Abstract
Military medical facilities treating patients injured in Iraq and Afghanistan have identified a large number of multidrug-resistant (MDR) Acinetobacter baumannii isolates. In order to anticipate the impact of these pathogens on patient care, we analyzed the antibiotic resistance genes responsible for the MDR phenotype in Acinetobacter sp. isolates collected from patients at the Walter Reed Army Medical Center (WRAMC). Susceptibility testing, PCR amplification of the genetic determinants of resistance, and clonality were determined. Seventy-five unique patient isolates were included in this study: 53% were from bloodstream infections, 89% were resistant to at least three classes of antibiotics, and 15% were resistant to all nine antibiotics tested. Thirty-seven percent of the isolates were recovered from patients nosocomially infected or colonized at the WRAMC. Sixteen unique resistance genes or gene families and four mobile genetic elements were detected. In addition, this is the first report of blaOXA-58-like and blaPER-like genes in the U.S. MDR A. baumannii isolates with at least eight identified resistance determinants were recovered from 49 of the 75 patients. Molecular typing revealed multiple clones, with eight major clonal types being nosocomially acquired and with more than 60% of the isolates being related to three pan-European types. This report gives a “snapshot” of the complex genetic background responsible for antimicrobial resistance in Acinetobacter spp. from the WRAMC. Identifying genes associated with the MDR phenotype and defining patterns of transmission serve as a starting point for devising strategies to limit the clinical impact of these serious infections.
The emergence and rapid spread of multidrug-resistant (MDR) isolates of Acinetobacter spp. causing nosocomial infections are of great concern worldwide (1, 23). The Centers for Disease Control and Prevention recently highlighted the enormity and gravity of MDR Acinetobacter baumannii infections in military medical facilities treating civilians and service personnel injured in Iraq/Kuwait and Afghanistan (7). Antimicrobial susceptibility testing of these isolates originally reported that more than half were resistant to three or more classes of antibiotics. This resistance profile leaves clinicians with limited therapeutic options. Another significant problem is the large number of military personnel that may be colonized by MDR Acinetobacter spp. (16). This reservoir would serve to widely disseminate MDR Acinetobacter spp. As has been seen with methicillin-resistant Staphylococcus aureus isolates and vancomycin-resistant enterococci (VRE), patients who are colonized can spread MDR pathogens (5, 6, 30).
Here, we report an analysis of the antibiotic susceptibility profile and genetic determinants of antibiotic resistance in 75 isolates of Acinetobacter spp. obtained from military and civilian patients at the Walter Reed Army Medical Center (WRAMC). This hospital was the major U.S. site receiving casualties from the conflict in Iraq/Kuwait and Afghanistan. Understanding the genetic background of antibiotic resistance is an important step in defining the impact of MDR Acinetobacter spp. in military and civilian patients.
MATERIALS AND METHODS
Study population, bacterial isolate identification, and susceptibility testing.
The Acinetobacter sp. isolates were collected from 75 unique civilian and military patients treated at the WRAMC in Washington, D.C. A medical record review was performed for each case. For the purpose of this particular study, (i) clinical infections were defined using standard criteria set by the National Nosocomial Infections Surveillance System (40) and (ii) hospital-acquired infection was defined as infection that occurred at least 72 h after arrival at the WRAMC. The injury severity score (values from 0 to 75) was calculated based upon the method of Baker et al. (3). Institutional Review Board approval for this retrospective medical chart review was obtained at the WRAMC.
All isolates were collected from March 2003 through February 2005. Fifty-two of the 75 isolates examined were collected from February 2004 to February 2005 and represent 67% of all isolates of Acinetobacter spp. collected at the WRAMC during that time period. An additional 9% of Acinetobacter sp. isolates from that time period were received but eliminated from this study due to insufficient patient information or being duplicate patient samples. In the case of duplicate patient samples, the first collected Acinetobacter sp. isolate was chosen to eliminate bias.
Antimicrobial susceptibility testing was performed on the 75 isolates according to the method established by the CLSI (formerly NCCLS) and interpreted with criteria published in 2005 (9, 31). Isolates were tested using cation-adjusted Mueller-Hinton agar (BBL MH II; Becton Dickinson Microbiology Systems, Sparks, MD). Meropenem, imipenem, ampicillin, ampicillin-sulbactam, ciprofloxacin, ceftazidime, cefepime, amikacin, and tobramycin disks (Becton Dickinson Microbiology Systems) were used.
Multidrug resistance was defined in this analysis as resistance to three or more representatives of the following classes of antibiotics: quinolones (ciprofloxacin), extended-spectrum cephalosporins (ceftazidime and cefepime), β-lactam/β-lactamase inhibitor combination (ampicillin-sulbactam), aminoglycosides (amikacin and tobramycin), and carbapenems (imipenem and meropenem).
PCR amplification, cloning, DNA sequencing, and restriction endonuclease digestion.
All target genes and corresponding primers used for PCR amplification are listed in Table 1. Positive controls were used for β-lactamase gene amplification as follows: blaSHV-1 cloned into pBC SK(−) (Stratagene, La Jolla, CA), blaTEM-1 cloned into the commercially available pBR322 (New England BioLabs, Beverly, MA), blaADC-7 cloned into pBC SK(+), blaCTX-M-2 cloned into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA), RGN238 blaOXA-1 construct, the isolate containing blaIMP-2 (a kind gift from G. Rossolini), and Pseudomonas aeruginosa PU21 possessing blaPER-1 (a kind gift from F. Danel) (11, 21, 38, 39, 45). Positive control reactions for blaVIM, blaGIM, and blaVEB-1 were not performed. For OXA set A to D, positive controls were also not used. Presumptive positive PCR products for β-lactamase genes were identified according to size and either cloning into the pCR-XL-TOPO vector followed by sequencing or direct sequencing of the PCR product. PCR product identification via pCR-XL-TOPO vector cloning and sequencing was carried out for representative blaADC, blaOXA-23-like, blaOXA-69-like, blaOXA-58-like, and blaPER positive amplicons. Direct sequencing of select positive amplicons was carried out for blaSHV and blaTEM. For all other PCR amplifications, the products obtained were considered presumptive positives based on amplicon size. These include PCR amplification reactions for all aminoglycoside-modifying enzyme (AME) genes, all quinolone resistance-determining regions (QRDRs) of gyrA and parC genes, adeE, adeR, carO, and int genes, variable regions of class 1 integrons, and insertion (IS) elements. In the case of a negative PCR, in which a positive control was not used, PCR amplifications were repeated at least twice for these genes. A negative control was run with every PCR.
