Abstract
The population dynamics of bifidobacteria in human feces during raffinose administration were investigated at the species level by using fluorescence in situ hybridization (FISH) coupled with flow cytometry (FCM) analysis. Although double-staining FISH-FCM using both fluorescein isothiocyanate (FITC) and indodicarbocyanine (Cy5) as labeling dyes for fecal samples has been reported, the analysis was interfered with by strong autofluorescence at the FITC fluorescence region because of the presence of autofluorescence particles/debris in the fecal samples. We circumvented this problem by using only Cy5 fluorescent dye in the FISH-FCM analysis. Thirteen subjects received 2 g of raffinose twice a day for 4 weeks. Fecal samples were collected, and the bifidobacterial populations were monitored using the established FISH-FCM method. The results showed an increase in bifidobacteria from about 12.5% of total bacteria in the prefeeding period to about 28.7 and 37.2% after the 2-week and 4-week feeding periods, respectively. Bifidobacterium adolescentis, the Bifidobacterium catenulatum group, and Bifidobacterium longum were the major species, in that order, at the prefeeding period, and these bacteria were found to increase nearly in parallel during the raffinose administration. During the feeding periods, indigenous bifidobacterial populations became more diverse, such that minor species in human adults, such as Bifidobacterium breve, Bifidobacterium bifidum, Bifidobacterium dentium, and Bifidobacterium angulatum, proliferated. Four weeks after raffinose administration was stopped, the proportion of each major bifidobacterial species, as well as that of total bifidobacteria, returned to approximately the original values for the prefeeding period, whereas that of each minor species appeared to differ considerably from its original value. To the best of our knowledge, these results provide the first clear demonstration of the population dynamics of indigenous bifidobacteria at the species level in response to raffinose administration.
Raffinose [β-d-fructofuranosyl-O-α-d-galactopyranosyl-(1,6)-α-d-glucopyranoside] is a nondigestible oligosaccharide that is widely distributed in many plants, such as sugar beet, cane, cabbage, potato, grape, wheat, barley, corn, and the seeds of many legumes (20, 21). In Hokkaido, Japan, raffinose is an important agricultural product that is extracted from sugar beets as a by-product of sugar processing. Several prebiotic effects in humans have been reported for this oligosaccharide, including reduction of fecal ammonia and indole (18), improvement of defecation frequency (18), and increased cell numbers among indigenous bifidobacteria (5). Increases in bifidobacterial populations in rats (8) and the suppression of T helper 2 cell-mediated immune responses in mice (19) have also been reported.
To obtain a scientific basis for the use of raffinose as a prebiotic, a precise determination of its effects on microbiotic composition in the human intestine, especially on the bifidobacterial population at the species level, is required. Because the majority of microbiota in the human intestine is not yet cultured (26), it is difficult to obtain reliable and quantitative results at the species level by using culture-dependent methods. For this purpose, molecular ecological methods are required to be applied to investigate the population dynamics of bifidobacteria.
Among the many molecular ecological methods for analyzing microbiota, fluorescence in situ hybridization (FISH) is a widely used method for monitoring microorganisms in complex ecosystems (9, 11, 14). In FISH analysis, sets of group-, genus-, or species-specific rRNA-targeted oligonucleotide probes are applied for the identification and quantification of microorganisms. In our previous study (8), we successfully applied FISH analysis to evaluate the effect of raffinose administration on the modulation of rat cecal microbiota and demonstrated a significant increase in the population of Bifidobacterium animalis, an indigenous bifidobacterial species in rats, up to 20.5% of the total bacterial population, compared to 0.2% observed in the basal-diet group. Although FISH analysis is considered suitable for the enumeration of bacterial cells in complex microbial communities, the manual counting of detected bacteria is time-consuming and laborious. Several attempts have therefore been made to minimize the counting effort by combining the analysis with a flow cytometry (FCM) technique for high-throughput analysis. Double-staining FISH-FCM analysis using both fluorescein isothiocyanate (FITC) and indodicarbocyanine (Cy5) as labeling dyes has been successfully applied to enumerate bacterial populations in human fecal samples collected in European countries (12, 13, 17, 22, 23, 24, 30). In our case, however, this approach was not directly applicable because of strong autofluorescence materials found in the feces of volunteers (mostly Asian people living in Japan). We therefore modified the FISH-FCM procedure to solve this problem and applied this modified method to investigate the population dynamics of bifidobacteria at the species level in the human intestine upon the administration of raffinose.
MATERIALS AND METHODS
Bacterial strain and medium.
Bifidobacterium breve JCM 1192T was obtained from the Japan Collection of Microorganisms (JCM, Wako, Japan). This bacterium was cultured in GAM broth (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) at 37°C for 12 h under anaerobic conditions, using mixed gas N2-CO2-H2 (8:1:1).
Design of human trial.
Fecal samples were collected from 13 healthy adults (11 males and 2 females, 23 to 57 years old) who originated from Japan (11 people), Indonesia (1 person), and Brazil (1 person). All subjects had lived in Japan for at least 6 months before the trial, and they consumed their usual diets, without restrictions on daily food consumption. Two grams of raffinose (Nippon Beet Sugar Manufacturing Co., Ltd., Tokyo, Japan) was introduced twice per day (total, 4 g/day) to all subjects for 4 weeks. Fecal samplings were conducted at 1 week before raffinose consumption (0W), at the 14th day (2W) and the 28th day (4W) of raffinose intake, and 4 weeks after raffinose intake was stopped (8W). This study was approved by the Ethics Committee of the Research Faculty of Agriculture, Hokkaido University, Japan.
Sample collection and preparation.
