Abstract
Soils from the hyperarid Atacama Desert of northern Chile were sampled along an east-west elevational transect (23.75 to 24.70°S) through the driest sector to compare the relative structure of bacterial communities. Analysis of denaturing gradient gel electrophoresis (DGGE) profiles from each of the samples revealed that microbial communities from the extreme hyperarid core of the desert clustered separately from all of the remaining communities. Bands sequenced from DGGE profiles of two samples taken at a 22-month interval from this core region revealed the presence of similar populations dominated by bacteria from the Gemmatimonadetes and Planctomycetes phyla.
The Atacama Desert of northern Chile stretches for more than 1,000 km along the narrow coastal plateau between the Rio Copiapó (27.32°S) and the town of Arica (18.48°S) near the Peruvian border. The interior of the desert, between the coastal escarpment and the foot of the Andes, has been described as “the most barren region imaginable,” devoid of plant life, receiving only a few millimeters of precipitation every few years (35), and potentially approaching the dry limit of microbial life (32). This study represents a further characterization of our recently reported observation that the hyperarid core of the Atacama Desert harbors both viable bacteria and recoverable DNA (26).
The objective of this work was to perform a general comparison of bacterial community structures along an elevational transect representing the unique, extreme conditions of the driest expanse of the central Atacama Desert. In addition, a single sample location was selected in the hyperarid core for a more complete characterization of bacterial populations.
Transect description.
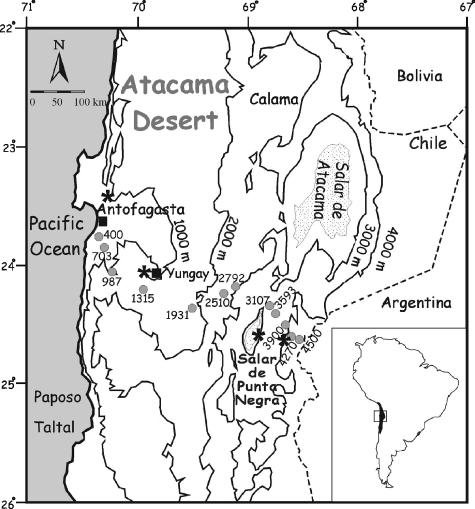
The Punta Negra transect that we sampled crosses the full extent of the Atacama Desert from the barren coastal range above Antofagasta (23.65°S, 70.24°W) at an elevation of 400 m to the slopes of the Volcán de Llullaillaco (24.72°S, 68.55°W) in the Andes at 4,500 m above sea level (Fig. 1). The hyperaridity along this transect restricts perennial vegetation to between 3,500 and 4,800 m, where unreliable precipitation events in both summer and winter support a vegetation belt of continuous but sparse and species-poor desert scrub and grassland with low beta-diversity and few endemics (2, 27, 37). Below this vegetation belt is an impressive, Mars-like expanse of absolute desert that has been largely devoid of rain and vascular plants for at least the last million years (22). Mean annual precipitation (MAP) and mean annual temperature (MAT) data were obtained from published studies and meteorological stations located in close proximity to the Punta Negra transect (Fig. 1).
FIG. 1.
Map of the study area indicating the locations of sampling sites along the Punta Negra transect. Filled circles (•) indicate sampling sites, filled squares (▪) represent towns along the transect, and asterisks (*) indicate locations of the following four weather stations along the transect: Antofagasta (23.41°S, 70.25°W; 10-m elevation; MAT = 17°C; MAP = 2.0 mm) (28), Yungay (24.08°S, 69.97°W; 950-m elevation; MAT = 16.5°C; MAP = 0.6 mm) (29), Salar Punta Negra (24.6°S, 68.92°W; 3,000-m elevation; MAT = 7°C; MAP = 7 mm) (24), and Llullaillaco (24.63°S, 68.67°W; 4,200-m elevation; MAT = 1.7°C; MAP = 35.7 mm) (24). Throughout the Central Atacama desert, MATs decrease linearly with increasing elevation beginning at 17°C (y = −0.0038 + 18.9; r2 = 0.94).
Soil sampling and analysis.