TABLE 1.
Primers for amplification of genes from Acinetobacter sp. isolates
| Primer name | Primer sequence (5′ to 3′)a | Annealing temp (°C) | Target gene(s) | Source or reference |
|---|---|---|---|---|
| SHV PROX | ATGCGTTATATTCGCCTGTG | 60 | All blaSHV genes | 33 |
| SHV DIS | TGCTTTGTTATTCGGGCCAA | |||
| TEM 285 | AAACGCTGGTGAAAGTA | 45 | All blaTEM genes | 33 |
| TEM 2023 | AGCGATCTGTCTAT | |||
| CTX-M2-1 | ATGATGACTCAGAGCATTCGCCGCT | 70 | blaCTX-M-2, blaCTX-M-4, blaCTX-M-5, | 33 |
| CTX-M2-2 | TCAGAAACCGTGGGTTACGATTTTCG | blaCTX-M-20, blaCTX-M-31, blaCTX-M-35, blaTOHO-1 | ||
| VEB-1 FOR | ATGAAAATCGTAAAAAGGATATT | 47 | blaVEB-1, blaVEB-1A, blaVEB-1B, blaVEB-2, | This study |
| VEB-1 REV | TTATTTATTCAAATAGTAATTCC | blaVEB-3 | ||
| PER F-1 | ATGAATGTCATTATAAAAG | 44 | blaPER-1 | This study |
| PER R-2 | TTGGGCTTAGGGCAG | |||
| IMP FOR | GTTTATGTTCATACWTCG | 45 | blaIMP-1, blaIMP-2, blaIMP-4, blaIMP-5, | This study |
| IMP REV | GGTTTAAYAAAACAACCAC | blaIMP-6, blaIMP-7, blaIMP-8, blaIMP-9, blaIMP-10, blaIMP-11, blaIMP-15, blaIMP-19, blaIMP-20, blaIMP-21 | ||
| VIM FOR | TTTGGTCGCATATCGCAACG | 66 | blaVIM-1, blaVIM-2, blaVIM-4, blaVIM-5, | This study |
| VIM REV | CCATTCAGCCAGATCGGCAT | blaVIM-6, blaVIM-8, blaVIM-9, blaVIM-10, blaVIM-11, blaVIM-12 | ||
| GIM FOR | ATATTACTTGTAGCGTTGCCAGC | 61 | blaGIM | This study |
| GIM REV | TTAATCAGCCGACGCTTCAG | |||
| ADC-7 FOR | ATGCGATTTAAAAAAATTTCTTGT | 50 | blaADC-1, blaADC-2, blaADC-3, blaADC-4, | 21 |
| ADC-7 REV | TTATTTCTTTATTGCATTCAG | blaADC-5, blaADC-6, blaADC-7 | ||
| OXA SET A FOR | ATGAAAAAATTTATACTTCC | 47 | blaOXA-24, blaOXA-25, blaOXA-26, blaOXA-33, | This study |
| OXA SET A REV | TTAAATGATTCCAAGATTTTC | blaOXA-40, blaOXA-72 | ||
| OXA SET B FOR | TCTGGTTGTACGGTTCAGC | 51 | blaOXA-23, blaOXA-27, blaOXA-49 | This study |
| OXA SET B REV | AGTCTTTCCAAAAATTTTG | |||
| OXA SET C FOR | ACAGAARTATTTAAGTGGG | 47 | blaOXA-51, blaOXA-58, blaOXA-64, blaOXA-69, | This study |
| OXA SET C REV | GGTCTACAKCCMWTCCCCA | blaOXA-70, blaOXA-71, blaOXA-75, blaOXA-78 | ||
| OXA SET D FOR | TAGCACTGCTTTTCTCAGCTG | 51 | blaOXA-20, blaOXA-37 | This study |
| OXA SET D REV | TTGACGGATTGAAGAATAGCACG | |||
| OXA1F | ACACAATACATATCAACTTCGC | 50 | blaOXA-1, blaOXA-4, blaOXA-30, blaOXA-31, | This study |
| OXA1R | GTGTGTTTAGAATGGTGATC | blaOXA-47 | ||
| OXA58-5′ | ATGAAATTATTAAAAATATTGAGTTTAG | 53 | blaOXA-58, blaOXA-96 | This study |
| OXA58-3′ | TTATAAATAATGAAAAACACCCAAC | |||
| Hep35 | TGCGGGTYAARGATBTKGATTT | 49 | Conserved region intI1, intI2, intI3 | 53 |
| Hep36 | CARCACATGCGTRTARAT | |||
| Hep58 | TCATGGCTTGTTATGACTGT | 55 | Class 1 integron cassette array | 53 |
| Hep59 | GTAGGGCTTATTATGCACGC | |||
| ISABA1a | ATGCAGCGCTTCTTTGCAGG | 55 | ISABA1 | 19 |
| ISABA1b | AATGATTGGTGACAATGAAG | |||
| IS1133 for | AGTACAAAAGCTGTGAGATTTCAG | 58 | IS1133 | This study |
| IS1133 rev | GATATTCATGAGCGCAATATTGGCT | |||
| adeR-5′ | ATGTTTGATCATTCTTTTTCTTTTG | 46 | adeR (regulatory gene of AdeABC | This study |
| adeR-3′ | TTAATTAACATTTGAAATATG | efflux gene cluster) | ||
| adeE-5′ | ATGTTGTCGAGTTTTTTTATTGCACG | 60 | adeE (efflux gene of AdeDE) | This study |
| adeE-3′ | TCATTTTGCCTTTTTGGTATTTAAC | |||
| aacA4 FOR | ATGACTGAGCATGACCTTGCG | 65 | aacA4 | This study |
| aacA4 REV | TTAGGCATCACTGCGTGTTCG | |||
| aacC1-5′ | ATGGGCATCATTCGCACATGTAGG | 64 | aacC1 | This study |
| aacC1-3′ | TTAGGTGGCGGTACTTGGGTC | |||
| aadA1-5′ | ATGAGGGAAGCGGTGATCG | 62 | aadA1 | This study |
| aadA1-3′ | TTATTTGCCGACTACCTTGGTG | |||
| aadB-5′ | ATGGACACAACGCAGGTCGC | 68 | aadB | This study |
| aadB-3′ | TTAGGCCGCATATCGCGACC | |||
| aphA6 FOR | ATGGAATTGCCCAATATTATTC | 55 | aphA6 | This study |
| aphA6 REV | TCAATTCAATTCATCAAGTTTTA | |||
| aacC2 FOR | ATGCATACGCGGAAGGCAATAAC | 65 | aacC2 | This study |
| aacC2 REV | CTAACCGGAAGGCTCGCAAG | |||
| gyrA-1 | AAATCTGCCCGTGTCGTTGGT | 63 | gyrA (QRDRb) | 52 |
| gyrA-2 | GCCATACCTACGGCGATACC | |||
| parC-1 | AAACCTGTTCAGCGCCGCATT | 58 | parC (QRDR) | 51 |
| parC-2 | AAAGTTGTCTTGCCATTCACT | |||
| QnrA | GGGTATGGATATTATTGATAAAG | 52 | qnrA | 27 |
| QnrB | CTAATCCGGCAGCACTATTA | |||
| CarO 5′ | ATGAAAGTATTACGTGTTTTAGTG | 50 | carO | This study |
| CarO 3′ | TTACCAGTAGAAGTTTACACC |
Y = C or T; M = A or T; R = A or G; B = C, G, or T; W = A or T; K = G or T.
