Abstract
Single-stranded gaps at the 3′ ends of Streptomyces linear replicons are patched by DNA synthesis primed by terminal proteins (TP) during replication. We devised an in vitro system that specifically incorporated dCMP, the first nucleotide at the 5′ ends, onto a threonine residue of the TP of Streptomyces coelicolor.
Chromosomes of soil bacteria Streptomyces spp. are notable for their linear structure (13), with covalently attached terminal protein (TP) and terminal inverted repeats of various lengths. Linear plasmids with the same structural features are also abundant in Streptomyces spp. Unlike the cases for the well-characterized TP-capped linear genomes of φ29 phage (reviewed in reference 15) and adenoviruses (reviewed in reference 14), which initiate replication at both ends by using the TP as the primer, replication of the linear chromosomes and plasmids of Streptomyces is initiated from an internal origin and proceeds to the termini (6, 16). This leaves single-stranded gaps of 200 to 300 nucleotides at the 3′ ends on various Streptomyces linear plasmids and chromosomes (6; C.-H. Huang, unpublished results) and presumably on the Streptomyces chromosomes. These telomere sequences contain extensive palindromes with potential to form complex and thermodynamically stable secondary structures, which presumably are important for structural integrity and for the patching of the single-strand gaps (12). Several mechanisms have been proposed for the end patching (7). Experimental evidence suggests a patching DNA synthesis using the TP as a primer (18).
The TPs of several linear Streptomyces chromosomes and plasmids have been isolated and sequenced (see, for example, references 3 and 20). They are conserved in amino acid sequences and sizes (184 or 185 amino acids) and contain a putative helix domain that resembles part of the DNA-binding “thumb” domain of human immunodeficiency virus reverse transcriptase and a putative amphiphilic beta-sheet that may be involved in the observed self-aggregation of the TP and/or in membrane binding. In addition, these proteins are rich in positively charged residues, which give rise to very high predicted pI values (11 to 12). There is no clear similarity between the TPs of Streptomyces chromosomes and those of φ29 phage, adenoviruses, or other TP-capped linear replicons.
In the Streptomyces chromosomes and some (but not all) linear plasmids, the gene encoding TP (tpg) is located downstream from tap, which encodes an 80-kDa telomere-associated protein, Tap (1). Both Tpg and Tap proteins are essential for replication of linear chromosomes (1, 3). Tap interacts with Tpg and specific motifs in the 3′ overhangs and is proposed to function in recruiting TP to the replication intermediates (1).
To investigate the mechanism of end patching, we devised an in vitro assay in which TP from Streptomyces coelicolor was specifically labeled with [32P]dCMP, the first nucleotide at the 5′ ends of the Streptomyces linear replicon. For a substrate for such deoxynucleotidylation, a TP expression vector was constructed by inserting a PCR-amplified TP gene of S. coelicolor (tpgsco) into the expression vector pRSET A (Invitrogen) under the control of the T7 promoter without a His tag, and transferred into Escherichia coli BL21-CodonPlus (DE3) (Stratagene). E. coli harboring pRSET A::tpgsco was cultured in LB at 37°C to log phase, harvested, and sonicated in TENG buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 20 mM NaCl, 10% glycerol) supplemented with 10 mM of β-mercaptoethanol. After centrifugation, the supernatant containing Tpgsco was used in the deoxynucleotidylation reaction. The chosen template of the deoxynucleotidylation was recombinant linear pLUS968 (20), which contained an autonomously replicating sequence of linear plasmid pSLA2 (18) and the 365-bp terminal sequence of the S. lividans chromosome (92% identical to that of the S. coelicolor chromosome in the terminal 167 bp). S. coelicolor M145 (4) containing pLUS968 was cultured to log phase in thiostrepton-supplemented tryptic soy broth medium (Difco) at 30°C to log phase, harvested, washed, and resuspended in two volumes of TENG buffer. After sonication, the lysate was centrifuged, and the supernatant, containing 3 to 5 mg/ml of protein, was used as the template and enzyme source for deoxynucleotidylation.
A typical reaction mixture contained 15 μl of the S. coelicolor(pLUS968) extract, 1 μl of Tpgsco-containing supernatant of E. coli extract, 50 mM Tris-HCl (pH 7.4), 10 mM Mg2+, 1 mM dithiothreitol, 3 mM ATP, and 3.3 pmol [α-32P]deoxynucleoside triphosphate (dNTP) (10 μCi; Ampharmacia) in a total volume of 30 μl. The reaction was carried out at 25°C for 30 min. To remove products of intrinsic DNA synthesis, the reaction product was treated with 10 units of DNase I at 37°C for 30 min. The final products were collected by trichloroacetic acid (TCA) precipitation or immunoprecipitation using rabbit antibody against polypeptide QRTVERYVKNEIKPR (residues 49 to 63 of Tpgsco) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by autoradiography.