Soil samples were taken using sterile tools at a depth of 25 to 30 cm at regular intervals along the Punta Negra transect in October 2002 and stored in sterile sealed polycarbonate tubes at 4°C (Table 1). In addition, a second sample was taken at 989 m in July 2004 for comparative population analysis. The sample depth was selected to target permanent populations rather than transient populations carried in blowing dust. Interplant regions were sampled when possible to avoid localized rhizosphere effects. Soil sample characteristics were analyzed by the University of Arizona Water Quality Center Laboratory (Tucson, AZ), and viable bacterial counts are reported in Table 2.
TABLE 1.
Characterization of the soil sampling sites along the Punta Negra transect
| Sample (elevation [m]) | Latitude (°S) | Longitude (°W) | Sample date (mo/yr) | Biomea | Plant cover (%) | Perennial vegetation |
|---|---|---|---|---|---|---|
| 400 | 23.74938 | 70.35822 | 10/2002 | Absolute desert | —b | Nolana spp. along dry wash |
| 703 | 23.95695 | 70.28595 | 10/2002 | Absolute desert | 0 | None |
| 987 | 24.07528 | 70.20925 | 10/2002 | Absolute desert | 0 | None |
| 989 | 24.07065 | 70.20177 | 07/2004 | Absolute desert | 0 | None |
| 1,315 | 24.36310 | 69.94595 | 10/2002 | Absolute desert | 0 | None |
| 1,931 | 24.46892 | 69.40787 | 10/2002 | Absolute desert | 0 | None |
| 2,510 | 24.33925 | 69.23507 | 10/2002 | Absolute desert | —b | Adesmia atacamensis |
| 2,792 | 24.26348 | 69.19138 | 10/2002 | Absolute desert | —b | Adesmia atacamensis, Oxalis |
| 3,107 | 24.44883 | 68.83053 | 10/2002 | Absolute desert | —b | Isolated Acantholippia deserticola 50-100 m from site |
| 3,593 | 24.52562 | 68.73160 | 10/2002 | Pre-Puna | 1-3 | Cristaria andicola, Sysimbrium phillipiani |
| 3,900 | 24.55812 | 68.66685 | 10/2002 | Puna | 1-3 | Stipa frigida, Fabiana bryoides, Artemisia copa, Happlopappus rigidus, C. andicola, Adesmia hystrix, Opuntia conoidea |
| 4,270 | 24.60033 | 68.58547 | 10/2002 | High Andean Steppe | 4-10 | Stipa frigida, Deyeuxia curvula, |
| 4,500 | 24.69612 | 68.62468 | 10/2002 | High Andean Steppe | 2-4 | Mulinum crassifolium, |
| Monschopsis monocephala, | ||||||
| Chaethantera revoluta, Perezia | ||||||
| atacamanensis, Viola spp. |
Absolute desert refers to the near absence of plant cover. Other biome designations were described previously by Villagrán et al. (37).
Too few plants were present to quantify the percent plant cover. The isolated plants observed were separated by expanses of unvegetated terrain.
TABLE 2.
Chemical and microbiological characteristics of the Punta Negra transect soil samples
| Sample (elevation [m]) | pHa | ECa,b (dS m−1) | TOCc (%) | Culturable countd (CFU g−1) |
|---|---|---|---|---|
| 400 | 7.75 | 0.81 | 0.02 | 1.62 × 105 |
| 703 | 6.43 | 5.40 | 0.01 | BQDL |
| 987 | 7.84 | 5.70 | 0.01 | 1.36 × 105 |
| 1,315 | 7.12 | 0.10 | 0.02 | 5.40 × 103 |
| 1,931 | 6.92 | 2.30 | 0.02 | 9.11 × 104 |
| 2,510 | 7.54 | 0.01 | 0.03 | 1.54 × 104 |
| 2,792 | 7.01 | 0.05 | 0.02 | 9.77 × 105 |
| 3,107 | 7.52 | 0.35 | 0.02 | 4.46 × 106 |
| 3,593 | 7.81 | 0.49 | 0.05 | 7.10 × 106 |
| 3,900 | 7.89 | 0.22 | 0.08 | 4.30 × 107 |
| 4,270 | 7.62 | 0.10 | 0.08 | 2.07 × 107 |
| 4,500 | 7.30 | 0.08 | 0.09 | 1.96 × 106 |
Soil-to-deionized water ratio of 1:1.
EC, electrical conductivity.
TOC, total organic carbon determined by high-temperature combustion using an NCS analyzer (Carlo Erba model Na1500) (detection limit, 0.01%).