QRDR, quinolone resistance-determining region.
For PCR, a 1:10 dilution of an overnight culture was boiled for 10 min. Amplification was then performed with 10 μl of this dilution as the DNA template. PCR conditions included 30 cycles of amplification under the following conditions: denaturation at 95°C for 30 s, annealing for 1 min at primer set-specific temperatures (Table 1), and extension at 72°C for 1 min/kb product. Cycling was followed by a final extension at 72°C for 10 min. PCR products were resolved on 1.0% agarose gels, stained with ethidium bromide, and photographed with UV illumination. Either the 1-kb DNA ladder or the 100-bp DNA ladder (Promega, Madison, WI) was used to assess PCR product size.
Amplification and HinfI (Promega) digestion of the QRDRs of topoisomerase IV parC and gyrA genes in Acinetobacter spp. were performed as follows. For the restriction endonuclease digestion, 40 μl of PCR amplification product was purified using a QIAquick PCR purification kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. The eluant was incubated at 37°C for 2.5 h with 10 U of HinfI. The digests were then separated by electrophoresis in 1.5% NuSieve (FMC BioProducts, Rockland, ME) plus 1.0% agarose gel stained with ethidium bromide and photographed with UV illumination. To serve as a control, the remaining 10 μl of undigested PCR product from each sample was resolved on the same agarose gel.
PCR products generated and cloned into pCR-XL-TOPO were sequenced using an ALF Express II automated DNA sequencer (Amersham Biosciences, Piscataway, NJ) with a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Biosciences) and Cy5-labeled M13 reverse and M13 universal primers (21). pCR-XL-TOPO clones containing the bla genes were cycle sequenced under the following conditions: DNA was heated to 94°C for 1 min, followed by 30 cycles of 94°C for 30 s, 55°C for 1 min, and 60°C for 2 min. Direct sequencing was performed for blaTEM and blaSHV genes by using previously described Cy5-labeled TEM104 reverse, TEM164 forward, and A forward primers under the same cycling conditions as those described above (20, 21).
Clonal determination by PCR ESI-MS.
To determine clonal relatedness by PCR electrospray ionization mass spectroscopy (PCR ESI-MS), the conserved regions of six bacterial housekeeping genes (efp, trpE, adk, mutY, fumC, and ppa) were amplified from each isolate by using eight sets of previously validated primers (14). The amplification products were then desalted and purified, and the mass spectra were determined using previously established protocols (13). These base compositions were compared to those of 23 isolates representing three major A. baumannii clone types (I, II, and III) that have spread throughout Europe (50). In addition, 26 American Type Culture Collection (ATCC) Acinetobacter sp. reference strains were used for Acinetobacter species verification. The European isolates and ATCC control strains used for this analysis were recently described by Ecker et al. (14).
RESULTS
Characteristics of the study population.
Acinetobacter sp. isolates were recovered from 75 patients: 67 men (89%) and 8 (11%) women. Patients in this study included active duty soldiers or civilians (e.g., contractors injured in Iraq) sent to Iraq/Kuwait or Afghanistan and civilians (family members of active duty soldiers and retirees eligible for care) hospitalized at the WRAMC. One patient was deployed to Afghanistan and 57 (76%) to Iraq/Kuwait, while 17 (23%) were nondeployed. More than half (63%) of the patients had positive Acinetobacter sp. cultures less than 3 days from the time of admittance.
The relevant descriptive statistics of the study population are summarized in Table 2. The mean age (± SD) of the population was 35 (± 18) years, with a range of 5 to 86 years. When Acinetobacter spp. were isolated, 53 patients (71%) were in an intensive care unit (ICU) or step-down unit, 51 (68%) were mechanically ventilated, and 66 patients (88%) had an indwelling central venous catheter. Sixty-seven patients (89%) had clinical infections and eight (11%) were colonized. Overall, five deaths (7% mortality rate) were observed in patients with A. baumannii infection (Table 3). Taken as a unique group, these five patients were older and were immunosuppressed. The death rate of the nondeployed patients was 29%. It is also notable that these isolates were from patients hospitalized for a prolonged period of time (Table 3).
TABLE 2.
Characteristics of the study populationa
| Parameter (units unless otherwise specified) | No. (%) of patients | Mean ± SD (range [minimum, maximum]) |
|---|---|---|
| Duration of hospitalization (days) | 75 | 53 ± 48 (3, 205) |
| Duration of ICU stay (days) | 75 | 19 ± 35 (0, 205) |
| Time febrile (days) | 75 | 11 ± 11 (0, 51) |
| Total surgeries (no.) | 75 | 6 ± 6 (0, 23) |
| Different antibiotics used (no.) | 75 | 6 ± 4 (1, 17) |
| Duration of antibiotic use (days) | 75 | 38 ± 37 (1, 205) |
| Changes to antibiotic regimen (no.) | 75 | 5 ± 5 (0, 21) |
| Debridement procedures (no.) | 75 | 4 ± 5 (0, 22) |
| Injury severity score | 58 | 21 ± 12 (4, 59) |
| APACHE II scoreb | 17 | 15 ± 10 (0, 30) |
| Hospital locations | ||
| Surgical ICU | 24 (32) | |
| Medical ICU | 26 (35) | |
| ICU step-down | 3 (4) | |
| ONPc | 11 (15) | |
| Surgery | 4 (5) | |
| Internal medicine | 4 (5) | |
| Emergency room | 3 (4) |
Patients in this study were civilians eligible for care (contractors injured in Iraq, family members of active duty soldiers, or retirees) and active duty military personnel hospitalized at the WRAMC.
