Abstract
Glycoside hydrolases are organized into glycoside hydrolase families (GHFs) and within this larger group, the β-galactosidases are members of four families: 1, 2, 35, and 42. Most genes encoding GHF 42 enzymes are from prokaryotes unlikely to encounter lactose, suggesting a different substrate for these enzymes. In search of this substrate, we analyzed genes neighboring GHF 42 genes in databases and detected an arrangement implying that these enzymes might hydrolyze oligosaccharides released by GHF 53 enzymes from arabinogalactan type I, a pectic plant polysaccharide. Because Bacillus subtilis has adjacent GHF 42 and GHF 53 genes, we used it to test the hypothesis that a GHF 42 enzyme (LacA) could act on the oligosaccharides released by a GHF 53 enzyme (GalA) from galactan. We cloned these genes, plus a second GHF 42 gene from B. subtilis, yesZ, into Escherichia coli and demonstrated that cells expressing LacA with GalA gained the ability to use galactan as a carbon source. We constructed B. subtilis mutants and showed that the increased β-galactosidase activity generated in response to the addition of galactan was eliminated by inactivating lacA or galA but unaffected by the inactivation of yesZ. As further demonstration, we overexpressed the LacA and GalA proteins in E. coli and demonstrated that these enzymes degrade galactan in vitro as assayed by thin-layer chromatography. Our work provides the first in vivo evidence for a function of some GHF 42 β-galactosidases. Similar functions for other β-galactosidases in both GHFs 2 and 42 are suggested by genomic data.
β-Galactosidases, such as the LacZ β-galactosidase of Escherichia coli, are used as molecular biology tools, industrial enzymes, and models for exploring structure-function relationships. However, these uses do not often yield insight into the physiological roles of these enzymes. Although β-galactosidases (EC 3.2.1.23) are also found in glycoside hydrolase families (GHFs) 1, 2, and 35 (groups differing in secondary structure and other attributes [20, 21]), we were especially curious about the in vivo function of GHF 42 β-galactosidases because they occur in organisms isolated from diverse habitats where lactose would not be present, e.g., hot springs (33, 47, 65), hypersaline environments (27, 54), and soil (Bacillus and Streptomyces spp.).
Over a dozen GHF 42 enzymes have been characterized, and most are β-galactosidases (as shown by maximal activity with the β-linked galactosidic substrate o-nitrophenyl-β-d-galactopyranoside [ONPG] or p-nitrophenyl-β-d-galactopyranoside [PNPG]) with less than 10% relative activity on nongalactosidic chromogens (with two exceptions [27, 34]). However, experimental data supporting lactose hydrolysis by GHF 42 β-galactosidases are absent (34, 44, 48) or weak. Several prokaryotes possessing a GHF 42 gene are unable to grow on lactose as a sole carbon source (1, 9, 27), and at least two GHF 42 β-galactosidases do not cleave lactose in vitro (27, 64). The determination of growth on lactose can be complicated by the presence of multiple β-galactosidases (7, 16, 25) because not all of the β-galactosidases may be participating equally (or at all) as lactases in vivo. Studies comparing the inductions or activities of different β-galactosidases in response to lactose within the same microorganism detected higher responses from a β-galactosidase other than a GHF 42 enzyme (16, 25, 28-30, 33). Lactose is thus not likely the substrate for these enzymes, and although we found reports of β-galactosidases (belonging to GHF 35) active on other substrates (plant and bacterial exo-β-1,3-galactanases [31, 35, 57, 66]), a plant exo-β-1,4-galactanase active on pectic galactan (55), and an archaean exo-β-d-glucosaminidase (56), these substrates were all too large to be directly accessible by the intracellular GHF 42 β-galactosidases. This suggests the existence of at least one unknown substrate for the GHF 42 enzymes.
We used a bioinformatic approach to search for clues regarding GHF 42 functions. We extensively analyzed the GHF 42 phylogenies and their adjacent genes from sequence databases and identified a conserved gene association, or gene synteny, suggesting a function in the degradation of a pectic plant polysaccharide, arabinogalactan type I. The presence of genes homologous to those encoding maltodextrin transporters suggested that oligosaccharide products resulting from the activity of an extracellular enzyme were being imported as substrates for the intracellular β-galactosidases. An example of this gene arrangement is found in Bacillus subtilis surrounding lacA (BSU34130) (12, 14), one of two nonadjacent GHF 42-encoding genes (the other being yesZ [BSU07080] [70]). We therefore used B. subtilis as a model to explore our hypothesized function. We first confirmed an increase in B. subtilis β-galactosidase activity in the presence of galactan. We then cloned the relevant genes from B. subtilis and examined (i) the effects of the heterologously expressed proteins on the physiology of E. coli, (ii) the consequences of interrupting these genes in B. subtilis, and, (iii) by using thin-layer chromatography (TLC), the effects of the enzyme activities in vitro. A recently published biochemical study also substantiates the ability of a GHF 42 enzyme to hydrolyze galactooligomers in vitro (23). In this way, we provide evidence that the GHF 42 enzyme from B. subtilis, LacA, likely acts as a β-1,4-galactooligomerase in vivo, contributing to the utilization of the galactan backbone of arabinogalactan type I. The presence of β-galactosidases encoded adjacent to GHF 53 genes in other organisms suggests that this function is not unique to LacA of B. subtilis. This information supports a possible environmental role, such as plant polysaccharide degradation, for these enzymes.
MATERIALS AND METHODS
Analyses of genomes and relevant phylogenies.