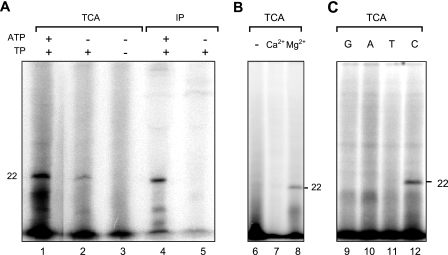
DNase I treatment eliminated radioactive products of a wide range of molecular masses, leaving a radioactively labeled product of about 22 kDa, which reacted with anti-TP antibody (Fig. 1A). The use of the antibody to recover the product actually gave a cleaner background than TCA precipitation. No labeled 22-kDa product was detected if the extrinsic TP was omitted (Fig. 1A). These results indicated that the product was dCMP-labeled Tpgsco.
FIG. 1.
In vitro incorporation of dCMP into Tpgsco. (A) Identification of the TP-dCMP adduct. A typical reaction using [α-32P]dCTP produced a labeled product of 22-kDa that was collected by TCA precipitation (lane 1) or immunoprecipitation (IP; lane 4). Omission of ATP resulted in a lower level of dCMP incorporation (lanes 2 and 5). Omission of extrinsic TP resulted in the complete absence of the labeled 22-kDa product (lane 3). (B) Requirement of magnesium for deoxynucleotidylation. Omission of MgCl2 (−; lane 6) or replacement of MgCl2 with CaCl2 (lane 7) resulted in a loss of dCMP incorporation. (C) Nucleotide discrimination in the deoxynucleotidylation reaction. Typical deoxynucleotidylation reactions were performed with the modifications described below. The products were treated with DNase I and collected by TCA precipitation for analysis. Different [α-32P]dNTPs (G, dGTP [lane 92]; A, dATP [lane 10]; T, dTTP [lane 11]; C, dCTP [lane 12]) were used in the reaction.
The inclusion of ATP boosted the deoxynucleotidylation in some, but not all, cell extract preparations (Fig. 1A), suggesting the need for this energy source for deoxynucleotidylation and variability of ATP levels among different cell extract preparations. The deoxynucleotidylation of Tpgsco required added Mg2+. No TP-dCMP complex was detected when Mg2+ was omitted or replaced with Ca2+ (Fig. 1B). The S. coelicolor cell extract supposedly supplied the enzymes and template (pLUS968 plasmid) for the reaction. No deoxynucleotidylation was detected in the absence of the extract (data not shown). Similarly, in vitro deoxynucleotidylation of φ29 and adenovirus TP either requires or is strongly stimulated by TP-DNA templates (5, 8).
dCMP is the first nucleotide at the 5′ end of pLUS968 (and the S. coelicolor chromosome) covalently bound to Tpgsco. Our deoxynucleotidylation system specifically selected dCTP and did not incorporate the other three dNTPs into the 22-kDa product (Fig. 1C). Moreover, the addition of nonradioactive CTP at a 90-fold concentration did not significantly reduce dCMP incorporation into Tpgsco (data not shown). Moreover, when the S. coelicolor extract was treated with micrococcal nuclease before addition to the reaction, no TP-dCMP formation was detected (data not shown). This supports the notion that the nucleotide specificity is a function of the template on which the incorporation takes place.
To identify the amino acid in Tpgsco that is attached to dCMP, the [α-32P]dCMP-Tpgsco adduct was eluted from the gel and subjected to acid hydrolysis and for phosphoamino acid identification using the procedures of Pargellis et al. (17) and Garcia et al. (9). The hydrolysate of [α-32P]dCMP-Tpgsco was subjected to two-dimensional electrophoresis on a Polygram CEL 400 cellulose thin-layer plate (Macherey-Nagel, Germany). The labeled phosphoamino acid was located by autoradiography, and the internal standards (nonradioactive phosphothreonine, phosphoserine, and phosphotyrosine) were determined by ninhydrin (0.25% in acetone) staining. The result (Fig. 2) revealed a labeled phosphothreonine, indicating that the dCMP is linked to a Thr residue on Tpgsco. This is consistent with the previously observed alkali lability of the phosphodiester bond between the TP and the linear Streptomyces DNA (see, for example, references 13 and 20), although the more alkali-labile Ser has been previously suggested to provide the linkage to DNA (20). There are 11 threonine residues in Tpgsco. Our preliminary proteolytic mapping results (data not shown) indicated that the dCMP was attached to a Thr in the C-terminal region, which contains no predicted functional or structural motif. In comparison, deoxynucleotidylation of φ29 (11) and adenoviruses (19) also takes place in the C-terminal region of TP but at a Ser residue instead.
FIG. 2.
