Abstract
An erm(B) gene carried on the Lactobacillus johnsonii G41 chromosome and the upstream and downstream regions were fully sequenced. Apparently, a 1,495-bp segment of pRE25 from Enterococcus faecalis carrying the erm(B) gene became inserted, by an unknown mechanism, into the L. johnsonii chromosome.
The possession of no acquired resistance to antibiotics is a key safety criterion that candidate probiotic microorganisms should fulfill besides functional and technological properties (6). A collection of intestinal lactic acid bacteria and bifidobacteria of human origin was investigated for antibiotic resistance during a survey for new probiotic strains (1). Among them, one Lactobacillus johnsonii strain (G41) was suspected of being resistant to erythromycin. Characterization of antibiotic resistance genes and the elements involved in their transfer might help to reduce the spread of antibiotic resistance by the food chain (2, 8). Besides mutations in the 23S rRNA molecule, more than 40 genes encoding efflux proteins, methylases, and inactivating enzymes have been described previously (4).
The MIC of erythromycin for strain G41 was found to be 256 μg ml−1 by Etest (AB Biodisk, Solna, Sweden). A MIC of 4 μg ml−1 has been recently proposed as the microbiological breakpoint to separate susceptible from resistant strains (2). In addition, the MIC of clindamycin for this strain was also 256 μg ml−1.
Identification and location of the erm(B) determinant.
The genes ermA, erm(B), erm(C), erm(F), and mef(A) are widely distributed among gram-positive and gram-negative organisms (3, 5). Using as a template total DNA from L. johnsonii G41, positive amplification was only obtained for erm(B); the primers and PCR conditions used were reported elsewhere (5). The sequence of this amplicon proved to be identical to erm(B) sequences from Streptococcus, Enterococcus, and Lactobacillus sp. strains.
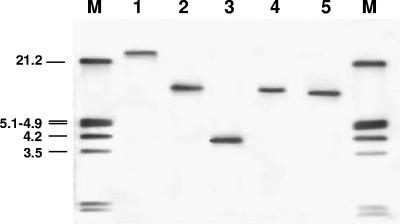
As no plasmid DNA was detectable in L. johnsonii G41, hybridization experiments, using as a probe a digoxigenin-labeled erm(B) internal segment and high-stringency conditions, located the erm(B) determinant on the bacterial chromosome (Fig. 1).
FIG. 1.
Southern blot analysis of total genomic DNA from L. johnsonii G41 left undigested or digested with EcoRI, BlgII, PstI, or ClaI (lanes 1 through 5, respectively) and hybridized with an internal segment of the erm(B) gene obtained by PCR and labeled with digoxigenin (Roche [Hoffmann-La Roche Ltd., Basel, Switzerland]). Lanes M, molecular size marker (digoxigenin-labeled EcoRI-HindIII-digested lambda DNA [Roche]). The values on the left are molecular sizes in kilobase pairs.
Sequence and analysis of the erm(B) gene and the surrounding regions.
Total DNA from L. johnsonii was digested with EcoRI and HindIII, self-ligated, and used as a template for inverse PCR with primers erm(B)1f (5′-CATCAAGCAATGAAACACG-3′) and emrB2r (5′-GTCTGTTTCAAAACAGTAGATG-3′), which are based on erm(B) internal sequences. An amplicon of around 5.5 kbp was obtained with the EcoRI-digested ligation, from which the complete sequence of erm(B) and the upstream and downstream regions were obtained (GenBank accession no. DQ518904).
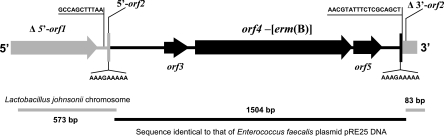
Two partial open reading frames (ORFs) with high homology to two contiguous genes on the L. johnsonii NCC 533 chromosome (GenBank accession no. NC_005362) were observed at the extremes of the segment (Fig. 2). The first (Δ5′-orf1 in Fig. 2) encoded the C-terminal part of a phosphoenolpyruvate phosphotransferase component (protein identification no. ABF70486.1). A second ORF split into two parts by the erm(B) region (5′-orf2 and Δ3′-orf2 in Fig. 2) may code for a hypothetical protein (protein identification no. AAS09591.1).
FIG. 2.
Diagram of the erm(B) gene of L. johnsonii G41 and the sequences and structures at its chromosomal integration position. Gray and black bars indicate the segment homologous to the L. johnsonii NC 533 chromosome (GenBank accession no. NC_005362) and that identical to the erm(B) locus in E. faecalis plasmid pRE25 (positions 12,527 to 14,022 of the sequence with GenBank accession no. X92945), respectively. ORFs and sequences are as indicated in the text. This diagram is not drawn to scale.
The inserted sequence comprised 1,495 bp identical to those corresponding to the erm(B) locus in Enterococcus faecalis plasmid pRE25 (7) (positions 12,577 to 14,022 of the sequence with GenBank accession no. X92945). This segment included three complete ORFs related to the MLS phenotype [orf3, encoding the putative MLS leader peptide; the erm(B) gene (orf4); and orf5, a second small ORF present at most erm(B) loci (3, 5)] and some adjacent pRE25 sequences.
The insertion is flanked by a duplicated 9-bp sequence (AAAGAAAAA) (Fig. 2), which is originally present once at the start of corresponding orf2 in the L. johnsonii NCC 533 genome and twice and in the same place and orientation in the pRE25 sequence. Chromosomal and plasmid sequences might provide homology for the integration of pRE25 in a manner similar to phage integration by attB and attP sites. However, two sequences with no homology to other DNA sequences appeared around the junction points (Fig. 2): one at the 3′ end (AACGTATTTCTCGCAGCT), immediately downstream of the 9-bp sequence, and a second one at the 5′ end (GCCAGCTTTAA), 75 bp upstream of the 9-bp sequence. These sequences suggest further DNA rearrangement during or after integration.
In conclusion, this work reports on the molecular characterization of an erm(B) gene encoding erythromycin and clindamycin resistance in L. johnsonii G41. The gene was found to be identical to that present in many gram-positive bacteria. Analysis of the surrounding regions suggested that a segment of the erm(B) locus from enterococcal plasmid pRE25 was integrated and rearranged into the L. johnsonii genome via unknown mechanisms.
Acknowledgments
This work was supported by an EU project within the sixth Framework Programme (ACE-ART, reference no. 506214). M. S. Ammor was awarded a postdoctoral fellowship from the Secretaría de Estado de Universidades e Investigación of the Spanish Ministry of Education and Science (reference no. SB2004-0165).
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Delgado, S., A. B. Flórez, and B. Mayo. 2005. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 50:202-207. [DOI] [PubMed] [Google Scholar]
- 2.European Commission. 2005. Opinion of the Scientific Panel on Additives and Products in Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human and veterinary importance. EFSA J. 223:1-12. [Google Scholar]
- 3.Jensen, L. B., N. Frimodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 4.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 5.Roberts, M. C., W. O. Chung, D. Roe, M. Xia, C. Marquez, G. Borthagaray, W. L. Whittington, and K. K. Holmes. 1999. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob. Agents Chemother. 43:1367-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saarela, M., G. Mogensen, R. Fonden, J. Mättö, and T. Mattila-Sandholm. 2000. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84:197-215. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 8.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]