Determined following a 14-day preincubation (room temperature) of soil with sterile, deionized water (10% [vol/wt] approximating field capacity) by serially diluting 0.5 g (dry weight) of soil in distilled water and plating onto R2A agar (Difco Laboratories, Detroit, MI) amended with 10 mg liter−1 cycloheximide to inhibit fungal growth. BQDL, below the quantitative detection limit of 1,000 CFU g−1 soil.
Soil bacterial community DNA extraction, amplification, and analysis.
Total genomic DNA was extracted from dry soil samples via direct lysis using the Fast DNA SPIN kit for soil (Qbiogene, Carlsbad, CA). Extraction blanks were processed in parallel throughout the full procedure as negative controls to evaluate potential DNA contamination from reagents. The V9 variable region of the 16S rRNA gene was PCR amplified from each extract using Bacteria primer 1070F (5′-ATG GCT GTC GTC AGC T-3′) and universal primer 1392R (5′-ACG GGC GGT GTG TAC-3′) with a 40-bp GC clamp (13). Amplification conditions followed the protocol of Colores et al. (7), with a slight modification.
Community structure was evaluated by denaturing gradient gel electrophoresis (DGGE) analysis of the 16S rRNA gene products using a D-Code Universal Mutation detection system (Bio-Rad Laboratories, Hercules, CA). Acrylamide gels (6%) were prepared with a 50 to 80% urea-formamide denaturing gradient. Lanes were loaded with either 20 μl (400-, 2,510-, 2,792-, 3,107-, 3,593-, 3,900-, 4,270-, and 4,500-m samples) or 40 μl (703-, 987-, 1,315-, and 1,931-m samples) of PCR product and the corresponding negative controls, run at a constant voltage of 50 V for 15 h at 60°C, and stained for visualization and photography with SYBR Green I (Molecular Probes, Eugene, OR). The banding pattern of each lane in the DGGE gels was scored using a method described previously by Konopka et al. (20), and the resulting matrix of binary data was analyzed with Kruskal's isotonic multidimensional scaling analysis (KIMDSA) (36).
Population analysis.
Multiple DNA extractions were performed and consolidated from the Oct 2002 987-m (four extracts) and the July 2004 989-m (nine extracts) soil samples to obtain sufficient template DNA to generate PCR-DGGE profiles with extractable bands. All bands from each profile were excised for PCR amplification and incubated overnight at 37°C in a DNA elution buffer (0.5 M NH4OAc, 1 mM EDTA, pH 8.0) (3). Amplified PCR products were compared to the original profiles by DGGE analysis (55 to 65% gradient) to confirm band purity and identity. Two to three DGGE-PCR cycles were performed to purify each band for sequence analysis. Duplicate bands were excised along the gradient line from replicate profiles of each sample, and 100% identity was confirmed. Both forward and reverse sequences were generated using primers 1070F and 1392R to confirm sequence accuracy (University of Arizona Research Laboratory Genomic Analysis and Technology Core, Tucson, AZ). All unique sequences were identified using BLAST (1) and the RDP Sequence Match and Classifier (6) programs and then deposited in GenBank (Table 3).
TABLE 3.