Fecal samples were collected in sterile Falcon tubes and stored at 4°C under anaerobic conditions by using an anaerobic pouch (Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan) for a maximum of 4 h before processing. Sample preparations were conducted as reported previously (8). About 0.5 g of fecal sample was suspended in ice-cold phosphate-buffered saline (PBS; 130 mM NaCl, 10 mM sodium phosphate buffer, pH 7.2) and centrifuged at 200 × g (low speed) for 5 min to remove large fecal particles/debris. This step was repeated three times, and the supernatants were pooled. Fecal bacteria were then pelleted from the pooled supernatant by using high-speed centrifugation at 9,000 × g for 2 min and washed with PBS three times to remove materials inhibitory to the FISH reaction. Cultured B. breve JCM 1192T cells were collected by centrifugation at 9,000 × g for 2 min and washed twice with PBS. Fecal samples and cultured bacterial cells were fixed with 4% (wt/vol) paraformaldehyde in PBS for 24 h. Following fixation, the cells were washed with PBS and stored in a known volume of 50% (vol/vol) ethanol-PBS at −20°C until use.
FISH-FCM analysis.
For each hybridization, 50 μl of fixed cells was centrifuged for 2 min at 20,600 × g in a 1.5-ml Eppendorf tube and resuspended in a mixture of 40 μl of hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl, pH 7.2) and 5 μl of oligonucleotide probe (25 ng/μl; Tsukuba Oligo Service Co., Ltd., Tsukuba, Japan). Formamide was added to the mixture of hybridization buffer for probes Non338, Eub338, Bif164m, and PBR2 at the indicated concentrations (Table 1). In the case of probe PBR2, the unlabeled oligonucleotides (helpers) (Table 1) were added to the hybridization mixture at the same concentration as PBR2 to improve the accessibility of the probe (8). After hybridization at 46°C for 16 h, 150 μl of washing buffer (225 mM NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl, pH 7.2) was added, and cells were collected by centrifugation for 2 min at 20,600 × g. Cells were then resuspended in 300 μl of washing buffer and incubated at 48°C for 20 min to remove nonspecifically bound probes. Finally, hybridized cells were centrifuged and resuspended in 1 ml of PBS for FCM analysis. Analyses of FCM were conducted using a BD FACSCanto flow cytometer (BD Biosciences, San Jose, CA) equipped with a 20-mW solid-state blue laser (488 nm) and a 17-mW helium-neon (He-Ne) red laser (633 nm). The 488-nm laser was used to measure the forward angle scatter (FSC) (using a photodiode with a 488/10-nm band-pass filter), the side angle scatter (using a photomultiplier tube [PMT] with a 488/10-nm band-pass filter), and the green fluorescence intensity (using a PMT with a 530/30-nm band-pass filter) conferred by FITC-labeled probe. The He-Ne red laser was used to detect the red fluorescence conferred by Cy5-labeled probes (using a PMT with a 660/20-nm band-pass filter). The system threshold for FSC signals was set, and all bacterial analyses were performed at low-flow-rate settings (10 μl/min). A total of 100,000 events were stored in list mode files, and data were analyzed using BD FACSDiva Software (BD Biosciences, San Jose, CA). The entire hybridization and counting analysis were performed three times for each probe and each fecal sample.
TABLE 1.
The 16S rRNA-targeted oligonucleotide probes used for the molecular analysis of fecal samples
| Probe group and name | Probe sequence (5′→3′) | Target organism | Target sitea | Formamide (%) | Reference(s) |
|---|---|---|---|---|---|
| Labeled | |||||
| Non338 | ACATCCTACGGGAGGC | Negative probe | 20 | 22, 29 | |
| Eub338 | GCTGCCTCCCGTAGGAGT | Bacteria | 338 | 20 | 1 |
| Bif164m | CATCCGGYATTACCACCC | Genus Bifidobacterium | 164 | 20 | 8 |
| Bado434 | GCTCCCAGTCAAAAGCG | B. adolescentis | 434 | 0 | 28 |
| Bang198 | AATCTTTCCCAGACCACC | B. angulatum | 198 | 0 | 28 |
| Bbif186 | CCACAATCACATGCGATCATG | B. bifidum | 186 | 0 | 28 |
| Bcat187 | ACACCCCATGCGAGGAGT | B. catenulatum group | 187 | 0 | 28 |
| Bden82 | ACTCTCACCCGGAGGCGAA | B. dentium | 82 | 0 | 28 |
| Blon1004 | AGCCGTATCTCTACGACCGT | B. longum | 1004 | 0 | 28 |
| PBR2 | CCATGCGGTGTGATGGAGC | B. breve | 182 | 20 | 27 |
| Nonlabeled (helpers) | |||||
| PBR2 (first upper helper) | ATCCGGCATTACCACCCGT | B. breve | 163 | 20 | 8 |
| PBR2 (first lower helper) | CAAAGGCTTTCCCAACACA | B. breve | 201 | 20 | 8 |
| PBR2 (second upper helper) | TTCCAGGAGCTATTCCGGT | B. breve | 144 | 20 | 8 |
| PBR2 (second lower helper) | GCGACCCCATCCCATGCCG | B. breve | 220 | 20 | 8 |
Positions of target sites are indicated based on the Escherichia coli 16S rRNA sequence.
