Abstract
We investigated the β-lactam resistance mechanism(s) of an in vitro-selected amoxicillin-resistant Helicobacter pylori strain (AmoxR). Our results demonstrated that resistance is due to a combination of amino acid substitutions in penicillin binding protein 1 (PBP1), HopB, and HopC identified in AmoxR, resulting in decreased affinity of PBP1 for amoxicillin and decreased accumulation of penicillin.
Helicobacter pylori is the most common cause of gastric and duodenal ulcers and is strongly associated with the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (4). Treatment of H. pylori infection typically employs a triple-drug regimen using two antibiotics and a proton pump inhibitor or bismuth (4). However, resistance to the most commonly used antibiotics has already been observed worldwide (3, 8).
In a previous study (1), we isolated an amoxicillin-resistant mutant strain of H. pylori 43579 (designated AmoxR) with a MIC of 4 to 8 μg/ml by progressively culturing an amoxicillin-susceptible strain (AmoxS; MIC, 0.03 to 0.06 μg/ml) in increasing amounts of amoxicillin. The penicillin binding protein (PBP) profile of this strain showed a decreased affinity of PBP1 for amoxicillin, and AmoxR demonstrated a decrease in 14C-labeled penicillin G accumulation (1). In this study, we report the presence of pbp1 and hop mutations in the in vitro-selected amoxicillin-resistant mutant AmoxR and determine their effects on amoxicillin susceptibility.
The pbp1 genes from AmoxS and AmoxR were amplified by PCR using primer sequences derived from the published 26695 sequence (12). The PCR products were sequenced at the University of California (UC), Riverside, Genomics Institute, and sequence alignment was performed in Sequencher (Gene Codes Corporation, Ann Arbor, MI). Comparison of the two pbp1 sequences revealed a threonine-to-methionine conversion in the 438th amino acid position of PBP1 in AmoxR, located adjacent to an SLN motif, one of the penicillin binding domains (7). Others have also identified mutations localized around the SKN, SNN, and KTG motifs (7), which may be involved in β-lactam resistance (6, 9, 10, 11). To characterize the effect of this single T438M mutation in PBP1 on amoxicillin resistance, the gene was introduced into the well-characterized amoxicillin-susceptible (MIC, 0.06 μg/ml) H. pylori strain 700392, using electroporation as described by Wang et al. (13). Transformant selection was performed by plating electroporated cells onto brucella agar plates containing 5% sheep blood and amoxicillin at various concentrations. Several transformants (including PBP1-T) were randomly selected and found to have MICs for amoxicillin of 0.5 μg/ml, an eightfold increase compared to the parental strain (Table 1). Amplification and sequencing of the pbp1 gene from PBP1-T confirmed that the transformant possessed the mutant pbp1 from AmoxR. PBP1-T also had comparably increased MICs for ampicillin and penicillin G, although little or no increase in MIC levels was noted for the other β-lactams examined, suggesting that this particular pbp1 mutation in AmoxR affects the binding affinity of PBP1 for only a few specific antibiotics.
TABLE 1.
MICs of each β-lactam antibiotic for H. pylori 700392 and various transformantsa
| Strain | MIC
|
||||||
|---|---|---|---|---|---|---|---|
| Amoxicillin | Ampicillin | Penicillin G | Ceftriaxone | Cefuroxime | Mezlocillin | Characteristics | |
| 700392 | 0.06 | 0.06 | 0.06 | 0.5 | 0.5 | 1 | ATCC type strain |
| PBP1-T | 0.5 | 0.5 | 0.5 | 0.5 | 1 | 1 | PBP1 transformant |
| HopB-T | 0.25 | 0.5 | 1 | 1 | 1 | 2 | HopB transformant |
| HopC-T | 0.125 | 0.5 | 0.5 | 1 | 0.5 | 1 | HopC transformant |
| PBP1-T/HopB-T | 2 | 4 | 2 | 4 | 2 | 4 | PBP1/HopB double mutant |
| PBP1-T/HopC-T | 1 | 2 | 1 | 2 | 1 | 2 | PBP1/HopC double mutant |
| PBP1/HopB/HopC triple mutant | 4 | 8 | 2 | 2 | 2 | 4 | Triple mutant |
| AmoxR | 4-8 | 4-8 | 4-8 | 4-8 | 4-8 | 2-4 | In vitro-selected strain |
MICs were determined after 48 h of incubation at 37°C by a broth microdilution method as described previously (1). The data shown (in μg/ml) are the modes of at least three separate experiments.