Bacteria identified from DGGE profiles of 987-meter (2002) and 989-meter (2004) soil samples
| Band | GenBank accession no. | Phylum | Closest BLAST match (GenBank accession no.) | % Identity | Source |
|---|---|---|---|---|---|
| 987-5 | DQ648483 | Gemmatimonadetes | Uncultured bacterium clone AT425_EubC11 (AY053483) | 95 | Gas hydrate sediments (Gulf of Mexico) |
| Uncultured forest soil bacterium clone DUNssu177 (AY913247) | 95 | Forest soil (Germany) | |||
| 987-2 | DQ648479 | Gemmatimonadetes | Uncultured Gemmatimonadetes clone AKYH1514 (AY921705) | 96 | Agricultural soil (Minnesota) |
| 987-3b | DQ648481 | Actinobacteria | Uncultured soil bacterium clone 288-2 (AF423245) | 97 | Agricultural soil (Riverside, CA) |
| 987-5b2 | DQ648484 | Planctomycetes | Uncultured bacterium clone pGXAR2 (DQ256391) | 93 | Alkaline soil (China) |
| 987-4 | DQ648482 | Gemmatimonadetes | Uncultured Gemmatimonadetes clone AKYH1194 (AY921682) | 94 | Agricultural soil (Minnesota) |
| 987-3b2 | DQ648480 | Planctomycetes | Uncultured planctomycete (AJ431346) | 96 | Ikaite tufa columns (Greenland fjord) |
| 987-1b | DQ648478 | Thermomicrobia | Uncultured bacterium clone FBP267 (AY250872) | 96 | Rock surface (McMurdo Dry Valley, Antarctica) |
| 989-12b2 | DQ648485 | Gemmatimonadetes | Uncultured bacterium clone AT425_EubC11 (AY053483) | 94 | Gas hydrate sediments (Gulf of Mexico) |
| Uncultured forest soil bacterium clone DUNssu028 (AY913247) | 94 | Forest soil (Germany) | |||
| 989-9b2 | DQ648486 | Gemmatimonadetes | Uncultured Gemmatimonadetes clone AKYH1514 (AY921705) | 95 | Agricultural soil (Minnesota) |
| 989-8b2 | DQ648487 | Proteobacteria | Acidithiobacillus ferrooxidans 33020 (AJ278719) | 99 | Uranium waste piles (Germany) |
| Acidithiobacillus thiooxidans (AY495961) | 99 | Mine drainage (Wales, United Kingdom) | |||
| 989-7b | DQ648488 | Planctomycetes | Uncultured bacterium clone pGXAR2 (DQ256391) | 93 | Alkaline soil (China) |
| 989-11b2 | DQ648482a | Gemmatimonadetes | Uncultured Gemmatimonadetes clone AKYH1194 (AY921682) | 94 | Agricultural soil (Minnesota) |
The sequence of 989-11b2 is 100% similar to that of 987-4.
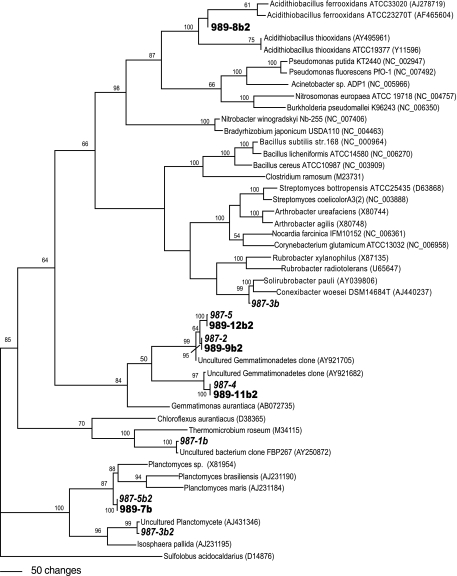
A base tree was constructed using nearly complete GenBank 16S rRNA gene sequences representing major phyla of Bacteria. Sequences were aligned using Clustal W (Wisconsin package version 10.3; Accelrys Inc., San Diego, CA), and the alignments were manually adjusted using MacClade v. 4.08 (25). Most parsimonious trees were constructed from the aligned sequences, and DGGE bands were individually incorporated to determine taxonomic affiliations (see Fig. 3).
FIG. 3.
One of seven most parsimonious trees generated from reference bacterial strain sequences (GenBank) and DGGE band sequences from 987-m (boldface italic type) and 989-m (boldface type) soil samples. A base tree was generated with nearly full-length sequences from the GenBank database using maximum parsimony analysis by heuristic search using tree bisection reconnection branch swapping on starting trees generated by random sequence addition as implemented by using PAUP 4.0 Beta 10 (34). DGGE band sequences were inserted individually into the base tree data set without realignment of base tree sequences. Sulfolobus acidocaldarius was used as the outgroup. Bootstrap values (1,000 replicates) from maximum parsimony analysis are given for nodes with ≥50% support.
Punta Negra transect results.
Viable bacteria were successfully cultured from all sites along the transect, including one of our soil samples (703 m) that yielded only one or two colonies per plate (Table 2). Counts were lowest at or below 2,510 m within the absolute desert region (Table 1) and highest from the 3,900- and 4,270-m soils of the Puna and High Andean Steppe biomes.