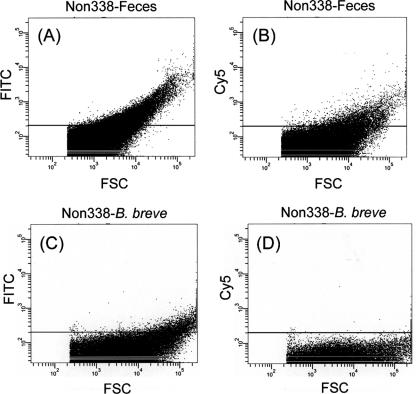
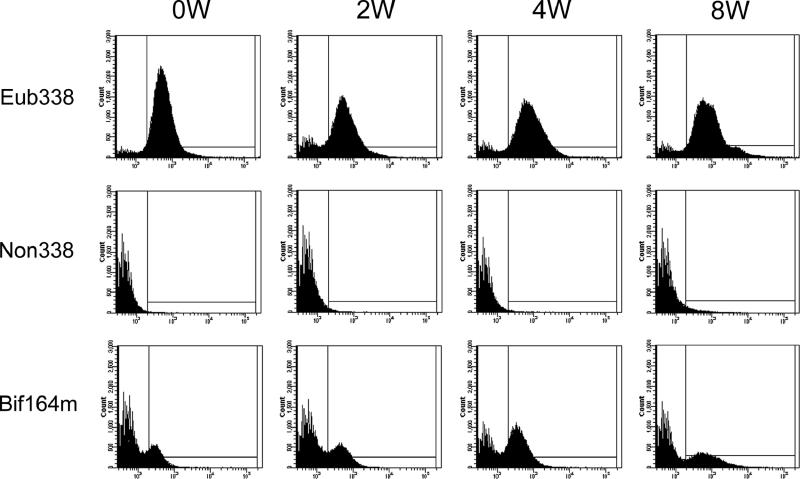
To select suitable fluorochrome for the analysis of fecal samples by FISH-FCM, the background fluorescence of a pure culture of B. breve JCM 1192T and a representative fecal sample were evaluated by FISH-FCM, using negative probes (Non338-FITC and Non338-Cy5) (see Fig. 2). In the measurements of microbial populations in the raffinose trials, fecal samples were hybridized with oligonucleotide probes labeled with a single fluorochrome (Cy5) for FISH-FCM analysis. Gating of the bacterial cells/particles was conducted, and counting was performed until 100,000 events were reached within the gated area. The proportions of target cells hybridized with Cy5-labeled probe (Table 1) having fluorescence intensities of >200 (see Fig. 3, right side of vertical line in each histogram) were calculated against 100,000 events. This proportion was then corrected by subtracting the background proportions, measured using Non338-Cy5 (Fig. 3B), to obtain the precise values for the fecal sample. The percentages for target bifidobacteria (at the genus or species level) were recalculated, taking the proportion of Eub338 obtained in this manner as total bacteria (100%).
FIG. 2.
FCM dot plots of fecal samples hybridized with Non338-FITC (A) and Non338-Cy5 (B) and plots of B. breve cells hybridized with Non338-FITC (C) and Non338-Cy5 (D), monitored using FCM. Fluorescence intensity is indicated on the vertical axis, and FSC intensity is indicated on the horizontal axis. The area above an intensity of 200 in FITC and Cy5 (solid line above horizontal axis in each panel) was evaluated as the occurrence of autofluorescence.
FIG. 3.
FCM histograms obtained from FISH analyses of fecal samples from one representative subject in the raffinose administration trial. Histograms are ordered vertically as samples that hybridized with total bacterial probe/Eub338-Cy5, negative probe/Non338-Cy5, or Bifidobacterium-specific probe/Bif164m-Cy5 and are ordered horizontally following the time periods for raffinose administration: prefeeding period (0W), 2-week feeding period (2W), 4-week feeding period (4W), and postfeeding period (8W).
FISH-microscopy analysis.
FISH-microscopy analyses were conducted as described previously (8) for the analysis of autofluorescence particles in fecal samples and the validation of FISH-FCM results. The total bacterial count was conducted by DAPI (4′,6-diamidino-2-phenylindole dihydrochloride n-hydrate) staining. Aliquots (3 μl) of fixed cells applied on Teflon printed glass slides (ADCELL, 12 wells, 5 mm in diameter; Erie Scientific Company, Portsmouth, NH) were hybridized by the addition of 8 μl of hybridization buffer with 1 μl of oligonucleotide probe (25 ng/μl) in a moist chamber at 46°C for 16 h. Washing of hybridized cells was conducted in prewarmed washing buffer for 20 min at 48°C. After drying, bacterial cells on the glass slides were stained with DAPI and the dried slides were mounted with VECTASHIELD mounting medium (Vector Laboratories, Inc., Burlingame, CA). To evaluate the occurrence of autofluorescence from debris/particles in feces, a representative fecal sample was analyzed without probe or after hybridization with Eub338 labeled with appropriate fluorochromes. For the validation of FISH-FCM results, several 16S rRNA-targeted oligonucleotide probes labeled with Cy3 and the helpers (Table 1) were used to enumerate the bifidobacterial populations at the genus and species levels. Bacterial cells were monitored using an Olympus BX50 epifluorescence microscope (Olympus Corporation, Tokyo, Japan) equipped with an Olympus DP30BW charge-coupled-device camera (Olympus Corporation) operated by MetaMorph Imaging System software (Molecular Devices Corporation, Sunnyvale, CA). DAPI and Cy3 signals were captured for at least 10 microscopic fields for each well and counted manually using Adobe Photoshop version 7.0 software (Adobe Systems Incorporated, San Jose, CA).
Measurement of fecal pH.
The pH of each fecal sample was measured by inserting the electrode of an ISFET pH meter KS701 (Shindengen Electric Manufacturing Co., Ltd., Tokyo, Japan) into the feces.
Data analyses.
The changes in bacterial proportions and pH values for fecal samples were analyzed statistically using SPSS software version 10.0.1 (SPSS, Inc., Chicago, IL). Bonferroni tests were performed for pair-wise multiple comparisons of the mean values for the control week (0W) and for the rest of the weeks (2W, 4W, and 8W).
RESULTS
Feasibility test for FISH-FCM method for enumeration of bifidobacterial populations in fecal samples.