The APACHE (Acute Physiology and Chronic Health Evaluation) II Score was calculated at the time of admission to ICU (medical and surgical).
ONP, orthopedics/neurology/physical medicine and rehabilitation.
TABLE 3.
Mortality associated with MDR A. baumannii infections in nonmilitary personnela
| Isolate | Age (yr) | Underlying condition(s) | Hospital day of death | Attributable mortality |
|---|---|---|---|---|
| AB001 | 71 | Chronic obstructive pulmonary disease | 27 | Ventilator-associated pneumonia |
| AB002 | 35 | Renal transplant | 44 | Pneumonia and lung abscess |
| AB029 | 85 | Abdominal aortic aneurysm | 88 | Ventilator-associated pneumonia |
| AB052 | 72 | Congestive heart failure, chronic renal failure | 45 | Pneumonia and sepsis |
| AB056 | 76 | Dementia | 93 | Anoxic brain injury and bacteremia |
The sixth death was a 21-year-old active duty soldier admitted due to a severe trauma blast. This patient died on hospital day 17 due to a pulmonary embolus. He previously had A. baumannii bacteremia (AB017) during his hospital course. All isolates except for AB029 were bloodstream infections and were acquired in the hospital.
Antimicrobial susceptibility.
Table 4 summarizes the sites of isolation and antibiotic resistance patterns found in these isolates compared to Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) data for Acinetobacter spp. in the United States during 2004. Isolates obtained at the WRAMC demonstrated significantly more resistance to ciprofloxacin, ceftazidime, cefepime, amikacin, tobramycin, imipenem, and meropenem (ampicillin-sulbactam was not available for comparison) than the U.S. Acinetobacter spp. collected in 2004.
TABLE 4.
Anatomical sites of isolation and antibiotic resistance patterns of isolates in this study
| Anatomical site | No. of isolates | % Resistance toa: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CIP | CAZ | FEP | SAM | AMK | TOB | IPM | MEM | ||
| All | 75 | 91 | 91 | 84 | 49 | 53 | 67 | 20 | 25 |
| Bloodstream | 40 | 90 | 90 | 88 | 58 | 50 | 70 | 23 | 25 |
| Skin | 18 | 89 | 94 | 78 | 56 | 61 | 72 | 17 | 28 |
| Wound | 8 | 100 | 100 | 75 | 25 | 50 | 75 | 13 | 13 |
| Respiratoryb | 5 | 80 | 60 | 80 | 40 | 60 | 60 | 20 | 20 |
| Urine | 2 | 100 | 100 | 100 | 100 | 100 | 50 | 50 | 50 |
| Intravenous catheter | 1 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 0 |
| Cerebrospinal fluid | 1 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 0 |
| MYSTICc | 44 | 42 | 32 | NRd | 14 | 13 | 8 | 8 | |
CIP, ciprofloxacin (quinolone); CAZ, ceftazidime (cephalosporin); FEP, cefepime (cephalosporin); SAM, ampicillin-sulbactam (β-lactam/β-lactamase inhibitor); AMK, amikacin (aminoglycoside); TOB, tobramycin (aminoglycoside); IPM, imipenem (carbapenem); MEM, meropenem (carbapenem).
Includes bronchoalveolar lavage fluid, trachea, and sputum.
Includes Acinetobacter spp. in the United States during 2004 in all specialties (http://www.mystic-data.org).
NR, not reported.
Overall, 89% of the isolates were resistant to at least three classes of antibiotics, hence meeting the criteria for multidrug resistance. More than 90% of the isolates were resistant to ciprofloxacin, and at least 80% were resistant to extended-spectrum cephalosporins. Nearly half (49%) of the isolates were also resistant to ampicillin-sulbactam. Notably, imipenem resistance was observed in 20% of isolates and meropenem resistance was detected in 25%. Eighty-one percent of isolates were resistant to at least one aminoglycoside (amikacin or tobramycin). Quite alarmingly, 11 isolates (15%) were resistant to the five classes of antibiotics tested.
Genotypic analysis: β-lactamase genes.
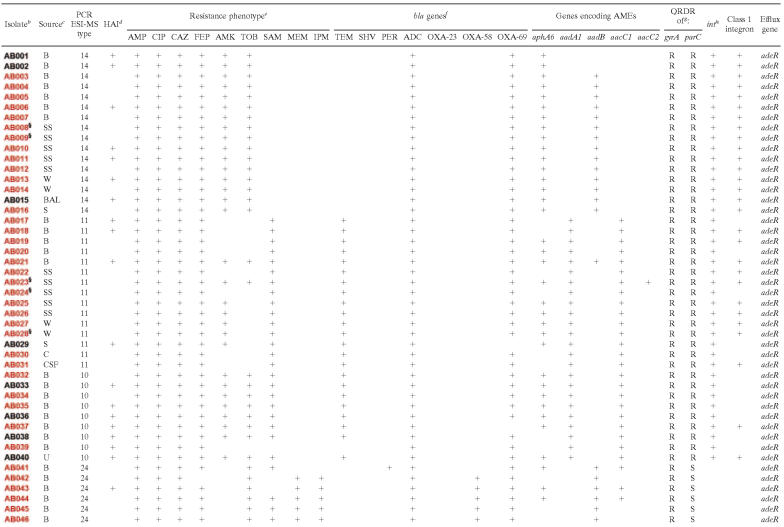
Table 5 lists the detected β-lactamase genes and the genes encoding the AMEs. Detailed genetic and phenotypic analyses of specific isolates can be found in Table 6. The ampC Acinetobacter-derived cephalosporinase, blaADC, was detected in 99% of the collection by PCR amplification (21). This cephalosporinase is responsible for resistance to ceftazidime and other extended-spectrum cephalosporins. Three distinct blaOXA-like genes were also identified. Using PCR, a blaOXA-69-like gene was found in 97% of the isolates (17). In two carbapenem-susceptible isolates (AB002 and AB006), ISABA1 was found to be proximal to the blaOXA-69-like gene. The blaOXA-23-like carbapenemase gene was detected in 11% of strains (12, 18), and the blaOXA-58-like carbapenemase gene was found in an additional 12% of the isolates (10, 35). Seventeen of 19 isolates (90%) that were imipenem and/or meropenem resistant by disk diffusion possessed evidence for the presence of a blaOXA-23-like or a blaOXA-58-like gene (Table 6). All eight isolates that contained a blaOXA-23-like gene were shown to have insertion sequence ISABA1 proximal to the gene, as previously reported by others (18, 49).