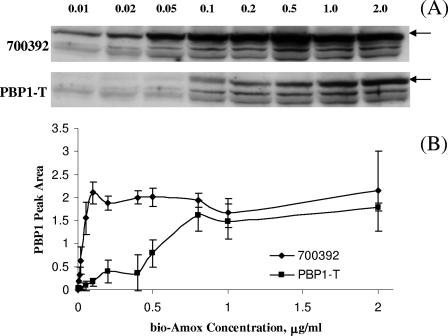
To compare the affinity of PBP1 for amoxicillin in 700392 and PBP1-T, we titrated the binding of PBP1 using biotinylated amoxicillin (bio-Amox) as described previously (1). In this study, equivalent amounts of membrane proteins were labeled for 10 min at room temperature with bio-Amox concentrations ranging from 0.001 to 2 μg/ml. At very low to moderate amoxicillin concentrations, the PBP1 in PBP1-T bound substantially less amoxicillin than the PBP1 in the susceptible parental strain (Fig. 1); it required more than eight times as much bio-Amox to achieve equivalent binding of PBP1 in PBP1-T as in 700392. Interestingly, the saturation of PBP1 with bio-Amox in both strains occurred at concentrations near their respective MICs (Fig. 1B). For strain 700392, saturation binding started between 0.05 and 0.1 μg/ml (MIC = 0.06), while in the transformant PBP1-T, saturation binding occurred at 0.8 μg/ml (MIC = 0.5). These data suggest a possible correlation between the degree of amoxicillin binding and the level of susceptibility to amoxicillin.
FIG. 1.
(A) Bio-Amox labeling of 700392 and PBP1-T PBPs. Equivalent amounts of membrane proteins were exposed to increasing amounts of bio-Amox to saturation. The numbers indicate bio-Amox concentrations (μg/ml) used to label PBP1. The arrows indicate the locations of the PBP1 band; the two lower bands correspond to PBP2 and PBP3. (B) Quantitative densitometric tracings of PBP1 bands. The data represent the means ± standard deviations based on four separate experiments (including the gel shown in panel A).
In our previous study, AmoxR accumulated >40% less 14C-labeled penicillin G than equal numbers of AmoxS bacteria (1). Since decreased permeability of antibiotics through the outer membrane of AmoxR could account for the lower accumulation of 14C-labeled penicillin G, we focused on H. pylori porin proteins (2, 5, 12). Sequence alignments of the genes hopA, hopD, and hopE from AmoxR, compared to their parental counterparts in AmoxS, did not reveal any amino acid substitutions. On the other hand, the hopB gene of AmoxR possesses a number of mutations localized in the amino acid 116 to 201 region, and the hopC gene possesses a stop codon at the 211th amino acid position (data not shown). When these genes were transformed into 700392, the resulting strains were observed to have increased MICs for amoxicillin of 0.25 μg/ml for HopB-T and 0.125 μg/ml for HopC-T (Table 1). When the MICs for other antibiotics were tested, both transformants had elevated MICs for ampicillin and penicillin G but little or no increase for the other β-lactams tested.
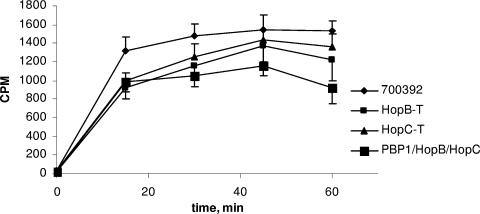
Subsequent experiments revealed decreased 14C-labeled penicillin G accumulation in the hop mutants, approximately 20% and 10% less than that in the parental strain for HopB-T and HopC-T, respectively, after 60 min of incubation. The most dramatic decrease was observed at the 15-min time point, when ∼30% less 14C-labeled penicillin G had accumulated in both strains (Fig. 2).