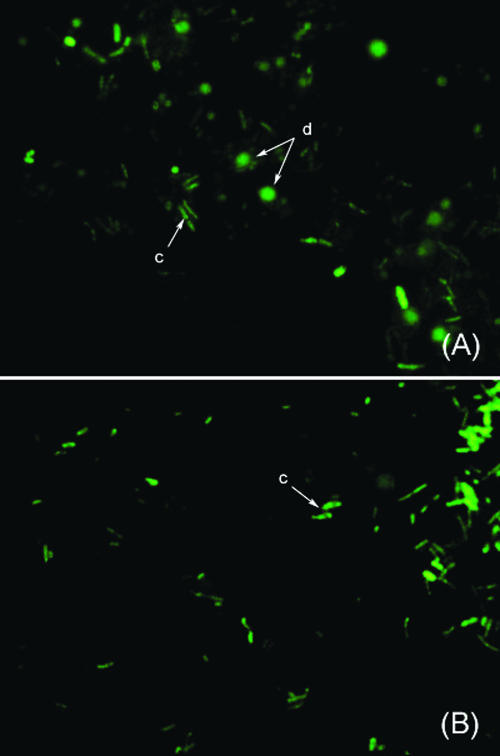
In the preliminary experiments for the enumeration of bacterial populations both in a pure culture of B. breve JCM 1192T and in fecal samples, we applied a previously reported FISH-FCM method in which FITC and Cy5 were used as fluorescent dyes to double stain the bacterial cells (30). Although satisfactory results were obtained for pure culture samples (data not shown), measurements of the fecal samples were not successful because of interference from high background fluorescence observed when Non338-FITC was used as a negative probe. We assumed that these phenomena were caused by the presence of autofluorescence materials in the fecal samples. We checked the fecal samples by using epifluorescence microscopy to confirm this assumption and found that autofluorescence particles could be seen in fecal samples even without hybridization probes (data not shown) when the detection filter for FITC was selected but not when the other filters (for Cy3 and Cy5) were selected. In addition, the autofluorescence fecal particles were found to be similar in size to bacterial cells (Fig. 1A) when the fecal sample was observed after hybridization with Eub338-FITC. In contrast, only bacterial cells were detected when the sample was hybridized with Eub338-Cy5 (Fig. 1B). From these results, Cy5 was expected to be a suitable fluorochrome for FISH-FCM analysis of fecal samples. To confirm this, FISH-FCM analysis of a representative fecal sample was conducted by hybridization with negative probes (Non338-FITC or Non338-Cy5). As expected, very high background fluorescence was observed when the fecal sample was hybridized with Non338-FITC (Fig. 2A), whereas relatively low background fluorescence was observed when Non338-Cy5 was used as a negative probe (Fig. 2B). In contrast, very low background fluorescence was detected when the pure culture of B. breve JCM 1192T was hybridized with either Non338-FITC (Fig. 2C) or Non338-Cy5 (Fig. 2D), indicating that autofluorescence particles were derived from fecal materials. Based on these results, Cy5 was selected for labeling all the oligonucleotide probes for FISH-FCM analysis of bacterial populations in fecal samples. When we applied this single-staining procedure to fecal samples, the results for bifidobacterial enumeration were very similar to those obtained by FISH-manual counting analysis (examples are shown in Table 3). A practical FISH-FCM method using Cy5 as a single fluorochrome was therefore successfully established for monitoring bifidobacterial populations in fecal samples having high-autofluorescence particles.
FIG. 1.
Microscopic images of a fecal sample hybridized with Eub338-FITC (A) and Eub338-Cy5 (B), monitored using epifluorescence microscopy. Bacterial cells (c) and fecal particle/debris (d) are indicated with arrows.
TABLE 3.
Bifidobacterium populations in fecal samples from two subjects determined using FISH-FCM and FISH-microscopya
| Subject and microorganism(s) | % of microbiota in fecal samples for indicated period and analysis
|
|||||||
|---|---|---|---|---|---|---|---|---|
| FISH-FCM
|
FISH-microscopy
|
|||||||
| Prefeeding (0W) | Feeding (2W) | Feeding (4W) | Postfeeding (8W) | Prefeeding (0W) | Feeding (2W) | Feeding (4W) | Postfeeding (8W) | |
| Subject 1 | ||||||||
| Total bacterial cellsb | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Bifidobacterium spp. | 16.7 (1.7) | 38.5 (3.5) | 42.7 (0.5) | 32.6 (4.9) | 19.9 (3.9) | 40.8 (5.1) | 45.5 (5.2) | 26.9 (5.7) |
| B. adolescentis | 11.6 (1.3) | 22.5 (1.2) | 27.5 (1.3) | 12.3 (5.3) | 10.7 (4.0) | 21.4 (3.3) | 35.6 (1.4) | 14.3 (3.1) |
| B. catenulatum group | 2.7 (0.7) | 3.5 (1.5) | 1.1 (0.0) | 2.2 (1.6) | 2.5 (0.7) | 5.5 (1.4) | 2.6 (0.5) | 2.6 (0.9) |
| B. longum | 8.5 (1.8) | 10.5 (2.9) | 9.1 (0.2) | NDc | 5.7 (1.8) | 11.6 (3.1) | 6.6 (0.6) | 0.5 (0.5) |
| B. breve | ND | 0.6 (0.9) | ND | ND | ND | 0.1 (0.1) | ND | ND |
| B. bifidum | ND | 3.9 (0.1) | 0.3 (0.3) | ND | ND | 2.4 (1.4) | 0.2 (0.5) | ND |
| B. dentium | ND | 1.9 (0.6) | ND | ND | ND | 1.3 (1.2) | 0.0 (0.1) | ND |
| B. angulatum | ND | 2.8 (1.8) | 0.7 (0.4) | ND | ND | 1.0 (0.7) | 0.6 (0.6) | ND |
| Sum of detected spp. | 22.8 | 45.7 | 38.7 | 14.5 | 18.9 | 43.3 | 45.6 | 17.4 |
| Subject 2 | ||||||||
| Total bacterial cells | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Bifidobacterium spp. | 6.7 (0.9) | 32.0 (1.2) | 36.5 (2.1) | 26.0 (0.6) | 6.9 (0.7) | 24.5 (7.5) | 31.4 (0.9) | 18.2 (1.3) |
| B. adolescentis | ND | 0.7 (0.2) | 2.6 (1.5) | 1.9 (1.0) | ND | 0.7 (0.6) | 2.5 (1.2) | 1.2 (1.1) |