TABLE 5.
Percentage of each antibiotic resistance gene detected in the collection of Acinetobacter sp. isolates from the WRAMC
| Gene | % Detection in isolates |
|---|---|
| Genes encoding β-lactamases | |
| blaADC | 99 |
| blaOXA-69-likea | 97 |
| blaOXA-23-likeb | 11 |
| blaOXA-58-likec | 12 |
| blaTEM | 40 |
| blaSHV | 1 |
| blaPER | 3 |
| Genes encoding AMEsd | |
| aacC1 | 56 |
| aacC2 | 5 |
| aadA1 | 39 |
| aadB | 48 |
| aphA6 | 71 |
Includes blaOXA-51-, blaOXA-58-, blaOXA-64-, blaOXA-69-, blaOXA-70-, blaOXA-71-, blaOXA-75-, and blaOXA-78-like genes.
Includes blaOXA-23-, blaOXA-27-, and blaOXA-49-like genes.
Includes blaOXA-58- and blaOXA-96-like genes.
aacC1 confers gentamicin resistance. aacC2 confers tobramycin and gentamicin resistance. aadA1 confers streptomycin and spectinomycin resistance. aadB confers tobramycin, gentamicin, and kanamycin resistance. aphA6 confers amikacin, gentamicin, kanamycin, and neomycin resistance.
TABLE 6.
Summary of resistance: phenotypic and genetic characteristics of the isolatesa
+, presence of indicated gene or resistance phenotype.
All isolates are A. baumannii, except AG073 (Acinetobacter genome species 3) and AJ075 (Acinetobacter johnsonii). Red, deployed to Iraq/Kuwait; blue, deployed to Afghanistan; black, nondeployed; §, colonization (all others are clinically significant infections).
B, blood; SS, skin survey; W, wound; BAL, bronchoalveolar lavage fluid; S, sputum; C, intravenous catheter; CSF, cerebrospinal fluid; U, urine; TA, trachea aspirate.
HAI, hospital-acquired infection.
AMP, ampicillin; CIP, ciprofloxacin; CAZ, ceftazidime; FEP, cefepime; AMK, amikacin; TOB, tobramycin; SAM, ampicillin-sulbactam; MEM, meropenem; IPM, imipenem.
OXA-23, blaOXA-23-like gene; OXA-58, blaOXA-58-like gene; OXA-69, blaOXA-69-like gene; ADC, Acinetobacter-derived cephalosporinase.
Resistance (R) and susceptibility (S) were based on HinfI endonuclease restriction digests.
int, integrase genes intI1, intI2, and intI3.
PCR amplification of class A β-lactamase genes revealed that 40% (30/75) of the isolates contained blaTEM, only one isolate contained blaSHV, and two isolates contained blaPER (24, 34). This is also the first description of blaPER in the United States. Evidence for other common Acinetobacter β-lactamase genes was absent (blaCTX-M and other blaOXA genes), including the carbapenem-hydrolyzing metallo-β-lactamase genes blaIMP, blaVIM, and blaGIM.
DNA topoisomerase mutations and quinolone resistance.
More than 90% of our isolates were resistant to ciprofloxacin. HinfI digestion of the QRDR amplification products of gyrA and parC in these isolates indicated that 88% contain a point mutation at Ser83 in gyrA or Ser80 in parC leading to quinolone resistance. Except for two isolates, this analysis correlated with susceptibility studies (Table 6). Our primers did not detect the presence of the plasmid-mediated quinolone resistance gene qnrA in these 75 isolates (27, 42).
Genes encoding AMEs.
Screening for genes encoding AMEs demonstrated that 97% of the isolates that are amikacin resistant contained the phosphotransferase gene aphA6 (Tables 5 and 6). Other genes encoding AMEs included the adenylyltransferase genes aadA1 (39%) and aadB (48%) and the acetyltransferase genes aacC1 (56%) and aacC2 (5%). Seventy-eight percent (40/51) of the tobramycin-resistant isolates contained either the aacC2 or aadB gene.
Genes encoding efflux pumps.
An important resistance determinant in A. baumannii is the AdeABC efflux pump (26, 28). A. baumannii isolates demonstrating enhanced expression of this pump are resistant to aminoglycosides, quinolones, tetracyclines, and trimethoprim. The AdeABC operon possesses two genes that encode proteins that act as a sensor (adeS) and a regulator (adeR) of the pump. This regulatory gene was identified in 95% of the isolates. The expression of the AdeABC efflux pump is being further evaluated.
We also detected the efflux pump gene adeE (part of the AdeDE pump in Acinetobacter genome species 3) (8). This amplicon was found in two isolates that did not contain the AdeABC pump (Table 6). By PCR ESI-MS, these two isolates were determined not to be A. baumannii (see below).
Mobile genetic elements.
Integrons are genetic elements harboring multiple antibiotic resistance genes. Central to the genetic organization of the integron is the integrase gene (int). The isolates were screened for intI1, intI2, and intI3 genes, class 1 integron specific gene cassettes, and the insertion sequence elements (IS1133 and ISABA1). Sixty-one percent of the isolates demonstrated evidence of int genes. Nearly 70% of these contained class 1 integron gene cassettes of various sizes (data not shown).
Ninety-five percent of the isolates contained ISABA1 and 5% of the isolates contained IS1133, another IS element associated with increased expression of the ADC β-lactamase in A. baumannii (19, 44). These two IS elements were found together in four isolates. We did not uncover evidence that the IS elements were located 5′ to the blaADC genes, but we are continuing to map the location of the IS elements. We also showed that in certain isolates, ISABA1 is proximal to the blaOXA-69-like gene and to the blaOXA-23-like resistance determinant as described above.
Species identification and clonality by PCR ESI-MS.