The genes and nucleotide sequences near GHF 42 genes in genomes (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and from smaller sequencing projects (Carbohydrate Active EnZYmes database [8]) were examined, and when possible, at least three open reading frames (ORFs) up- and downstream of the GHF 42 gene were analyzed. The orientation and size of the ORFs were also considered. The theoretical amino acid sequences were used to search the Conserved Domain Database of the National Center of Biotechnology Information (NCBI) by using CDsearch (40) in parallel with searches of the NCBI protein database (http://www.ncbi.nlm.nih.gov) by using BLAST (2). Frequently, significant alignments with conserved domains were detected (in the pfam database [3] and/or clusters of orthologous groups database [59, 60]), and the matches with the lowest E values were recorded. If neither of these was identified or if the alignment did not match a majority of the conserved domain, BLAST results and the general trend of putative function were recorded instead. Different colors were assigned to genes with different functions, producing color-coded alignment diagrams whereby conserved patterns could more easily be discerned. The N-terminal sequences of each GHF 42 were checked using the Signalp server (4) to detect signal sequences. The conceptual translations of GHF 42 and GHF 53 genes were initially aligned using Clustal W in BioEdit (17, 18, 61) and visually inspected to correct errors. The aligned sequences were trimmed to avoid bias in the N- and C-terminal regions and covered regions homologous to BSU34130 (from amino acid positions 16 to 684 for GHF 42) and BSU34120 (from positions 53 to 337 for GHF 53). Bootstrapped neighbor-joining phylogenetic trees were produced using MEGA (37).
Initial screen for increased β-galactosidase activity.
B. subtilis strain PLBS338 is a prototrophic derivative of 168 (67). Cells of this strain, hereforth referred to as wild type, were screened for increased β-galactosidase activity by using a homogenous solution of cells washed in M9 minimal medium (43) containing no carbon to inoculate a semiquantitative screen using 48-well plates containing Spizizen's minimal medium (19) containing 0.2% of a sugar (glucose, galactose, ribose, xylose, mannose, fucose, fructose, sorbose, rhamnose, sucrose, trehalose, salicin, cellobiose, melibiose, maltose, raffinose, lactose, sorbitol, gentiobiose, N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, d-galacturonic acid, d-glucuronic acid, lyxose, or lactobionic acid) or 0.2% of polysaccharide (cellulose, dextran, dextran sulfate, starch, inulin, glycogen, xanthan gum, laminarin, locust gum, chitin, arabinogalactan type II [from larch], gum arabic, polygalacturonic acid, apple pectin, citrus pectin, phytone, xylan, gellan gum, soy flour, or galactan [arabinogalactan type I from lupin from which most of the arabinose has been removed {41}]). After 24 h at 30°C, ONPG was added to 2.2 mM and the plates returned to 30°C for 24 h before 0.5 M Na2CO3 was added to stop the reaction and intensify the color. The upper half of the liquid was removed from 96-well plates to avoid precipitates and read in a Molecular Devices Thermo Max microplate reader at 420 nm at room temperature. The results were interpreted as no effect (A420, <0.7), modest increase (A420, >0.7), and clear increase (A420, ≥1.5).
Construction of vectors.
A library was created in E. coli to obtain 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-hydrolyzing transformants expected to carry one of the two β-galactosidase genes (both GHF 42) of B. subtilis, either the yesZ gene or a portion of the yvf gene region, including the genes yvfN, historically renamed lacA, and yvfO, which we renamed galA. Genomic DNA was harvested from lysozyme-treated B. subtilis cells using the Puregene genomic DNA isolation kit (Gentra Systems, Inc.) (with the modification of lysis at 85°C for 10 min) and restricted with BglII, and the pΔα18 plasmid (62) DNA (lacZ−, Ampr) with BamHI, and genomic fragments of 6.6 to 9.5 kb were purified using gel extraction. The restricted plasmid was treated with calf-intestinal phosphatase, and the genomic DNA fragments and vector were ligated (Fast-Link; Epicentre). These constructs were used to transform E. coli ER2585F′ (lacY+ lacZ thi Tetr) treated with the Z competent kit (Zymo Research). This host-vector combination does not produce the native β-galactosidase (LacZ) of E. coli. The resulting genomic library was screened for X-Gal (0.1 mg ml−1) hydrolysis on LB (Luria-Bertani) with ampicillin (0.1 mg ml−1) and IPTG (isopropyl-β-d-thiogalactoside) (100 μM) at 37°C to detect β-galactosidases. Other media used the same concentrations of X-Gal, ampicillin, and IPTG. The transformants pYes and pYvf were subcloned, yielding constructs pYess and pYvfs, respectively. The pYvfs plasmid was subcloned to yield pLacA and pGalA. Plasmids expressing both GalA (pGalA) and either YesZ (pYesZsG) or LacZ (E. coli) were also created, yielding pGalAYesZsG and pGalALacZ. The lacZ gene was also placed into pΔα18 (pLacZ). The lacZ gene was from a pET-28a(+) construct (6), and an XbaI site was created in a noncoding 5′ region using the QuikChange site-directed mutagenesis kit (Stratagene) to obtain constructs without a six-His tag (Table 1).
TABLE 1.