| B. catenulatum group | 1.7 (0.3) | 11.0 (0.9) | 12.5 (0.6) | 7.2 (0.2) | 1.4 (0.7) | 8.7 (2.7) | 9.6 (3.2) | 5.2 (3.3) |
| B. longum | 2.3 (0.1) | 2.5 (0.2) | 6.4 (0.3) | 6.8 (0.3) | 3.0 (0.9) | 3.2 (1.0) | 5.5 (1.1) | 5.4 (1.8) |
| B. breve | ND | 1.9 (1.2) | 1.0 (0.1) | 0.1 (0.2) | ND | 1.3 (1.2) | 0.8 (0.7) | ND |
| B. bifidum | ND | 0.2 (0.1) | ND | ND | ND | 0.2 (0.4) | ND | ND |
| B. dentium | ND | 0.7 (0.1) | 0.1 (0.1) | 0.1 (0.1) | ND | 0.4 (0.8) | ND | ND |
| B. angulatum | ND | 0.2 (0.2) | 0.7 (0.4) | 1.3 (0.6) | ND | 0.2 (0.6) | 0.4 (0.5) | 0.5 (0.7) |
| Sum of detected spp. | 4.0 | 17.2 | 23.3 | 17.4 | 4.4 | 14.7 | 18.8 | 12.3 |
The values are means, with standard deviations in parentheses, representing the proportions of bifidobacterial cells against those of total bacterial cells (Eub338). In FISH-FCM, the measurements were conducted in triplicate (three independent hybridizations), while 10 microscopic fields were counted in FISH-microscopy monitoring.
Total bacterial cells as represented by total cells hybridized with probe Eub338.
ND, not detected.
Effect of raffinose administration on bacterial population and pH values in human feces. (i) Total bacterial count by DAPI staining and fecal pH.
Total DAPI counts did not change significantly during and after the raffinose administration relative to the initial values (0W). Total counts (means ± standard errors of means) at 0W were 1.4 × 1011 ± 0.2 × 1011 cells/g wet feces. The average counts were reduced slightly to 1.2 × 1011 ± 0.2 × 1011 and 1.0 × 1011 ± 0.1 × 1011 cells/g wet feces after 2W and 4W, respectively. At 8W, the total count recovered to 1.2 × 1011 ± 0.1 × 1011 cells/g wet feces. The pHs for fecal samples (means ± standard errors of means) at 0W, 2W, 4W, and 8W were 7.0 ± 0.2, 6.6 ± 0.2, 6.6 ± 0.2, and 6.9 ± 0.2, respectively. Although there was a tendency toward decreases in pH during raffinose administration, these differences were not statistically significant because of the high variation in pH among subjects.
(ii) Population dynamics of bifidobacteria.
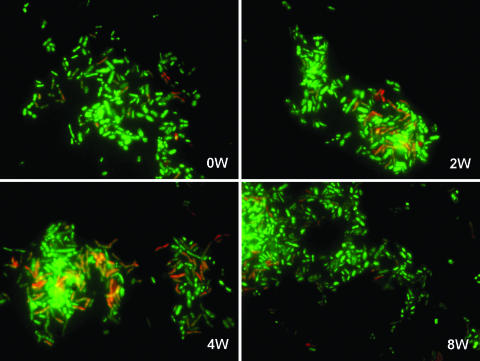
The newly developed FISH-FCM method described above was used for the analysis of fecal samples to enumerate the proliferation of bifidobacteria in response to raffinose administration. Typical histograms for FISH-FCM analysis of a representative subject are shown in Fig. 3, and the results for the average total and species-level bifidobacterial populations obtained from the 13 volunteers are summarized in Table 2. At 0W, the average total bifidobacteria (Bif164m) accounted for 12.5% of the total bacteria (Eub338). During raffinose administration, the averages for total bifidobacteria dramatically increased to 28.7 and 37.2% of total bacteria at 2W and 4W, respectively. At 8W, the population of bifidobacteria decreased to 16.1% of total bacteria. A similar tendency was confirmed by enumeration using FISH-microscopy analysis (examples are shown in Table 3), in which the stimulation effect of raffinose on bifidobacterial growth was clearly demonstrated (Fig. 4). In the species-level analysis, three bifidobacterial species, Bifidobacterium adolescentis, the Bifidobacterium catenulatum group, and Bifidobacterium longum, predominated at 0W and accounted on average for 4.3, 1.8, and 1.6% of total bacteria, respectively, whereas B. breve and Bifidobacterium bifidum were detected at the low levels of 0.4 and 0.2% of total bacteria, respectively. The species Bifidobacterium dentium and Bifidobacterium angulatum were not detected at 0W. During raffinose administration, populations of all the Bifidobacterium species, including B. dentium and B. angulatum, increased at 2W. However, at 4W, only B. adolescentis, the B. catenulatum group, and B. longum, the predominant species, continued to proliferate, whereas the populations of minor species (B. breve, B. bifidum, B. dentium, and B. angulatum) decreased. At 8W, the populations of all Bifidobacterium species were reduced. Although the populations of the major group (B. adolescentis, the B. catenulatum group, and B. longum) returned to approximately the initial 0W values, the populations of the minor group were quite variable. For instance, B. breve, which was detected at 0.4% at 0W and proliferated up to 1.7% at 2W, was reduced to 0.04% at 8W, whereas the previously undetected B. angulatum and B. dentium appeared to persist at the considerable levels of 0.5% and 0.05%, respectively, at 8W. These results not only confirm the previously reported growth stimulation effect of raffinose on indigenous bifidobacteria, but also clarify for the first time the population dynamics of bifidobacteria at the species level.