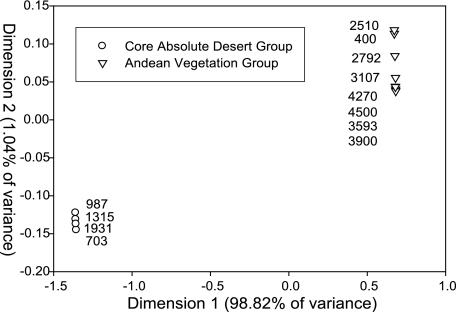
16S rRNA genes were also successfully amplified from all soil DNA extracts. Analysis of 16S rRNA gene DGGE profiles from transect samples revealed two interesting clusters suggesting the presence of two distinct bacterial community structures (Fig. 2). The first cluster included soils from 703, 987, 1,315, and 1,931 m in the core absolute desert, where hyperaridity has prohibited the growth of vascular plants for millions of years (12) and precipitation events occur only once every 20 to 50 years (estimated from gullying of tailings at abandoned nitrate mines). The second cluster, referred to here as the Andean vegetation group, included all of the remaining elevations (400, 2,510, 2,792, 3,107, 3,593, 3,900, 4,270, and 4,500 m), although at the time of sampling, vegetation was observed only at elevations of 3,593 m and above. Total plant species richness increased from 2 at 3,593 m to 7 at and above 3,900 m. In terms of plant cover, the 4,270-m elevation was the highest, ranging from 4 to 10%. Both above and below this elevation, plant cover declined to between 1 and 4% (Table 1).
FIG. 2.
KIMDSA of the DGGE profiles from the Punta Negra transect analyzed in three dimensions with 99.35% of the variance explained, a stress factor of 0.0344, and a P value of 0.04. Analysis was done using the statistical software package R (R Foundation for Statistical Computing, Vienna, Austria). The permutation test used confirmed the significance of the classification of the groups established by KIMDSA.
The sharp separation of the bacterial communities along the Punta Negra transect into two distinct groups suggests that bacterial community profiles could serve as more effective indicators of extreme hyperaridity in the Atacama Desert than the presence of perennial vegetation. This hypothesis is based on the fact that samples from 400-, 2,510-, 2,792-, and 3,107-m elevations clustered with the Andean vegetation group rather than the core absolute desert group, despite the virtual absence of perennial vegetation at these elevations (Fig. 2 and Table 1). None of the soil properties reported in Table 2 explain these unexpected results. Houston and Hartley (17) previously categorized the region of the Atacama Desert below 2,300 m as a zone of extreme hyperaridity, although they explained that significant variations in the intensity of aridity occur within the zone. Due to the limited availability of weather data along the Punta Negra transect, one can only speculate that the observed transition in bacterial community affiliation from the core absolute desert group to the Andean vegetation group at 2,510 m represents a significant variation in moisture availability.
Several observations support the speculation that the community profiles reflect the frequency and history of precipitation or exposure to moisture. In October 2002, we observed evidence of past vegetation extending 500 m or more below the present 3,593-m lower vegetation limit (Table 1). Occasional root fragments were excavated from extensive fields of tuco-tuco (Ctenomys fulvus: Rodentia: Ctenomydae) burrows (8) located across what is now unvegetated terrain at ∼3,100 m. Although we have not dated these root fragments or systematically mapped the extent of such tuco-tuco fields in the vast areas that lack perennial plants, it appears that tuco-tucos are capable of tracking winter and summer annual blooms that presumably result from precipitation events in previous years. Second, recent surveys of fossil rodent middens spanning a broad sector of the central Atacama Desert suggest that north of 24°S, wet summers occasionally yield patches of summer-flowering annuals at elevations down to 2,500 m; south of 24°S, the same holds true for winter annuals (4, 21, 22, 23, 27). These potential historical expansions of winter and summer annuals below the 3,593-m lower perennial vegetation limit of October 2002 hint at the occurrence of past precipitation events that influenced microbial populations at the fringe, 2,510-m, 2,792-m, and 3,107-m sample locations. Samples at 2,510 and 2,792 m were also located on an alluvial fan in areas potentially exposed to runoff from regular precipitation events in the Cordillera Domeyko and Andes mountain ranges (12).
The 400-m sample location is separated from the remaining sites of the Andean vegetation group by the large expanse of absolute desert in the Central Valley. This sample location is the only elevation in the transect located below the crest of the Cordillera de la Costa escarpment. In some coastal regions such as Paposo (Fig. 1), a semipermanent fog zone develops where the coastal escarpment is massive (11). Although this stratus layer dissipates in areas such as Antofagasta, where the coastal topography levels off, the presence of Pacific moisture may be sufficient to affect microbially diverse populations in this region. Thus, minor differences in the frequencies of soil moisture exposure, above and below the absolute desert, resulting from isolated rainfall events, runoff exposure, or Pacific moisture may explain the distinct separation of the bacterial community profiles into two different community structures.