FIG. 2.
14C-labeled penicillin G accumulation of H. pylori 700392 and hop transformants. Log-phase cultures were incubated with 14C-labeled penicillin G, and 1-ml aliquots were taken at 15, 30, 45, and 60 minutes, centrifuged, and washed, and radioactivity was measured in a scintillation counter. The error bars represent the means ± standard errors of the means based on three separate experiments.
Double mutants possessing pbp1-T438M and altered hop genes were constructed and showed increased MICs for all β-lactams tested in an additive fashion, particularly the PBP1-T/HopB mutant (Table 1), indicating that the PBP1 and HopB alterations work in concert to decrease the effectiveness of the antibiotics. When both hop genes were transformed into PBP1-T, creating a triple mutant, a further decrease in 14C-labeled penicillin G accumulation (Fig. 2), as well as a concurrent increase in the MIC, was observed (Table 1). The MIC level for amoxicillin observed in the triple mutant was 4 μg/ml, comparable to that seen in AmoxR (4 to 8 μg/ml). In addition, the 14C-labeled penicillin G accumulation levels in this triple mutant reflected about a 40% decrease compared to the parental strain at the 60-min time point (Fig. 2), comparable to that observed with AmoxR (1). These experiments suggest that the amino acid substitutions affecting PBP1, HopB, and HopC account for all the resistance mechanisms found in AmoxR.
Acknowledgments
This research was supported in part by grants from UC MEXUS and the UC Cancer Research Coordinating Committee. E.-M. A. Co acknowledges support provided by a Graduate Division Dissertation Grant at UCR.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.DeLoney, C. R., and N. L. Schiller. 2000. Characterization of an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3368-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doig, P., M. M. Exner, R. E. W. Hancock, and T. J. Trust. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duck, W. M., J. Sobel, J. M. Pruckler, Q. Song, D. Swedlow, C. Friedman, A. Sulka, B. Swaminathan, T. Taylor, M. Hoekstra, P. Griffin, D. Smoot, R. Peek, D. C. Metz, P. B. Bloom, S. Goldschmid, J. Parsonnet, G. Triadafilopoulos, G. I. Perez-Perez, N. Vakil, P. Ernst, S. Czinn, D. Dunne, and B. D. Gold. 2004. Antimicrobial resistance incidence and risk factors among Helicobacter pylori-infected persons, United States. Emerg. Infect. Dis. 10:1088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Exner, M. M., P. Doig, T. J. Trust, and R. E. W. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrits, M. M., D. Schuijffel, A. A. van Zwet, E. J. Kuipers, C. M. J. E. Vanderbrouke-Grauls, and J. G. Kursters. 2002. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2229-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris, A. G., S. L. Hazell, and A. G. Netting. 2000. Use of digoxigenin-labelled ampicillin in the identification of penicillin-binding proteins in Helicobacter pylori. J. Antimicrob. Chemother. 45:591-598. [DOI] [PubMed] [Google Scholar]
- 8.Kursters, J. G., and E. J. Kuipers. 2001. Antibiotic resistance of Helicobacter pylori. J. Appl. Microbiol. 90:134s-144s. [DOI] [PubMed] [Google Scholar]
- 9.Kwon, D. H., M. P. Dore, J. J. Kim, M. Kato, M. Lee, J. Y. Wu, and D. Y. Graham. 2003. High-level β-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto, T., H. Yoshiyama, T. Nakazawa, I.-D. Park, M.-W. Chang, H. Yanai, K. Okita, and M. Shirai. 2002. A change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J. Antimicrob. Chemother. 50:849-856. [DOI] [PubMed] [Google Scholar]
- 11.Paul, R., S. Postius, K. Melchers, and K. P. Schafer. 2001. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob. Agents Chemother. 45:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quakenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-553. [DOI] [PubMed] [Google Scholar]
- 13.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]