TABLE 2.
The average genus and species-level bifidobacterial populations in human feces monitored using FISH-FCM analysis
| Microorganism | Probe | % of microbiota in fecal samples for indicated periodb
|
|||
|---|---|---|---|---|---|
| Prefeeding (0W) | Feeding (2W) | Feeding (4W) | Postfeeding (8W) | ||
| Total bacterial cellsa | Eub338 | 100 | 100 | 100 | 100 |
| Bifidobacterium genus | Bif164m | 12.5 (1.8) | 28.7 (4.5)d | 37.2 (4.2)d | 16.1 (2.6) |
| B. adolescentis | Bado434 | 4.3 (1.1) | 6.9 (1.9) | 9.7 (2.6) | 3.7 (1.2) |
| B. catenulatum group | Bcat187 | 1.8 (0.6) | 3.8 (1.5) | 4.7 (1.7) | 1.7 (0.7) |
| B. longum | Blon1004 | 1.6 (0.7) | 3.3 (0.8) | 5.3 (1.0)d | 1.8 (0.6) |
| B. breve | PBR2 | 0.4 (0.3) | 1.7 (0.4)d | 0.5 (0.2) | 0.04 (0.03) |
| B. bifidum | Bbif186 | 0.2 (0.1) | 0.7 (0.3) | 0.3 (0.2) | 0.1 (0.0) |
| B. dentium | Bden82 | NDc | 0.6 (0.2)d | 0.2 (0.1) | 0.05 (0.03) |
| B. angulatum | Bang198 | ND | 1.0 (0.2)d | 0.7 (0.2)d | 0.5 (0.2) |
| Sum of detected species | 8.3 | 18.0 | 21.4 | 7.9 | |
Total bacterial cells are represented as total cells hybridized with probe Eub338.
Percentages of bifidobacterial cells in total bacterial cells in fecal samples were enumerated using FISH-FCM analysis. The values are means, with standard errors of means indicated in parentheses (n = 13).
ND, not detected.
Statistically significant difference (P < 0.05) compared with data for the prefeeding period (0W).
FIG. 4.
Population dynamics of the bifidobacteria for one representative subject in the raffinose administration trial as determined using FISH-microscopy analysis. Bacterial cells from fecal samples were stained with DAPI (green) and hybridized with a Bifidobacterium genus-specific oligonucleotide probe (Bif164m) (red) in FISH analyses. Images are for each sampling time period: prefeeding (0W), 2 weeks (2W), 4 weeks (4W), and postfeeding (8W).
Discrepancies between total bifidobacteria and sums of bifidobacterial species in FISH-FCM analysis.
We found discrepancies between the proportion of total bifidobacteria (Bif164m) and the sum of the proportions of each bifidobacterial species detected using species-specific probes. On average, the species-specific probes used in this study detected 49 to 66% of the bifidobacterial population in fecal samples (Table 2). The undetected percentage differed depending on the individuals and the time of sampling. Almost no discrepancy was found in subject 1 at 0W, 2W, and 4W, whereas subject 2 showed lower proportions of detected Bifidobacterium species throughout the experiments (Table 3). To further examine these discrepancies, we enumerated fecal samples from both subjects by using FISH-microscopy. The results were quite similar to those obtained by FISH-FCM (Table 3), suggesting that the discrepancy described above was not caused by the FCM methodology.
DISCUSSION
We established a practical high-throughput FISH-FCM method for monitoring bacterial populations of fecal samples, particularly those containing autofluorescence particles. The double-staining method using FITC and Cy5 was not applicable because of interference from significant background fluorescence originating from autofluorescence materials contained in the fecal samples collected in our study. Biological molecules such as NADH, riboflavin, and flavin coenzymes contained in plant and animal cells are responsible for the autofluorescence in FCM analysis, where the peak of the autofluorescence emission heavily overlaps with the FITC fluorescence region (2, 6). It seems that the autofluorescence particles could be residues of plant fibers because we frequently observed highly autofluorescent fiber-shaped particles under epifluorescence microscopy when the detection filter for FITC was selected (data not shown). It has also been noticed that autofluorescence arises in plant tissue from chlorophylls, alkaloids, and flavonoids in addition to the aforementioned fluorescent molecules (2). It is important to note that interference by autofluorescence in FISH-FCM analysis using FITC dye has also been experienced for fecal samples collected in European countries (E. E. Vaughan, Wageningen University, The Netherlands, personal communication). However, judging from the relatively low background signals observed in FISH-FCM using unhybridized fecal samples (30), and the fact that dual staining using both FITC and Cy5 as fluorochromes has been successfully applied to the analysis of fecal samples (12, 17, 22), we assumed that fecal samples from European countries generally contained fewer autofluorescence particles than those from Japan.