Primers used in this study
| Primer name | 5′ to 3′ primer pair sequencesa | Purpose |
|---|---|---|
| TAX | GGCACAATGACTGGGAAAG | Detection of Cmr insertion in lacA and in galA |
| GCACCTTCACAGCTTGTTTC | ||
| YES | CGACCGTGGAATATGAAC | Detection of Cmr insertion in yesZ |
| GCTGTCCTGATAACCATTTG | ||
| ZX | CAGCAGCGGTCTAGAGCCGCGCGGCAGC | “QuikChange” addition of XbaI site (for lacZ use) |
| GCTGCCGCGCGGCTCTAGACCGCTGCTG | ||
| LACM | GCTAAGAGAACAAGGAGGAGACATATGATGTCAAAGCTTG | “QuikChange” addition of NdeI site for pETLacM |
| CAAGCTTTGACATCATATGTCTCCTCCTTGTTCTCTTAGC | ||
| GALD | CGGAGTCAGGACATATTCCAAAGGATCCCTGAAACAAAAAAATCC | “QuikChange” addition of BamHI site for pETGalD |
| GGATTTTTTTGTTTCAGGGATCCTTTGGAATATGTCCTGACTCCG |
For each primer, the forward primer is on top and the reverse primer is on the bottom.
Studies with E. coli.
The colony sizes of E. coli strains expressing different β-galactosidases with (pYvfs, pGalAYesZsG, and pGalALacZ) or without (pLacA, pGalA, pYesZs, and pLacZ) simultaneous expression of the GalA galactanase were measured after 5 days of growth at 30°C on minimal media (M9 containing ampicillin and vitamins [Basal Medium Eagle vitamin solution]) containing no carbon source or 0.2% galactose, lactose, or galactan, using cells grown on the galactose minimal medium, and washed and resuspended in M9 without carbon as inoculum. The growth was compared to that of E. coli carrying pΔα18 on the same media, with the growth of this strain on galactose used as a positive control and growth on lactose used as a negative control. Within E. coli, GalA was expected to be exported because the Signalp server (4) predicted a signal peptide in the N-terminal sequence even when the gram-negative option was selected, although the predicted cleavage site was slightly different.
Creation and studies of B. subtilis mutants.
To interrupt the genes of interest, the Cmr determinant (chloramphenicol acetyltransferase [CAT] cassette) of pC194 (13) (ptrpBGI-PLK [42]) was blunt-end ligated into lacA, galA, and yesZ using SrfI, PmlI, and SmaI sites, respectively, to create placA::Cmr, pgalA::Cmr, and pyesZ::Cmr. E. coli with placA::Cmr or pyesZ::Cmr constructs was detected by screening for the loss of β-galactosidase activity on LB medium with IPTG and X-Gal. A modified plasmid restriction pattern was used to screen for pgalA::Cmr. These interrupted-gene constructs were transformed into wild-type B. subtilis. Cells were made competent via a two-step transformation procedure (19), omitting the freezing step. Recombinants were selected using chloramphenicol (5 μg ml−1) in Trypticase soy agar without dextrose (Difco). PCRs using ready-to-go PCR beads (GE Healthcare) and primer pairs, where one primer annealed outside of the relevant cloned region (Table 1), were used to confirm correct CAT cassette insertion.
β-Galactosidase activity of mutant and wild-type strains was observed on Schaffer agar (53) containing X-Gal with or without arabinose (0.05%) and with 0.2% galactose or galactan. The ONPG plate assay was repeated using wild-type and mutant strains with glucose, arabinose, xylose, mannose, rhamnose, lactose, starch, cellulose, arabinogalactan type II, gentiobiose, polygalacturonic acid, citrus pectin, apple pectin, phytone, xylan, gellan gum, galactan, and soy flour.
LacA β-galactosidase purification.
Using the QuikChange kit, an NdeI site was formed at the start site of lacA, allowing its insertion and ligation (as an NdeI-SacI fragment) into the vector pET28a+. In E. coli MC1061(DE3) cells, this construct, pETHLacA, allows inducible expression of the LacA protein with an N-terminal six-histidine tag (H-LacA). This strain was grown in terrific broth (58) with 30 μg ml−1 of kanamycin at 37°C to an optical density of 0.4; it was cooled to 18°C, IPTG was added, and it was incubated at 18°C for 15 h. The cell pellet resulting from centrifugation (6,370 × g at 4°C for 11 min) was resuspended (with the inclusion of a Complete EDTA-free protease inhibitor cocktail tablet [Roche]) at 3 ml/g of Za buffer (a modification of Z buffer [43] without β-mercaptoethanol, consisting of 0.1 M phosphate buffer, pH 6.5, with 10 mM KCl and 1 mM MgSO4), passed through a French pressure cell (18,000 lb/in2), and centrifuged (30,996 × g at 4°C for 30 min), and the clarified lysate was applied to nickel-iminodiacetate beads at 4°C. H-LacA was then batch elution purified using centrifugation (1,239 × g at 10°C for 5 min). The material was triple washed with an initial wash (with Zm buffer [Za buffer at pH 7], 300 mM NaCl, 5 mM imidazole) and a secondary wash (with Zm buffer, 300 mM NaCl, 20 mM imidazole) prior to elution (with Zm buffer, 300 mM NaCl, 150 mM imidazole). Eluted aliquots were dialyzed overnight at 4°C in 1 liter of Za buffer, followed by a 2-h dialysis, and used for enzyme characterization.
LacA β-galactosidase characterization.