Identification of 32P-labeled amino acids. The [α-32P]dCMP-labeled TP was isolated by immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis and hydrolyzed in acid. The hydrolysate was vacuum dried in the presence of NaOH and mixed with the three phosphoamino acid standards phosphoserine (Ser-P), phosphothreonine (Thr-P), and phosphotyrosine (Tyr-P) and analyzed on a cellulose thin-layer sheet by two-dimensional electrophoresis. The radioactive phosphoamino acid detected by autoradiography was superimposed on the standards, which were stained with ninhydrin (dotted areas). The origin is indicated by a circle.
The DNA polymerases involved in TP-primed DNA synthesis in φ29 and adenoviruses are of B-type superfamily (also referred to as α-like polymerases). No such DNA polymerase was found in the complete genome sequences of S. coelicolor and S. avermitilis. It is not clear which DNA polymerase of Streptomyces is involved in end patching. Bao and Cohen (2), using Tap as a scaffold, identified DNA polymerase I (PolI) as a component of the Streptomyces telomere complex. They further demonstrated that PolI possessed a reverse transcription activity and that the polA gene was essential in Streptomyces. We, however, could create a polA null mutation in two strains of S. coelicolor (3454 and 3456) by using the REDIRECT procedure (10), and the chromosomes in these mutants remained linear (T.-W. Huang and C. W. Chen, unpublished data). Moreover, cell extract from two independent polA null mutants (HT1-3 and HT1-4) of 3456 could support [32P]dCMP incorporation as efficiently as the polA+ extracts (data not shown), indicating that PolI either is not essential for deoxynucleotidylation or is surrogated by another polymerase activity in the extracts.
Acknowledgments
We thank David Hopwood for critical reading of the manuscript and suggestions for improvement.
This study is funded by research grants from National Science Council, Republic of China, to C.W.C. (NSC93-2321-B010-004, NSC94-2321-B010-005) and to C.-C.Y. (NSC93-2311-B-033-001, NSC94-2311-B-033-002) and a National Lectureship Award from the Ministry of Education (2001-2004), Republic of China, to C.W.C.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Bao, K., and S. N. Cohen. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2004. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc. Natl. Acad. Sci. USA 101:14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, L., A. Bernad, J. A. Esteban, and M. Salas. 1992. DNA-independent deoxynucleotidylation of the ϕ29 terminal protein by the ϕ29 DNA polymerase. J. Biol. Chem. 267:1225-1230. [PubMed] [Google Scholar]
- 6.Chang, P. C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12:192-196. [DOI] [PubMed] [Google Scholar]
- 8.Chowrira, B. M., J. S. Zhao, and L. A. Lucher. 1991. Formation in vitro of the pTP-dCMP initiation complex of human adenovirus type 12. J. Gen. Virol. 72:427-430. [DOI] [PubMed] [Google Scholar]
- 9.García, P., J. M. Hermoso, J. A. García, E. García, R. López, and M. Salas. 1986. Formation of a covalent complex between the terminal protein of pneumococcal bacteriophage Cp-1 and 5′-dAMP. J. Virol. 58:31-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermoso, J. M., E. Mendez, F. Soriano, and M. Salas. 1985. Location of the serine residue involved in the linkage between the terminal protein and the DNA of phage ϕ29. Nucleic Acids Res. 13:7715-7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, C.-H., Y.-S. Lin, Y.-L. Yang, S.-W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28:905-926. [DOI] [PubMed] [Google Scholar]
- 13.Lin, Y.-S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 14.Liu, H., J. H. Naismith, and R. T. Hay. 2003. Adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 272:131-164. [DOI] [PubMed] [Google Scholar]
- 15.Meijer, W. J., J. A. Horcajadas, and M. Salas. 2001. ϕ29 family of phages. Microbiol. Mol. Biol. Rev. 65:261-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musialowski, M. S., F. Flett, G. B. Scott, G. Hobbs, C. P. Smith, and S. G. Oliver. 1994. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J. Bacteriol. 176:5123-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pargellis, C. A., S. E. Nunes-Duby, L. M. de Vargas, and A. Landy. 1988. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J. Biol. Chem. 263:7678-7685. [PubMed] [Google Scholar]
- 18.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-904. [DOI] [PubMed] [Google Scholar]
- 19.Smart, J. E., and B. W. Stillman. 1982. Adenovirus terminal protein precursor. Partial amino acid sequence and the site of covalent linkage to virus DNA. J. Biol. Chem. 257:13499-13506. [PubMed] [Google Scholar]
- 20.Yang, C.-C., C.-H. Huang, C.-Y. Li, Y.-G. Tsay, S.-C. Lee, and C. W. Chen. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: A novel class of replication priming proteins. Mol. Microbiol. 43:297-305. [PubMed] [Google Scholar]