PCR ESI-MS is a form of high-throughput multilocus sequence typing that is able to distinguish Acinetobacter isolates at the species level as well as to determine clonality (13). Using this technique, we identified at the genetic level 73 A. baumannii isolates, 1 Acinetobacter genome species 3 isolate, and 1 Acinetobacter johnsonii isolate. There was a distribution of 16 different clone types identified within the collection. Further analysis revealed eight main clone types. All eight main types were involved in nosocomial acquisition (28 patients, 37% of isolates) (Table 6). It is notable that carbapenem-resistant isolates were composed of four distinct clones while three of these types possessed blaOXA-23-like genes; the fourth, ESI-MS type 24, contained the blaOXA-58-like gene.
Interestingly, the base composition analysis of 21 of 23 reference strains representing European clones I, II, and III matched 32% of the isolates exactly for all six housekeeping loci tested (efp, trpE, adk, mutY, fumC, and ppa). Related clones of European type I (ESI-MS types 16 and 46) and II (ESI-MS types 1, 10, and 11) are found in an additional 37% of the isolates (Table 7). These isolates are identical in five of the six genetic loci examined. Sequence variations occurred only within the mutY locus, suggesting that these isolates are variants of the well-characterized European strains. Consistent with previous studies comparing the relatedness and aminoglycoside resistance of these pan-European strains, clone type III (ESI-MS type 14) remains the most uniform in its antibiogram and genetic profiles, while clones I and II have slightly diverse susceptibility and genetic patterns within each group (32, 50).
TABLE 7.
Distribution of Acinetobacter sp. PCR ESI-MS clone types isolated at the WRAMCa
| European type | PCR ESI-MS type | No. of isolates | Hospital location(s) | Time course |
|---|---|---|---|---|
| I | 15b | 4 | SICU, MICU, ICU step-down | May 2003-August 2004 |
| I | 16 | 1 | ONP | July 2004 |
| I | 46 | 2 | MICU, ONP | April 2003-May 2003 |
| II | 1 | 1 | MICU | October 2003 |
| II | 10 | 9 | SICU, ONP, surgery, ICU step-down, internal medicine, ER | August 2003-February 2005 |
| II | 11 | 15 | SICU, MICU, ONP | May 2003-November 2004 |
| II | 12b | 4 | SICU, MICU, internal medicine | August 2004-December 2004 |
| III | 14b | 16 | SICU, MICU, ONP, surgery, ICU step-down | May 2003-December 2004 |
| § | 3 | 6 | SICU, MICU, ONP | July 2003-February 2005 |
| § | 9 | 1 | MICU | October 2004 |
| § | 24 | 11 | SICU, MICU, ONP | June 2004-February 2005 |
| § | 39 | 1 | MICU | September 2003 |
| § | 47 | 1 | SICU | January 2004 |
| § | 48 | 1 | SICU | April 2004 |
| § | Acinetobacter genome sp. 3 | 1 | ER | August 2003 |
| § | A. johnsonii | 1 | ER | January 2004 |
All isolates are A. baumannii except Acinetobacter genome sp. 3 and A. johnsonii. §, Isolates not related to European strains. SICU, surgical intensive care unit; MICU, medical intensive care unit; ONP, orthopedics/neurology/physical medicine and rehabilitation; ER, emergency room.
Exact matches to European reference strains.
DISCUSSION
The goal of our study was to link the resistance phenotypes and the genetic determinants of resistance in A. baumannii, giving a “snapshot” of the complex nature of this endemic situation. From these data, it is clear that the potential impact of A. baumannii infections on military and civilian personnel in receiving hospitals is significant.
A unique characteristic of our study is that bloodstream infections were the most common clinical source of Acinetobacter spp. This may reflect the nature and severity of injury among these patients: more than two-thirds of the patients were critically ill, had central venous catheters, and were on mechanical ventilation when their isolates were obtained. Hence, in light of our susceptibility testing, empirical monotherapy for bloodstream infections would be inappropriate. Only two isolates in this collection are resistant to both amikacin and imipenem-cilistatin, and therefore, combination therapy with these antibiotics would ensure that at least one agent is effective for the majority of isolates. It is notable these isolates (from the WRAMC) have a broader range of resistance than those (from the MYSTIC) typically found in U.S. hospitals (Table 4).
A second striking feature of this study is the large number of antibiotic resistance genes found in these isolates. Forty-nine A. baumannii isolates had eight or more resistance determinants. Our genetic analysis revealed that the blaADC and blaOXA-69-like genes were present in nearly all of the strains (17). More significantly, we have also detected for the first time in the United States the presence of the blaOXA-58-like gene (4, 10, 12, 18, 29, 35, 37, 47, 49). OXA-23- and OXA-58-like β-lactamases contribute to meropenem and imipenem resistance in 90% of the carbapenem-resistant isolates in this collection. We also detected the presence of an IS element, ISABA1, located proximal to the blaOXA-23-like genes in our isolates. This IS element may act as a strong promoter and be linked to higher levels of carbapenem resistance by increasing β-lactamase expression. This analysis is similar to contemporary studies by Turton et al. (49), wherein IS elements (ISABA1 upstream of blaOXA-23-like genes) have been found in carbapenem-resistant A. baumannii isolates in the United Kingdom. Insertion elements have also been shown to flank blaOXA-58 and enhance its expression (36). Interestingly, this is the first description of blaPER in the United States.
It is possible that the carbapenem-resistant isolates also have reduced permeability of the outer membrane, altered penicillin binding proteins, or upregulated efflux pumps. We did not find evidence for common metallo-β-lactamases (IMP, VIM, and GIM type) that confer high levels of carbapenem resistance. Investigations analyzing the outer membrane proteins in carbapenem-resistant isolates are in progress.
Many of our isolates were resistant to quinolones due to mutations in the QRDR. Although two isolates do not have these mutations, an upregulated efflux pump might account for their quinolone resistance.
Aminoglycoside resistance (secondary to the presence of genes encoding AMEs) was also prevalent. Many of these genes are widespread in Pseudomonas aeruginosa and A. baumannii and mirror those described in a collection of MDR A. baumannii isolates from Europe (clone types I, II, and III) (22, 32). Unlike the European strains, we did not always find the aminoglycoside gene aadA1 in conjunction with aacC1. As previously shown, a significant proportion of the genes encoding AMEs are contained in class 1 integrons. There was a 93% correlation with the presence of aadA1 and int genes as previously described (2, 43, 48, 54).