Specific activity was measured in 1.2 ml of Za buffer with 2.2 mM ONPG incubated at 40°C for 15 min before the reaction was started with 10 μl of diluted purified enzyme. The reaction was stopped after 5 min with 0.5 ml of 0.5 M Na2CO3, and the activity, defined as the release of 1 μmol of o-nitrophenol (ONP) (or p-nitrophenol [PNP]) per min, was measured at 420 nm. Specific activity was expressed as units per milligram of protein. Protein concentrations were determined using the protocol for Bio-Rad (Hercules, CA) protein assay dye reagent. All assays were performed at least in triplicate, and the activity for all characterizations was measured as described above. The thermodependence of activity was assayed between 27 and 60°C for 5 min. Thermostability assays were performed by incubating aliquots of enzyme at 40, 50, 55, or 60°C and removing them at incremental times to assay at 40°C for 5 min. Optimal pH values were determined by assaying in citric acid buffers (0.1 M) (pH 5 to 6) and Za buffer (0.1 M) (pH 6 to 8). The effect of metal ions was examined in a solution of 0.1 M MOPS (morpholinepropanesulfonic acid) (pH 7) with or without either 1 mM MgCl2, MnCl2, CaCl2, or CuCl2 or 10 mM NaCl or KCl. Substrate preference was examined using ONP and PNP substrates (ONPG, PNPG, PNP-α-d-galactopyranoside, PNP-α-l-arabinopyranoside [PNPAp], PNP-α-l-arabinofuranoside, ONP-β-d-glucopyranoside, PNP-β-d-glucopyranoside, PNP-α-d-glucopyranoside, ONP-β-d-fucopyranoside, PNP-β-d-mannopyranoside, and ONP-β-d-xylopyranoside) at 2.2 mM at 40°C after 5 min.
In vitro galactan degradation and thin-layer chromatography.
Similar to pETHLacA construction and H-LacA expression, a BamHI site was placed in pGalA to aid in the construction of pETGalA-H for inducible expression of GalA with a C-terminal six-histidine tag (GalA-H), with E. coli MC1061(DE3) cells transformed, grown, and induced as they were for pETHLacA. The cells were then incubated at 37°C for 4 h, harvested, frozen overnight, and thawed, and a single extraction was performed using CelLytic B bacterial cell lysis extraction reagent (Sigma) (15 ml/g) as per the directions of the manufacturer. Activity (assayed using azo-galactan [Megazyme]) occurred mainly in the clarified lysate under these conditions and was not present in cells without the construct.
The effects of the lysate containing GalA-H and the purified H-LacA were examined singly and in combination by incubating them with galactan (final concentration, 1.5%) at 37°C for 3 h and then freezing them. Concentration was then achieved by lyophilization, followed by resuspension in 50% ethanol and removal of precipitated proteins and intact polysaccharide via centrifugation (3,000 × g, 10 min). The supernatants were applied to a silica gel TLC plate (Fisherband, GelG) alongside controls (galactose, lactose, and stachyose), and a single ascension performed using a solvent system of 1-butanol-acetic acid-water (3:2:2, vol/vol). The plate was sprayed with a solution of 14% (vol/vol) sulfuric acid in ethanol containing 0.07% naphthoresorcinol and incubated at room temperature overnight in order to visualize the products.
RESULTS
Examination of gene arrangements and genomes containing GHF 42 genes.
We undertook a database analysis of GHF 42 β-galactosidase gene arrangements with the hope that potential operons might provide clues to the function(s) of the encoded enzymes. Conceptual translations of ORFs near GHF 42 genes were analyzed, and we diagrammed each gene arrangement using a color scheme for general functions in order to look for recurring patterns. The phylogeny of GHF 42 (not shown) is comprised mainly of two robust divisions, one containing genes mostly from Firmicutes and Actinobacteria and the other consisting of sequences mainly from Proteobacteria and Thermus spp.
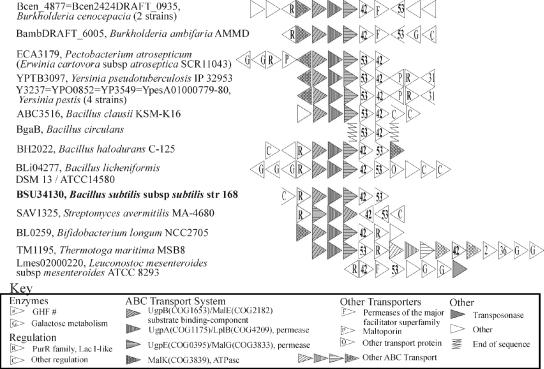
Most interesting was a set of arrangements found in both of these divisions that included COG 3867, arabinogalactan endo-1,4-β-galactosidases belonging to GHF 53 (Fig. 1). These enzymes hydrolyze the β-1,4-galactan backbone of a pectic substance (arabinogalactan type I) found in plants, such as soybeans and citrus fruit, into oligomers. These arrangements also included genes putatively encoding ATP-binding cassette (ABC) transporter subunits with homology to subunits known or predicted to be involved in the transport of maltodextrins, oligosaccharides, or sugars in general. Some arrangements appear to encode complete ABC systems, with encoded ATPases compatible with sugar-transport systems (COG3839 [MalK] genes). Several arrangements also have galactose metabolism genes; this result is expected if galactose is a product of GHF 42 activity. Interestingly, the GHF 42 enzymes that are encoded near a GHF 53 gene are not restricted to a single phylogenetic cluster (not shown). However, a phylogenetic tree of GHF 53 sequences (not shown) showed that the bacterial GHF 53 enzymes associated with GHF 42 genes are more related to each other than to GHF 53 genes without this association. Examining the other GHF 53 arrangements revealed that the GHF 53 genes of Bacteroides thetaiotamicron, Microbulbifer degradans, and some Xanthomonas spp. are adjacent to GHF 2 β-galactosidase genes.
FIG. 1.