Identification of specific populations from the hyperarid core region.
The four DGGE profiles from the core absolute desert group contained between four and seven bands each, and three of the bands were common to all four profiles. The 987-m sample was chosen from among these samples for further characterization because the culturable counts were the highest (Table 2) and because the DGGE profile had the greatest number of bands. The July 2004 sample was taken nearby at 989 m (Table 1) to determine the constancy of populations observed in the 2002 samples. DGGE analysis of the 989-m sample produced a profile similar to that of the 987-m sample from 2002. Seven bands were sequenced and identified from the 987-m sample (2002), and five were sequenced and identified from the 989-m sample (2004) (Table 3). A similarity distance analysis generated using PAUP4.0 Beta 10 indicated that four of the five bands from the 2004 sample (989 m) were of the same phylotype as bands identified from the 2002 sample (987 m) (sequences with similarity distances of <0.01 were classified as the same phylotype). Similarity distances for these bands are as follows: band 12b2 from the 989-m sample (989-12b2) and 987-5, 0.0059; 989-9b2 and 987-2, 0.0028; 989-11b2 and 987-4, 0.0000; and 989-7b and 987-5b2, 0.0030.
BLAST analysis indicated that the majority of the band sequences from both samples were most closely related to uncultured, unidentified bacterial clones (93 to 97%) (Table 3). The one exception was band 8b2 from the 989-m sample, which had 99% sequence identity to the Gammaproteobacteria Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans. Of the remaining bands, three bands that were common to both profiles were affiliated with Gemmatimonadetes, and one band was affiliated with Planctomycetes (Fig. 3 and Table 3). Although only one cultured Gemmatimonadetes bacterium has been described in the literature, numerous clones from diverse soils of five different continents, including the Tataouine Desert of Tunisia, have been identified (5, 15, 30, 38). Planctomycetes were originally associated with freshwater, marine, and hot spring environments, but these results combined with data from other recent studies identifying clones from soils and sediments indicate that these organisms may also be present in a diverse range of ecosystems (33). The remaining three bands from the 987-m sample were associated with Planctomycetes (987-3b2), Actinobacteria (987-3b), and Thermomicrobia (987-1b). The actinobacterium (987-3b) was assigned to the Rubrobacteraceae family with 98% confidence by the RDP Classifier, and the most closely related sequence was the unidentified clone 288-2 (GenBank accession no. AF423245) isolated from an arid Australian soil (16). The final band, band 987-1b, is most closely related to the uncultured bacterium clone FBP267 (accession no. AY250872) identified from a cryptoendolithic community extracted from Beacon sandstone from the McMurdo Dry Valleys region of South Victoria Land, Antarctica (10). Both 987-1b and FBP267 associate at a high bootstrap value (100) with Thermomicrobium roseum (Fig. 3) of the phylum Thermomicrobia (green nonsulfur bacteria).
As might be anticipated, the predominance of Gemmatimonadetes and Planctomycetes in the 987-m and 989-m samples is unique compared to typical soil populations. In a recent analysis of 32 clone libraries from a variety of surface soils, Janssen (18) found that 92% of the bacterial clones belonged to nine dominant phyla: Proteobacteria (39%), Acidobacteria (20%), Actinobacteria (13%), Verrumicrobia (7%), Bacteriodetes (5%), Chloroflexi (3%), Planctomycetes (2%), Gemmatimonadetes (2%), and Firmicutes (1.8%). In contrast, sandy subsurface soils sampled by Zhou et al. (39) in Virginia and Delaware at depths of 1.6 to 7.0 m revealed communities with much less diversity but that were still dominated by Proteobacteria (90%) accompanied by Acidobacteria (3%) and Firmicutes (3%). Nagy et al. (31) previously reported a shift in this distribution to 51% Acidobacteria, 15.5% Proteobacteria, 13.3% Flexibacteria and relatives, 6.7% Actinobacteria, 4.5% Planctomycetes, and 8.9% unknown for arid surface soils. This distribution shows a slight increase in the relative abundance of Planctomycetes in arid soils, but it does not reflect the relative diversity observed in the Atacama Desert soils.