Our analysis of the genetic relatedness of these 75 isolates by PCR ESI-MS against 26 ATCC reference strains and 23 European A. baumannii isolates firmly establishes the complexity of the transmission dynamics of this endemic situation. It is remarkable that more than 60% of the isolates were related to the pan-European strains that have been collected from more than 25 countries, including Spain, Greece, Portugal, Poland, France, South Africa, The Netherlands, and Italy, over the last 20 years (32, 50). We further demonstrate that isolates from each of the eight main clones (including all three pan-European types) were also responsible for the hospital-acquired infections at the WRAMC. During the 13-month period, January 2004 to February 2005, seven of the eight main clone types of A. baumannii simultaneously circulated at the WRAMC (Table 7). Although we identified a total of 16 unique clone types, one limitation of our study is that the isolates we analyzed may not have included all Acinetobacter sp. clone types present at the WRAMC during the entire collection period (March 2003 to February 2005).
Why are these MDR isolates spreading so readily? It has been suggested that the epidemic potential of A. baumannii may be linked to the presence of class 1 integrons that contain antibiotic resistance genes (25). In an environment in which antibiotics are frequently used, possessing integrons with multiple resistance determinants confers a strong selective advantage (46). We found genetic evidence for the integron-encoded integrase genes intI1, intI2, and intI3 in 60% of our collection (Table 6). Class 1 integron cassettes were found in 43% of our MDR isolates. As warned by Richet and Fournier, the major concern regarding A. baumannii is the large repertoire of resistance genes carried by a series of mobile genetic elements in the genome (15, 41).
The majority of our population were previously healthy military personnel that were wounded and became critically ill (58 patients sustained trauma and 53 patients were in an ICU). Despite the large number of bloodstream isolates, mortality rates were not high. Nevertheless, it is important to keep in mind that mortality rates were higher in immunosuppressed patients possessing MDR A. baumannii isolates (Table 3). Also alarming is that MDR A. baumannii isolates with at least eight resistance genes were frequently recovered from patients who were intubated.
This study highlights the need for careful epidemiological and molecular surveillance among receiving hospitals and renewed infection control policies with regard to Acinetobacter spp. It is significant that 37% of the isolates were nosocomial in origin (obtained from patients colonized or infected at the WRAMC). In light of these observations and the nature of the multiclonal spread, we suspect that incomplete disinfection procedures and environmental transmission are playing important roles in the dissemination of Acinetobacter spp. in military medical facilities. As with recounts of the genetic basis of multidrug resistance in hospital outbreaks reported by Turton et al., our paper describes isolates from patients that have been transported from the combat theater hospitals in Iraq and Afghanistan to the U.S. military medical facility in Landstuhl, Germany (47). In Germany, they are stabilized prior to further transport to the WRAMC for additional treatment (8,000 miles, three hospital settings). The various environments afford multiple opportunities for acquisition and spread of Acinetobacter spp. The next challenge is to discern the impact of resistance determinants on clinical outcomes.
Our comprehensive analysis is the first in depth study to link the MDR phenotypes and clonality to the genetic determinants of resistance to β-lactams, aminoglycosides, and quinolones in A. baumannii in the U.S. As we have learned with methicillin-resistant S. aureus isolates and vancomycin-resistant enterococci, many of these Acinetobacter sp. isolates may also serve as reservoirs for antibiotic resistance genes that can be transmitted to other pathogens. Hence, the potential introduction of MDR Acinetobacter sp. isolates into European and American civilian, military, and Veterans Affairs medical centers has far reaching consequences. The implementation of specific educational programs, strategies for prevention of primary and secondary acquisition, and interventions designed to control this emerging nosocomial pathogen needs to be an urgent imperative in the care of civilian and military personnel in both European and U.S. hospitals. The diversity and complexity of the resistance determinants found and the potential for widespread dissemination of these strains through Europe and North America could lead to important changes in the global epidemiology of A. baumannii-associated diseases.
Acknowledgments
The Veterans Affairs Merit Review Program and the National Institutes of Health (RO1 AI063517-01) supported these studies. J.M.T. is supported in part by NIH grant T32 GM07250 and the Case Medical Scientist Training Program.
The manuscript was reviewed at the Walter Reed Army Medical Center and is being published without objection. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of Defense, the U.S. Army, or other organizations listed.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Abbo, A., S. Navon-Venezia, O. Hammer-Muntz, T. Krichali, Y. Siegman-Igra, and Y. Carmeli. 2005. Multidrug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 11:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbott, Y., R. O'Mahony, N. Leonard, P. J. Quinn, T. van der Reijden, L. Dijkshoorn, and S. Fanning. 2005. Characterization of a 2.6 kbp variable region within a class 1 integron found in an Acinetobacter baumannii strain isolated from a horse. J. Antimicrob. Chemother. 55:367-370. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. P., B. O'Neill, W. Haddon, Jr., and W. B. Long. 1974. The injury severity score: a method for describing patients with multiple injuries and evaluating emergency care. J. Trauma 14:187-196. [PubMed] [Google Scholar]
- 4.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calfee, D. P., L. J. Durbin, T. P. Germanson, D. M. Toney, E. B. Smith, and B. M. Farr. 2003. Spread of methicillin-resistant Staphylococcus aureus (MRSA) among household contacts of individuals with nosocomially acquired MRSA. Infect. Control Hosp. Epidemiol. 24:422-426. [DOI] [PubMed] [Google Scholar]
- 6.Calfee, D. P., E. T. Giannetta, L. J. Durbin, T. P. Germanson, and B. M. Farr. 2003. Control of endemic vancomycin-resistant Enterococcus among inpatients at a university hospital. Clin. Infect. Dis. 37:326-332. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2004. Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004. Morb. Mortal. Wkly. Rep. 53:1063-1066. [PubMed] [Google Scholar]