Gene arrangements encoding GHF 42 and GHF 53 enzymes. Many arrangements also show ABC transporter genes and transcriptional regulators similar to LacI. Each locus tag refers to the relevant GHF 42 gene.
β-Galactosidase activity in B. subtilis.
We decided to test the effects of galactan on the model organism B. subtilis, which contains two GHF 42 β-galactosidase genes, one of which is adjacent to a GHF 53 homolog. The presence of an in vivo substrate would be expected to increase β-galactosidase activity. Therefore, B. subtilis cultures were screened for increased β-galactosidase activity in the presence of simple sugars and other more complex carbon sources (listed in Materials and Methods), such as galactan, by testing the amount of ONPG hydrolysis. The average background β-galactosidase activity was greater on polysaccharides (A420, 0.40 to 0.70) than on the simple sugars (none of which caused notable increases in β-galactosidase activity). However, soy flour (a known source of arabinogalactan type I) and galactan clearly increased β-galactosidase activity (A420, ≥1.5) above the polysaccharide control (cellulose). The ONPG hydrolysis was above background levels but less than that observed for soy flour or galactan with gum arabic, PGA, apple and citrus pectin, phytone, xylan, and gellan gum (A420, 0.71 to 1.35).
Construction of vectors and knockouts.
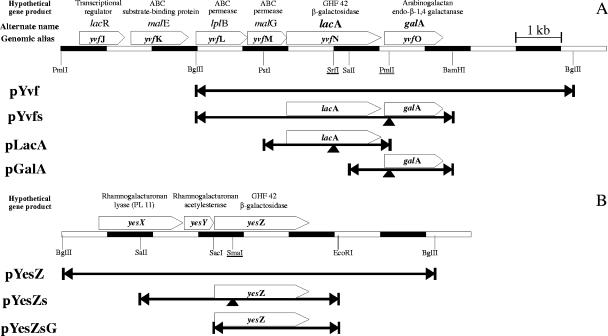
Because B. subtilis has two β-galactosidase genes (lacA and yesZ), we examined whether the expression of one or both was responsible for the increased ONPG hydrolysis. Using the B. subtilis genome sequence to determine appropriate restriction sites, genomic DNA from B. subtilis was used to construct libraries in E. coli. The X-Gal-hydrolyzing transformants were pYvf, which encoded LacA (the GHF 42 β-galactosidase) and GalA (the GHF 53 galactanase), or pYesZ, which encoded YesZ (the second GHF 42 β-galactosidase). These constructs were used to create subclones (pLacA, pGalA, and pYesZs) (Fig. 2). Then the constructs placA::Cmr, pgalA::Cmr, and pyesZ::Cmr, which carry the CAT cassette interrupting the lacA, galA, and yesZ genes, were made. In E. coli, the disruption of the plasmid-borne lacA or yesZ genes yielded colonies that did not hydrolyze X-Gal. The disruption of galA alone (in pYvfs) did not affect X-Gal hydrolysis by the intact preceding lacA gene. E. coli expressing GalA alone did not hydrolyze X-Gal.
FIG. 2.
Plasmid inserts from B. subtilis genome. Fragments of the B. subtilis genome encoding LacA (A) and YesZ (B) were obtained by the creation of genomic libraries. The fragments were subcloned using the restriction endonuclease sites indicated. Other sites (underlined) were used to insert CAT cassettes (at arrows) in order to disrupt the genes.
Physiological effects on E. coli.
The initial gene cloning allowed us to determine whether the expression of GalA, LacA, and/or YesZ could affect E. coli physiology, specifically its growth on lactose or galactan as a sole carbon source. The E. coli strain ER2585F′, which contains a chromosomal deletion of lacZ, did not grow on galactan and hydrolyzed X-Gal only when it expressed a cloned gene encoding LacA, YesZ, or LacZ (Table 2). The GHF 42 β-galactosidases encoded by lacA (pLacA) and yesZ (pGalAYesZg) were each expressed in (lacZ−) E. coli, but unlike the E. coli expressing LacZ, they were unable to immediately grow on lactose minimal medium (Table 2).
TABLE 2.
Physiological effects of lacZ, lacA, galA, and yesZ expression in E. coli
| Construct | Enzyme expressiona
|
Hydrolysis or growthb
|
||||||
|---|---|---|---|---|---|---|---|---|
| LacZ | YesZ | LacA | GalA | X-Gal | Galactose | Lactose | Galactan | |
| pΔα18 | − | − | − | − | − | + | − | − |
| PLacZ | + | − | − | + | + | + | − | |
| PLacA | − | − | + | − | + | + | − | − |
| PGalA | − | − | − | + | − | + | − | − |
| pYvfs (pGalALacA) | − | − | + | + | + | + | − | + |
| PGalAYesZg | − | + | − | + | + | + | − | − |
| pGalALacZ | + | − | − | + | + | + | + | − |
+, gene for enzyme present; −, gene for enzyme absent.
+, hydrolysis detected (hydrolysis of X-Gal determined by formation of blue product and hydrolysis of other compounds determined by growth on M9 media). −, no hydrolysis detected.
Of greater interest for testing our hypothesis was determining whether the expression of both GalA and a β-galactosidase (LacA, YesZ, or LacZ) would confer on E. coli the ability to grow on galactan minimal medium. The only E. coli strain that exhibited growth on galactan equivalent to that seen on the positive controls (galactose) was the one expressing the construct carrying both the galactanase gene (galA) and lacA (Table 2).
Creation of B. subtilis mutants and comparison with the wild type.