The results from this research provoke questions of interest for future study. Although this study is far from exhaustive in identifying the diversity of bacteria present in the driest regions of the Atacama Desert, it is evident that microbes are capable of enduring extremes of aridity that prevent the growth of vascular plants. The unique phylogenetic distribution of the organisms identified in this study compared to those of other arid soils would suggest that the hyperarid environment does select for bacteria in specific divisions. With the knowledge gained here regarding the predominant organisms present in these extremely arid soils, efforts can now be made to isolate these organisms by using recently identified techniques for culturing the recalcitrant members of phyla such as Gemmatimonadetes and Planctomycetes (9, 14, 19, 38).
Nucleotide sequence accession numbers.
All unique sequences in this work have been deposited in the GenBank database under accession numbers DQ648478 to DQ648488 (Table 3).
Acknowledgments
This research was supported by grants CHE-0133237 and ATM-0213657 from the National Science Foundation and grant 2 P42 ES04940-11 from the National Institute of Environmental Health Sciences Superfund Basic Research Program, NIH.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo, M. T. K., C. Castor, C. Marticorena, M. Muñoz, L. Cavieres, O. Mathei, F. Squeo, M. Grosjean, and R. Rodríguez. 1998. The flora of Llullaillaco National Park located in the transitional winter-summer rainfall area of the northern Chilean Andes. Gayana Botánica 55:93-110. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, p. 2.7.3-2.7.6. John Wiley & Sons, Inc., Hoboken, N.J.
- 4.Betancourt, J. L., C. Latorre, J. A. Rech, J. Quade, and K. A. Rylander. 2000. A 22,000-year record of monsoonal precipitation from northern Chile's Atacama Desert. Science 289:1542-1546. [DOI] [PubMed] [Google Scholar]
- 5.Chanal, A., V. Chapon, K. Benserara, M. Barakat, R. Christen, W. Achouak, F. Barras, and T. Heulin. 2006. The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ. Microbiol. 8:514-525. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colores, G. M., R. E. Macur, D. M. Ward, and W. P. Inskeep. 2000. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 66:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortés, A., E. Miranda, M. Rosenmann, and J. R. Rau. 2000. Thermal biology of the fossorial rodent Ctenomys fulvus from the Atacama Desert, northern Chile. J. Therm. Biol. 25:425-430. [DOI] [PubMed] [Google Scholar]
- 9.Davis, K. E. R., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Torre, J. R., B. M. Goebel, E. I. Friedmann, and N. R. Pace. 2003. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valley, Antarctica. Appl. Environ. Microbiol. 69:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon, M. O., and J. A. E. Hoffman. 1997. Lomas formations of the Atacama Desert, northern Chile. In S. D. Davis, V. H. Heywood, O. Herrera-MacBryde, J. Villa-Lobos, and A. C. Hamilton (ed.), Centres of plant diversity, a guide and strategy for their conservation, vol. 3. The Americas, p. 528-535. World Wildlife Federation, Information Press, Oxford, England. [Google Scholar]
- 12.Dunai, T. J., G. A. González López, and J. Juez-Larré. 2005. Oligocene-Miocene age of aridity in the Atacama Desert revealed by exposure dating of erosion-sensitive landforms. Geology 33:321-324. [Google Scholar]
- 13.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzymski, J. J., B. J. Carter, E. F. DeLong, R. A. Feldman, A. Ghadiri, and A. E. Murray. 2006. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl. Environ. Microbiol. 72:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, A. J., J. Bowyer, M. P. Holley, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Houston, J., and A. J. Hartley. 2003. The Central Andean west-slope rainshadow and its potential contribution to the origin of hyperaridity in the Atacama Desert. Int. J. Climatol. 23:1453-1464. [Google Scholar]
- 18.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konopka, A., T. Bercot, and C. Nakatsu. 1999. Bacterioplankton community diversity in a series of thermally stratified lakes. Microb. Ecol. 38:126-135. [DOI] [PubMed] [Google Scholar]
- 21.Latorre, C. L., J. L. Betancourt, J. A. Rech, J. Quade, C. Holmgren, C. P. Placzek, M. Vuille, and K. A. Rylander. 2005. Late quaternary history of the Atacama Desert. In M. Smith and P. Hesse (ed.), 23 degrees south: archaeology and environmental history of the Southern Deserts. National Museum of Australia, Canberra, Australia.