- 8.Chau, S.-L., Y.-W. Chu, and E. T. S. Houang. 2004. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob. Agents Chemother. 48:4054-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial disk susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 10.Coelho, J., N. Woodford, M. Afzal-Shah, and D. Livermore. 2006. Occurrence of OXA-58-like carbapenemases in Acinetobacter spp. collected over 10 years in three continents. Antimicrob. Agents Chemother. 50:756-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danel, F., L. M. Hall, D. Gur, H. E. Akalin, and D. M. Livermore. 1995. Transferable production of PER-1 beta-lactamase in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 35:281-294. [DOI] [PubMed] [Google Scholar]
- 12.Donald, H. M., W. Scaife, S. G. Amyes, and H.-K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ecker, D. J., R. Sampath, L. B. Blyn, M. W. Eshoo, C. Ivy, J. A. Ecker, B. Libby, V. Samant, K. A. Sannes-Lowery, R. E. Melton, K. Russell, N. Freed, C. Barrozo, J. Wu, K. Rudnick, A. Desai, E. Moradi, D. J. Knize, D. W. Robbins, J. C. Hannis, P. M. Harrell, C. Massire, T. A. Hall, Y. Jiang, R. Ranken, J. J. Drader, N. White, J. A. McNeil, S. T. Crooke, and S. A. Hofstadler. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. USA 102:8012-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. T. Pennella, C. Agasino Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddell, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLOS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith, M. E., J. M. Ceremuga, M. W. Ellis, C. H. Guymon, D. R. Hospenthal, and C. K. Murray. 2006. Acinetobacter skin colonization of US Army soldiers. Infect. Control Hosp. Epidemiol. 27:659-661. [DOI] [PubMed] [Google Scholar]
- 17.Heritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 20.Hujer, A. M., K. M. Hujer, and R. A. Bonomo. 2001. Mutagenesis of amino acid residues in the SHV-1 beta-lactamase: the premier role of Gly238Ser in penicillin and cephalosporin resistance. Biochim. Biophys. Acta 1547:37-50. [DOI] [PubMed] [Google Scholar]
- 21.Hujer, K. M., N. S. Hamza, A. M. Hujer, F. Perez, M. S. Helfand, C. R. Bethel, J. M. Thomson, V. E. Anderson, M. Barlow, L. B. Rice, F. C. Tenover, and R. A. Bonomo. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 β-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 49:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huys, G., M. Cnockaert, A. Nemec, L. Dijkshoorn, S. Brisse, M. Vaneechoutte, and J. Swings. 2005. Repetitive-DNA-element PCR fingerprinting and antibiotic resistance of pan-European multi-resistant Acinetobacter baumannii clone III strains. J. Med. Microbiol. 54:851-856. [DOI] [PubMed] [Google Scholar]
- 23.Jain, R., and L. H. Danziger. 2004. Multidrug-resistant Acinetobacter infections: an emerging challenge to clinicians. Ann. Pharmacother. 38:1449-1459. [DOI] [PubMed] [Google Scholar]
- 24.Jeong, S. H., I. K. Bae, S. B. Kwon, K. Lee, D. Yong, G. J. Woo, J. H. Lee, H. I. Jung, S. J. Jang, K. H. Sung, and S. H. Lee. 2005. Investigation of a nosocomial outbreak of Acinetobacter baumannii producing PER-1 extended-spectrum beta-lactamase in an intensive care unit. J. Hosp. Infect. 59:242-248. [DOI] [PubMed] [Google Scholar]
- 25.Koeleman, J. G., J. Stoof, M. W. Van Der Bijl, C. M. Vandenbroucke-Grauls, and P. H. M. Savelkoul. 2001. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 39:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marque, S., L. Poirel, C. Heritier, S. Brisse, M. D. Blasco, R. Filip, G. Coman, T. Naas, and P. Nordmann. 2005. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 43:4885-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascini, E. M., and M. J. Bonten. 2005. Vancomycin-resistant enterococci: consequences for therapy and infection control. Clin. Microbiol. Infect. 11(Suppl. 4):43-56. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests, 8th ed. Approved standard M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.Nemec, A., L. Dolzani, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233-1240. [DOI] [PubMed] [Google Scholar]
- 33.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum β-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel, L., S. Marqué, C. Héritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pournaras, S., A. Markogiannakis, A. Ikonomidis, L. Kondyli, K. Bethimouti, A. N. Maniatis, N. J. Legakis, and A. Tsakris. 2006. Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J. Antimicrob. Chemother. 57:557-561. [DOI] [PubMed] [Google Scholar]
- 38.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, L. B., L. L. Carias, A. M. Hujer, M. Bonafede, R. Hutton, C. Hoyen, and R. A. Bonomo. 2000. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 1999. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit. Care Med. 27:887-892. [DOI] [PubMed] [Google Scholar]
- 41.Richet, H., and P. E. Fournier. 2006. Nosocomial infections caused by Acinetobacter baumannii: a major threat worldwide. Infect. Control Hosp. Epidemiol. 27:645-646. [DOI] [PubMed] [Google Scholar]
- 42.Robicsek, A., D. F. Sahm, J. Strahilevitz, G. A. Jacoby, and D. C. Hooper. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 49:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz, J., M. M. Navia, C. Casals, J. M. Sierra, M. T. Jimenez De Anta, and J. Vila. 2003. Integron-mediated antibiotic multiresistance in Acinetobacter baumannii clinical isolates from Spain. Clin. Microbiol. Infect. 9:907-911. [DOI] [PubMed] [Google Scholar]
- 44.Segal, H., R. Thomas, and B. Gay Elisha. 2003. Characterization of class 1 integron resistance gene cassettes and the identification of a novel IS-like element in Acinetobacter baumannii. Plasmid 49:169-178. [DOI] [PubMed] [Google Scholar]
- 45.Sun, T., M. Nukaga, K. Mayama, E. H. Braswell, and J. R. Knox. 2003. Comparison of beta-lactamases of classes A and D: 1.5- Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 12:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 47.Turton, J. F., M. E. Kaufmann, M. J. Gill, R. Pike, P. T. Scott, J. Fishbain, D. Craft, G. Deye, S. Riddell, L. E. Lindler, and T. L. Pitt. 2006. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J. Clin. Microbiol. 44:2630-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turton, J. F., M. E. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 50.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 51.Vila, J., J. Ruiz, P. Goni, and T. Jimenez de Anta. 1997. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J. Antimicrob. Chemother. 39:757-762. [DOI] [PubMed] [Google Scholar]
- 52.Vila, J., J. Ruiz, P. Goni, A. Marcos, and T. Jimenez de Anta. 1995. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 39:1201-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, P. A., C. J. McIver, Y. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 54.Wu, T. L., L. Ma, J. C. Chang, L. H. Su, C. Chu, H. S. Leu, and L. K. Siu. 2004. Variable resistance patterns of integron-associated multidrug-resistant Acinetobacter baumannii isolates in a surgical intensive care unit. Microb. Drug Resist. 10:292-299. [DOI] [PubMed] [Google Scholar]