The transformation of B. subtilis with placA::Cmr, pgalA::Cmr, or pyesZ::Cmr led to Cmr colonies, indicating that marker replacement, double-crossover recombination had occurred as none of these vectors can replicate within B. subtilis. PCR amplification of genomic DNA from these recombinants yielded products larger (than that from the wild type) by 1.4 kb, indicating the presence of the Cmr cassette and the disruption of the genes of interest (data not shown).
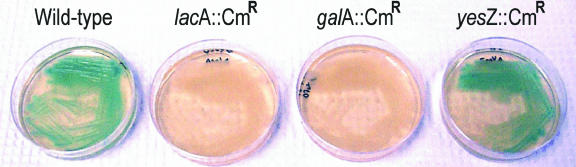
Wild-type B. subtilis is unable to grow on galactose as a sole carbon source because it relies on the arabinose transporter to import this sugar (36). Also, in its natural state, arabinogalactan possesses arabinose-containing side chains. Hence, arabinose might help regulate relevant transporters, other glycoside hydrolases, or interact with other relevant regulatory proteins. For this reason, arabinose was used in combination with other sugars to determine whether it affected β-galactosidase expression. The interruption of lacA, but not of yesZ, prevented increased activity on galactan, and a functional galA gene was required for the expression of LacA (Table 3; Fig. 3). The addition of arabinose enhanced the intensity of the blue color observed from both the wild-type and the mutant strains in the presence of galactan, but when arabinose was used alone or in combination with galactose, it did not cause increased β-galactosidase activity in wild-type colonies.
TABLE 3.
B. subtilis hydrolysis of X-Gal in the presence of various substratesa
| Construct | Arabinose | Arabinose + galactose | Galactan | Arabinose + galactan |
|---|---|---|---|---|
| Wild type | − | − | + | ++ |
| lacA::Cmr | − | NT | − | − |
| yesZ::Cmr | − | NT | + | ++ |
| galA::Cmr | − | NT | − | − |
+, hydrolysis of X-Gal detected; −, no visible hydrolysis of X-Gal; NT, not tested.
FIG. 3.
Effect of mutations on B. subtilis β-galactosidase expression. The increased β-galactosidase expression by wild-type cells when galactan is present as observed by X-Gal hydrolysis is eliminated where the lacA or galA gene has been interrupted by the insertion of the CAT cassette. Interruption of the yesZ gene had no effect on X-Gal hydrolysis.
To further observe the β-galactosidase expression of the mutants, the 48-well ONPG plate assay was repeated using a reduced number of substrates. The interruption of lacA reduced the β-galactosidase activity observed with polygalacturonic acid, citrus and apple pectins, galactan, and soy flour to a level observed with controls. An equivalent decrease in activity was not detected for the lacA::Cmr or yesZ::Cmr mutants on phytone, xylan, and gellan gum.
LacA β-galactosidase purification and characterization.
LacA is the first GHF 42 enzyme known to be encoded adjacent to a GHF 53 gene to be characterized. Enzyme from clarified lysate was initially examined for comparison (data not shown). Then H-LacA was purified using nickel-charged iminodiacetate column material. The purified protein had a specific activity of 51 units/mg. H-LacA had the highest activity at pH 6.0 to 6.5 and 50°C, and it was thermolabile above this temperature. These results agree with those for untagged LacA (not shown). None of the tested metal ions increased enzyme activity when added, and Zn2+, Co2+, and Cu2+ were inhibitory. Activity on PNPG was equal to that on ONPG. Trace activity was observed with ONP-fucose and PNPAp (Table 4), but activity with other substrates was not detectable.
TABLE 4.
Relative activity of the purified LacA enzyme on various chromogenic substrates
| Substrate | % Relative activitya |
|---|---|
| ONPG | 100 |
| PNPG | 104 |
| p-Nitrophenyl-α-l-arabinopyranoside | 4 |
| o-Nitrophenyl-β-d-fucopyranoside | 4 |
| p-Nitrophenyl-α-d-galactopyranoside | <0.1 |
| p-Nitrophenyl-β-d-glucopyranoside (PNPGlu) | <0.1 |
| o-Nitrophenyl-β-d-xylopyranoside | <0.1 |
| p-Nitrophenyl-β-d-mannopyranoside | <0.1 |
| p-Nitrophenyl-α-l-arabinofuranoside | <0.1 |
Activity on ONPG was taken as 100% and corresponds to a specific activity of 46 U/mg. Results are as measured by ONP or PNP release at 40°C.
Thin-layer chromatography.
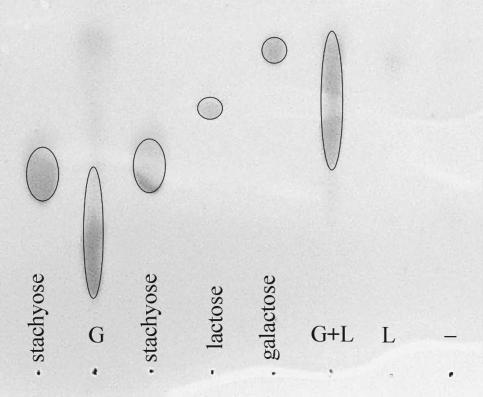
We assayed the ability of the LacA and GalA enzymes to degrade galactan and examined the reaction products using TLC. The sizes of these products were compared to those of a mono-, di-, and tetrasaccharide (galactose, lactose, and stachyose, respectively). Intact galactan (untreated or undegraded) was not very soluble under the conditions used and was thus not visible on the plate. Galactan treated with GalA-H yielded oligosaccharide products, but only treatment with both GalA and LacA resulted in significant release of monosaccharide-sized products, presumably galactose, as well as other small oligosaccharides (Fig. 4). Neither the control without added enzyme nor the treatment with LacA alone showed any significant galactan degradation.