- 22.Latorre, C., J. L. Betancourt, K. A. Rylander, and J. Quade. 2002. Vegetation invasions into absolute desert: a 45 000 yr rodent midden record from the Calama-Salar de Atacama basins, northern Chile. Geol. Soc. Am. Bull. 114:349-366. [Google Scholar]
- 23.Latorre, C., J. L. Betancourt, K. Rylander, J. Quade, and O. Matthei. 2003. A 13.5-kyr vegetation history from the arid prepuna of northern Chile (22-23°S), Palaeogr. Palaeoclimatol. Palaeoecol. 194:223-246. [Google Scholar]
- 24.Luebert, F., and R. Gajardo. 2000. Vegetación de los Andes áridos del norte de Chile. Lazaroa 21:111-130. [Google Scholar]
- 25.Maddison, D. R., and W. P. Maddison. 2001. MacClade 4: Analysis of Phylogeny and Character Evolution. Version 4.08. Sinauer Associates, Sunderland, Mass.
- 26.Maier, R. M., K. P. Drees, J. W. Neilson, J. Quade, D. A. Henderson, and J. L. Betancourt. 2004. Microbial life in the Atacama Desert. Science 306:1289-1290. [DOI] [PubMed] [Google Scholar]
- 27.Maldonado, A., J. L. Betancourt, C. L. Latorre, and C. Villagrán. 2005. Pollen analyses from a 50,000-yr rodent midden series in the southern Atacama Desert (25°30′S). J. Quat. Sci. 20:493-507. [Google Scholar]
- 28.Marquet, P. A., F. Bozinovic, G. A. Bradshaw, C. Cornelius, H. Gonzalez, J. R. Gutierrez, E. R. Hajek, J. A. Lagos, F. Lopez-Cortes, L. Nuñez, E. F. Rosello, C. Santoro, H. Samaniego, V. G. Standen, J. C. Torres-Mura, and F. M. Jaksic. 1998. Los ecosistemas del desierto de Atacama y area andina adyacente en el norte de Chile. Revista Chilena Hist. Nat. 71:593-617. [Google Scholar]
- 29.McKay, C. P., E. I. Friedmann, B. Gómez-Silva, L. Cáceres-Villanueva, D. T. Andersen, and R. Landheim. 2003. Temperature and moisture conditions for life in the extreme arid region of the Atacama Desert: four years of observations including the El Niño of 1997-1998. Astrobiology 3:393-406. [DOI] [PubMed] [Google Scholar]
- 30.Mummey, D. L., and P. D. Stahl. 2003. Candidate division BD: phylogeny, distribution and abundance in soil ecosystems. Syst. Appl. Microbiol. 26:228-235. [DOI] [PubMed] [Google Scholar]
- 31.Nagy, M. L., A. Pérez, and F. Garcia-Pichel. 2005. The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ). FEMS Microbiol. Ecol. 54:233-245. [DOI] [PubMed] [Google Scholar]
- 32.Navarro-González, R., F. A. Rainey, P. Molina, D. R. Bagaley, B. J. Hollen, J. de la Rosa, A. M. Small, R. C. Quinn, F. J. Grunthaner, L. Cáceres, B. Gomez-Silva, and C. P. McKay. 2003. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 302:1018-1021. [DOI] [PubMed] [Google Scholar]
- 33.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 34.Swofford, D. L. 2006. PAUP*: Phylogenetic Analysis Using Parsimony. Macintosh Beta v. 10. Sinauer Associates, Sunderland, Mass.
- 35.Trewartha, G. T. 1981. The earth's problem climates, 2nd ed., p. 23-40. University of Wisconsin Press, Madison, Wis.
- 36.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S, 4th ed., p. 385-389. Springer Statistics and Computing Series, New York, N.Y.
- 37.Villagrán, C., J. J. Armesto, and M. T. Kalin Arroyo. 1981. Vegetation in a high Andean transect between Turi and Cerro León in Northern Chile. Vegetation 48:3-16. [Google Scholar]
- 38.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, J., B. Xia, H. Huang, A. V. Palumbo, and J. M. Tiedje. 2004. Microbial diversity and heterogeneity in sandy subsurface soils. Appl. Environ. Microbiol. 70:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]