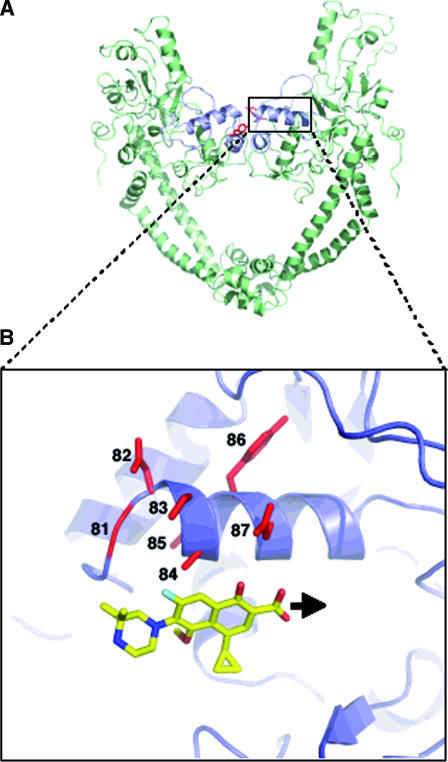
In E. coli, G81 is assumed to interact with the R7 fluoroquinolone ring substituent, while the remaining core of the quinolone interacts with amino acids 83 and 87, the positions strongly involved in quinolone resistance (16). The same observation has been made by Sindelar et al. (21), who suggested that for M. smegmatis, a rapidly growing mycobacteria, the R7 ring binds to gyrase near amino acid 81. It is conceivable that modifications of gyrase such as those observed in M. tuberculosis WT (A83) and the mutant at position 81 (A- or C-81) could create a steric hindrance, leading to a displacement of the binding of quinolones compared to that of E. coli as shown in Fig. 2. Consequently, the binding of molecules with a bulky group at position R7 decreases, while no change in binding would occur with molecules like nalidixic acid and flumequine, which have a linear substituent at that position. The results obtained for the enzymes carrying mutations at position 88 bring new insights into the role of the positioning of quinolones into the quinolone binding pocket of the M. tuberculosis DNA gyrase.
FIG. 2.
(A) Ribbon representation of the 59-kDa N terminal of GyrA adapted from the crystal structure of the breakage-reunion domain of the GyrA protein of E. coli (16). The QRDR (quinolone resistance-determining region) is in blue, and the active site (Tyr122) is in red. (B) Close-up of the region outlined by the broken box highlighting the schematic representation of the fluoroquinolone positioning in the GyrA α4 helix domain. The fluoroquinolone used for the model is gatifloxacin. The black arrow shows the displacement of the fluoroquinolone positioning in the M. tuberculosis GyrA α4 helix domain compared to the one proposed for E. coli as suggested by our study.
Acknowledgments
We thank Aurélie Chauffour for MIC determinations.
This work was supported by grants from Ministère de l'Education Nationale et de la Recherche (grant UPRES 1541) and Association Française Raoul Follereau.
Footnotes
Published ahead of print on 2 October 2006.
REFERENCES
- 1.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 48:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, A., N. Veziris, E. Cambau, C. Truffot-Pernot, V. Jarlier, and L. M. Fisher. 2006. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob. Agents Chemother. 50:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, A. A. Vernon, et al. 2003. Treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 4.Cambau, E., F. Bordon, E. Collatz, and L. Gutmann. 1993. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob. Agents Chemother. 37:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cambau, E., and V. Jarlier. 1996. Resistance to quinolones in mycobacteria. Res. Microbiol. 147:52-59. [DOI] [PubMed] [Google Scholar]
- 6.Cambau, E., W. Sougakoff, M. Besson, C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1994. Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J. Infect. Dis. 170:479-483. [DOI] [PubMed] [Google Scholar]
- 7.Chan, E. D., V. Laurel, M. J. Strand, J. F. Chan, M. L. Huynh, M. Goble, and M. D. Iseman. 2004. Treatment and outcome analysis of 205 patients with multidrug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 169:1103-1109. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, A. F., W. W. Ew, E. W. Chan, M. L. Chin, M. M. Hui, and R. C. Chan. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 11.Crofton, J., P. Chaulet, and D. Maher. 1997. Guidelines for the management of drug-resistant tuberculosis, p. 1-40. World Health Organization, Geneva, Switzerland.
- 12.Grimaldo, E. R., T. E. Tupasi, A. B. Rivera, M. I. Quelapio, R. C. Cardano, J. O. Derilo, and V. A. Belen. 2001. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant Mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int. J. Tuberc. Lung Dis. 5:546-550. [PubMed] [Google Scholar]
- 13.Ji, B., N. Lounis, C. Maslo, C. Truffot-Pernot, P. Bonnafous, and J. Grosset. 1998. In vitro and in vivo activities of moxifloxacin and clinafloxacin against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 15.Lindstedt, B. A., L. Aas, and G. Kapperud. 2004. Geographically dependent distribution of gyrA gene mutations at codons 83 and 87 in Salmonella Hadar, and a novel codon 81 Gly to His mutation in Salmonella enteritidis. APMIS 112:165-171. [DOI] [PubMed] [Google Scholar]
- 16.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 17.Perlman, D. C., W. M. El Sadr, L. B. Heifets, E. T. Nelson, J. P. Matts, K. Chirgwin, N. Salomon, E. E. Telzak, O. Klein, B. N. Kreiswirth, J. M. Musser, R. Hafner, et al. 1997. Susceptibility to levofloxacin of Mycobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. AIDS 11:1473-1478. [DOI] [PubMed] [Google Scholar]
- 18.Pitaksajjakul, P., W. Wongwit, W. Punprasit, B. Eampokalap, S. Peacock, and P. Ramasoota. 2005. Mutations in the gyrA and gyrB genes of fluoroquinolone-resistant Mycobacterium tuberculosis from TB patients in Thailand. Southeast Asian J. Trop. Med. Public Health 36(Suppl. 4):228-237. [PubMed] [Google Scholar]
- 19.Rafii, F., M. Park, and J. S. Novak. 2005. Alterations in DNA gyrase and topoisomerase IV in resistant mutants of Clostridium perfringens found after in vitro treatment with fluoroquinolones. Antimicrob. Agents Chemother. 49:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz, J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51:1109-1117. [DOI] [PubMed] [Google Scholar]
- 21.Sindelar, G., X. Zhao, A. Liew, Y. Dong, T. Lu, J. Zhou, J. Domagala, and K. Drlica. 2000. Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrob. Agents Chemother. 44:3337-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO. 2004. Antituberculosis drug resistance in the world. The WHO/IUATLD Global Project on Antituberculosis Drug Resistance Surveillance 1999-2002. Report no. 3/WHO/CDS/TB/2004. World Health Organization, Geneva, Switzerland.