FIG. 4.
Enzymatic degradation of galactan in vitro. Preparations containing either GalA-H (G) or H-LacA (L) were incubated separately or concurrently (G+L) with galactan for 3 h at 37°C, and the products were analyzed by TLC after precipitation and removal of the remaining whole polysaccharide. A negative control treatment using no enzymes (−) was also examined. The amount of product applied to the TLC from the G+L reaction was half that applied from the others (−, L, and G) in order to prevent overloading. Migration patterns of the products were compared to those for stachyose, lactose, and galactose.
DISCUSSION
Initial analyses.
An in-depth examination of the phylogeny of GHF 42 suggested that there might be more than one function for this group, with many observed gene arrangements and many organisms possessing two or more nonidentical GHF 42 genes. One frequent arrangement led us to hypothesize that arabinogalactan type I was a substrate for this system, with oligosaccharides released by the action of β-galactanases as the substrates for the GHF 42 enzymes. The presence in some arrangements of nearby galactose metabolism genes for further processing of this sugar and the increased likelihood of finding arabinogalactan type I from plant material in the native environment of microorganisms like B. subtilis were also supportive of this hypothesis.
We looked for previous examinations of GHF 42 activity with β-galactans and found two examples demonstrating a lack of activity on whole polysaccharide (26, 34), as expected for intracellular enzymes. In at least one of these examples, arabinogalactan type II was used (34). Arabinogalactan type II has a backbone-linked β-1,3, rather than a β-1,4 like arabinogalactan type I, indicating the importance in distinguishing between them. We also looked for evidence of our hypothesized function in reference to studies of galactanases. Increased β-galactanase and β-galactosidase activities in response to galactan (but not the specific genes) in B. subtilis were identified by the work of Labavitch et al. (38), but the degradation of the galactanase products was not studied further. Later, Errington and Vogt noticed the potential for a polycistronic operon in B. subtilis (15), with the same organization we have now observed in several other genomes. It was then shown that lacA (genome annotation, yvfN) is regulated by the nearby repressor lacR, although the function of the system was unclear as it was not activated by simple sugars (9).
The work of those authors suggested an unexplored functional association between LacA and GalA. Several endo-β-1,4 galactanases from Bacillus spp. yield galactotetramer and galactotrimer products (38, 51, 63, 69) which are more likely to be hydrolyzed by GHF 42 than whole polysaccharide. In addition, the solved structure for GHF 42 (from Thermus thermophilus A4) shows, in its monomeric form, a cleft-type active site that could be suitable for the degradation of an oligosaccharidic substrate (22). This enzyme can also form a trimer with a pocket-type active site suitable for the hydrolysis of smaller substrates such as lactose (22) or galactobiose. This supports the possibility that other GHF 42 enzymes, such as the LacA enzyme from B. subtilis, could accommodate both larger substrates like galactooligomers and the smaller galactobiose molecule, assuming that other GHF 42 structures are similar to that found for T. thermophilus sp. A4.
Therefore, we chose B. subtilis, which also has the advantages of a well-studied and easily manipulated genetic system, a sequenced genome, a lack of β-galactosidases in other GHFs, and an inability to use lactose as a sole carbon source, for testing the potential for the involvement of a β-galactosidase in the degradation of galactan.
Experiments involving genes from B. subtilis.
Using B. subtilis as a model, we wanted to show that arabinogalactan type I degradation is a likely role for the gene synteny we found, with GHF 42 enzymes having the specific role of degrading the oligosaccharides produced by the GHF 53 enzymes seen encoded in the arrangements. We showed that, unlike lactose, adding arabinogalactan type I (but not type II) to a medium caused increased β-galactosidase expression in, and allowed the growth of, B. subtilis. Using B. subtilis mutants with interrupted genes, we then demonstrated that the observed response was specific to the gene arrangement. LacA (GHF 42) (and not YesZ) was responsible for the observed β-galactosidase activity, and GalA (GHF 53) was also necessary for the increase in activity to occur. The lack of an effect of galA interruption on LacA expression in E. coli constructs (as observed by X-Gal hydrolysis) suggests that the loss of LacA expression in B. subtilis by the interruption of GalA is not a polar effect.
We also demonstrated the ability of these enzymes to confer a new physiological trait on E. coli. Only LacA in combination with GalA allowed E. coli to grow on galactan, indicating cooperative activity on the substrate to yield galactose. Also, neither YesZ nor LacA effectively allowed E. coli to grow on lactose, further supporting the hypothesis that this is not the natural substrate of either. Controls with E. coli expressing GalA together with β-galactosidases other than LacA (i.e., YesZ or LacZ) did not grow on galactan, showing that these are not functional substitutes and that not all β-galactosidases are galactooligomerases. Additionally, the work of Nakano et al. (46) indicates a failure of LacZ to hydrolyze galactooligomers even in vitro.
In our in vitro TLC experiments, we compared changes in the sizes of products yielded by the enzymes from the arabinogalactan type I to controls of different sizes. The galactanase yielded oligosaccharides larger than lactose, but when combined with the β-galactosidase, some of the products were monosaccharide sized. Further experiments were limited by the difficulty in finding a commercial source of larger β-1,4-galactooligomers. Because of this limitation and our interest in comparing our results with other research, our preliminary biochemical characterization of LacA used standard NP sugars. The characterizations of other GHF 42 enzymes, none of which are encoded adjacent to a GHF 53 gene, also used NP sugars, allowing us to determine that the B. subtilis LacA does not differ significantly.