For the most part, FISH-FCM has been applied to monitor bacterial populations at the genus or group level. The application of FISH-FCM in the analysis of fecal bacteria at the species level is still limited to Bacteroides species (23) and Clostridium species (13). We reported, for the first time, the FISH-FCM analysis of fecal bifidobacterial populations at the species level to clarify the effects of raffinose administration. The average proportion of bifidobacterial populations in human feces accounted for 12.5% of total bacteria at 0W (Table 2). This was higher than the average for people living in European countries, where it is generally about 3.5% of total bacteria (12, 17, 22). The difference in bifidobacterial proportions between these studies may be caused by differences in the diet of each subject as well as differences in common food materials between Japan and European countries. Based on the species-level analysis using FISH-FCM, the majority of bifidobacteria was composed of B. adolescentis, the B. catenulatum group, and B. longum, in that order (Table 2). This result is in agreement with that reported by Matsuki et al. (15), in which the same order was found using real-time PCR analysis for bifidobacterial cell numbers in fecal samples from human adults in Japan.
With the consumption of 4 g of raffinose per day, the average proportion of bifidobacteria in feces increased from 12.5% (0W) to 37.2% (4W) of total bacteria (Table 2). This result represents the first precise clarification of the effect of raffinose on the growth of indigenous bifidobacteria in the human intestine by a molecular approach. We have not conducted a crossover and/or parallel study, since at least the increases of bifidobacterial population by administration of raffinose have been established both in a rat experiment (8) and in a human study (5). A previous report found a higher proportion of bifidobacteria (58 to 80% of total culturable bacteria) in response to raffinose administration than we found (5). This may be because of the higher raffinose intake (15 g/day) and/or the application of a culture-dependent method for the enumeration of bacterial populations. In many cases, results obtained using culture-dependent methods for the evaluation of bacterial proportions in complex ecosystems tend to underestimate or overestimate.
Species-level analysis during and after raffinose intake revealed many interesting features of the population dynamics of bifidobacteria. The proportions of each species of the major group (B. adolescentis, the B. catenulatum group, and B. longum) continued to increase until the end of the administration period (4W), reaching two to three times the original (0W) levels, and then returned almost to the 0W proportions at 8W. Moreover, the orders of predominance at the species level were almost the same throughout the experimentation period (0W to 8W). These results may reflect the established niches of the predominant bifidobacteria in the human intestine. In contrast, the proportions of the minor members were not stable. Although the members of the minor group appeared to be boosted dramatically at 2W, they generally failed to establish niches at 4W, and their populations became more variable after raffinose administration was stopped (8W). For example, although the proportion of B. breve increased four times from 0W to 2W, it then decreased to 1/10 of the 0W proportion at 8W. In contrast, B. angulatum and B. dentium, which were not detected at all at 0W, persisted considerably at 8W. B. dentium was originally isolated from dental caries (25), and its presence has also been demonstrated in the human intestine at about 107 cells/g feces by real-time PCR (15). Although a relatively high proportion of this bacterium was detected in the present study, as much as 0.6% of the total bacteria (Table 2) (which may correspond to about 1010 cells/g feces), its impacts on the health of the human host are generally not well understood. The reason for instabilities in the population dynamics of the minor members of the bifidobacteria is not clear. However, these results at least indicate that raffinose not only increases the diversity of bifidobacterial populations, but also possibly establishes different bifidobacterial compositions in the human intestine after the administration is stopped.
A discrepancy was found between the total bifidobacterial proportion (Bif164m) and the sum of the proportions of each species of bifidobacteria in fecal samples (Table 2). The presence of other known bifidobacteria associated with fecal samples, such as Bifidobacterium lactis and Bifidobacterium gallicum, may be one reason for this discrepancy. Whereas we did not monitor these two species, the presence of these species in human feces is relatively rare (3, 16). The presence of other unidentified bifidobacteria in the feces was also considered a possibility. In another report (10), a 16S rRNA clone library derived from human fecal samples showed bacterial clones affiliated with uncultured Bifidobacterium species. Similarly, many uncultured, Bifidobacterium-related 16S rRNA clones have been found in fecal samples from adults and distinguished among live- and dead-cell fractions by using FCM with a sorting system (4). Our results suggest that there may still be unidentified bifidobacteria that have not yet been characterized in fecal samples. To test this suggestion, we conducted a matching analysis to compare probe sequences to 16S rRNA sequences stored in the Ribosomal Database Project (RDP-II) collection (7). It appeared that about 9% of the total Bifidobacterium-related 16S rRNA sequences were categorized as “unidentified uncultured bifidobacteria” (Table 4) by matching their sequences (>1,200 bp) to seven 16S rRNA sequences of Bifidobacterium species-specific oligonucleotide probes (Table 1). Based on our study and previous work (4, 10), the occurrence of certain unidentified Bifidobacterium species may be expected. This possibility must be analyzed in the future to clarify the more detailed population structure of bifidobacterial species.
TABLE 4.
Computer analysis of identification of uncultured bacterial 16S rRNA sequences using oligonucleotide probe sequences based on the Ribosomal Database Project II Release 9 collectiona
| Sequence group | No. (%) of sequences |
|---|---|
| Bifidobacteriab | 145 (100) |
| Isolates | 98 (68) |
| Uncultured bifidobacteria | 47 (32) |
| Identifiedc | 34 (23) |
| Unidentified | 13 (9) |
Sequences used in the analysis are ≥1,200 bp in size. Data collection was conducted in March 2006.
Sequences that match the probe Bif164m.
Identification was conducted by matching the sequences of seven 16S rRNA-targeted oligonucleotide probes (Bado434, Bang198, Bbif186, PBR2, Bcat187, Bden82, and Blon1004) to the sequences that match the Bif164m probe stored in the Ribosomal Database Project II Release 9 collection.
In conclusion, we established a feasible method of FISH-FCM for the high-throughput analysis of microbiota in a wide variety of fecal samples. Using this method, we demonstrated the effect of raffinose administration on the growth of indigenous bifidobacteria by showing their population dynamics not only at the genus level, but also at the species level. Although we cannot obtain a general conclusion by a human trial with the participation of only 13 volunteers, monitoring the population dynamics at the species level revealed many interesting features regarding the differences in growth responses to raffinose between major and minor groups of bifidobacteria. These findings contribute not only to the scientific characterization of raffinose effects, but also to a more comprehensive understanding of the bifidobacterial ecology in the human gastrointestinal tract.
Acknowledgments
We thank all the volunteers who participated in this study.
This work was supported by the Special Coordination Funds for Promoting Science and Technology, commissioned by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubin, J. E. 1979. Autofluorescence of viable cultured mammalian cells. J. Histochem. Cytochem. 27:36-43. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, S., E. J. Woodmansey, J. C. M. Paterson, M. E. T. McMurdo, and G. T. Macfarlane. 2005. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 40:28-37. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Amor, K., H. Heilig, H. Smidt, E. E. Vaughan, T. Abee, and W. M. de Vos. 2005. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benno, Y., K. Endo, N. Shiragami, K. Sayama, and T. Mitsuoka. 1987. Effects of raffinose intake on human fecal microflora. Bifidobacteria Microflora 6:59-63. [Google Scholar]
- 6.Benson, R. C., R. A. Meyer, M. E. Zaruba, and G. M. McKhann. 1979. Cellular autofluorescence—is it due to flavins? J. Histochem. Cytochem. 27:44-48. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinoto, A., A. Suksomcheep, S. Ishizuka, H. Kimura, S. Hanada, Y. Kamagata, K. Asano, F. Tomita, and A. Yokota. 2006. Modulation of rat cecal microbiota by administration of raffinose and encapsulated Bifidobacterium breve. Appl. Environ. Microbiol. 72:784-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 11.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lay, C., L. Rigottier-Gois, K. Holmstrøm, M. Rajilic, E. E. Vaughan, W. M. de Vos, M. D. Collins, R. Thiel, P. Namsolleck, M. Blaut, and J. Doré. 2005. Colonic microbiota signatures across five Northern European countries. Appl. Environ. Microbiol. 71:4153-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lay, C., M. Sutren, V. Rochet, K. Saunier, J. Doré, and L. Rigottier-Gois. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7:933-946. [DOI] [PubMed] [Google Scholar]
- 14.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 15.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller, S., K. Saunier, C. Hanisch, E. Norin, L. Alm, T. Midtvedt, A. Cresci, S. Silvi, C. Orpianesi, M. C. Verdenelli, T. Clavel, C. Koebnick, H. J. F. Zunft, J. Doré, and M. Blaut. 2006. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 72:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagura, T., H. Muraguchi, K. Uchino, T. Aritsuka, and Y. Benno. 1999. Effects of ingestion of raffinose-rich soup on the fecal flora and daily defecation in humans. J. Intest. Microbiol. 13:1-7. (In Japanese.) [Google Scholar]
- 19.Nagura, T., S. Hachimura, M. Hashiguchi, Y. Ueda, T. Kanno, H. Kikuchi, K. Sayama, and S. Kaminogawa. 2002. Suppressive effect of dietary raffinose on T-helper 2 cell-mediated immunity. Br. J. Nutr. 88:421-426. [DOI] [PubMed] [Google Scholar]
- 20.Rathbone, E. B. 1980. Raffinose and melezitose, p. 145-185. In C. K. Lee (ed.), Developments in food carbohydrate, vol. 2. Applied Science Publishers, Ltd., London, United Kingdom. [Google Scholar]
- 21.Reddy, N. R., S. K. Sathe, and D. K. Salunkhe. 1989. Carbohydrates, p. 51-74. In D. K. Salunkhe and S. S. Kadam (ed.), CRC handbook of world food legumes: nutritional chemistry, processing technology, and utilization, vol. 1. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 22.Rigottier-Gois, L., A. G. Le Bourhis, G. Gramet, V. Rochet, and J. Doré. 2003. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237-245. [DOI] [PubMed] [Google Scholar]
- 23.Rigottier-Gois, L., V. Rochet, N. Garrec, A. Suau, and J. Doré. 2003. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridisation combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 26:110-118. [DOI] [PubMed] [Google Scholar]
- 24.Rochet, V., L. Rigottier-Gois, S. Rabot, and J. Doré. 2004. Validation of fluorescent in situ hybridization combined with flow cytometry for assessing interindividual variation in the composition of human fecal microflora during long-term storage of samples. J. Microbiol. Methods 59:263-270. [DOI] [PubMed] [Google Scholar]
- 25.Scardovi, V., and F. Crociani. 1974. Bifidobacterium catenulatum, Bifidobacterium dentium, and Bifidobacterium angulatum: three new species and their deoxyribonucleic acid homology relationships. Int. J. Syst. Bacteriol. 24:6-20. [Google Scholar]
- 26.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suksomcheep, A., I. N. Sujaya, K. Saito, A. Yokota, K. Asano, and F. Tomita. 2001. Development of 16S rDNA targeted probes for detection of some human health beneficial strains of intestinal microorganisms. Biotechnol. Sust. Util. Biol. Resour. Trop. 15:111-125. [Google Scholar]
- 28.Takada, T., K. Matsumoto, and K. Nomoto. 2004. Development of multi-color FISH method for analysis of seven Bifidobacterium species in human feces. J. Microbiol. Methods 58:413-421. [DOI] [PubMed] [Google Scholar]
- 29.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 30.Zoetendal, E. G., K. Ben-Amor, H. J. M. Harmsen, F. Schut, A. D. L. Akkermans, and W. M. de Vos. 2002. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl. Environ. Microbiol. 68:4225-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]