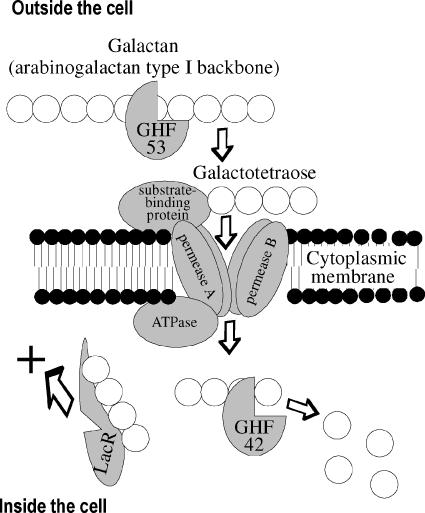
Our observations, including the failure of the combination of arabinose and galactose to increase β-galactosidase activity, combined with the requirement for GalA for β-galactosidase expression lead us to propose the following pathway (Fig. 5), where galactooligomers may be the natural inducers that release the repressor LacR. Although the substrate we used had most of the arabinose side chains associated with arabinogalactan type I removed, B. subtilis cells that degrade the native form of this substrate would probably also use arabinose as a carbon source. If the side chains do not interfere with the activity of GalA, then mixed oligomers may be transported into the cell, where they are acted on by both α-l-arabinofuranosidases and β-galactosidases.
FIG. 5.
Hypothesized functionality indicated by GHF 42/GHF 53 gene arrangements. Starting outside the cell, the GHF 53 galactanase releases oligomers, such as galactotetraose, from galactan. These enter the cell using an ABC transporter, which consists of several subunits. Once within the cell, the oligomers release repression (LacR) of the operon and are degraded by the GHF 42 β-galactosidase, releasing galactose.
The source of galactan (plant cell walls that also contain many other compounds) and the ability of B. subtilis to use a wide variety of polysaccharides make it unlikely that this substance is available as the sole carbon source in the environment. However, that GalA and LacA can (based on our TLC and E. coli experiments) degrade galactan and that the gene expression appears to be induced by its presence led us to believe that they are important in this process. Given the versatile nature of B. subtilis and the complexity of plant polysaccharides, it is also likely that these enzymes can contribute to the degradation of additional galactosidic polysaccharides in the environment. However, given that LacA does not function as a lactase, we propose that LacA be renamed to GalO to better acknowledge its function in oligosaccharide degradation.
Other aspects of B. subtilis and beyond.
The combination of these results, especially the gain of function shown in E. coli, indicates that LacA and GalA of B. subtilis can degrade galactan. It is unknown what specific or nonspecific transport system E. coli is using to import galactan oligomers, but the function of the ABC transporter genes (yvfK, yvfL, and yvfM) in B. subtilis is strongly implied by our work, contrasting the study of Kamionka and Dahl (32), who suggested that these proteins were irrelevant to the function of the β-galactosidase. The participation of a B. subtilis ATPase is also implied by our work. The MalK homolog YurJ (BSU32550) (as predicted by Quentin et al. [49]) or MsmX (BSU38810) may fulfill this role, much like MsmX of Streptococcus mutans (50) contributes to the transport of multiple substrates. These are the first results clearly supporting an in vivo function for a GHF 42 β-galactosidase and also suggest possible functions for adjacently encoded proteins. Similar gene arrangements in other organisms suggest that this is not an isolated example.
Additional examples of this functional arrangement (with or without the gene arrangement we observed) likely exist. It is also possible that a galactanase gene exists elsewhere in a given genome. A relationship between a β-galactosidase without lactase activity and a galactanase encoded elsewhere in the genome was recently proposed by Hinz et al. (24) based on galactooligomerase activity in the GHF 42 enzyme BgalII (Bifidobacterium adolescentis) (23). Although (inefficient) growth by Bifidobacterium longum on galactan has been observed in conjunction with increased β-galactosidase activity (10), the genes encoding the observed activity were not identified (B. longum has a GHF 2 gene as well as two GHF 42 genes) nor was the β-galactosidase activity shown to be necessary for growth. A comparable reaction may also be occurring in some fungi. In comparison to bacterial galactanases, most fungal galactanases (5, 11, 39, 45, 52, 68) yield β-1-4-galactobiose and some fungi have β-galactosidases with reasonable Km values (4.5 to 19.4 mM) on galactobiose (46). The association between GHF 53 and GHF 2 genes in a few bacteria also supports possible GHF 53-GHF 2 synergy.
As mentioned earlier, the GHF 42 enzymes may have more than one function. YesZ in B. subtilis is clearly not expressed under the same conditions as LacA and is only 43% homologous to LacA. The similarity of the hypothetical products of genes preceding yesZ to enzymes acting on rhamnogalacturonan (Fig. 2) suggests that this substrate may be relevant to the function of YesZ. Perhaps this GHF 42 acts on galactose-containing side chains or on a poly- or oligosaccharide that frequently coexists with rhamnogalacturonan. Other possible functions for GHF 42 β-galactosidases might be activity on aryl-galactosides, such as galactosylated anthocyanins.
Supplementary Material
Acknowledgments
This research was supported by Department of Energy grant DE-FG02-93ER20117, NSF grant MO-0347475, and Penn State Astrobiology Center NASA-Ames Cooperative Agreement no. NCC2-1057. Stephanie Shipkowski was partially supported by grant NSF/IGERT DGE-9972759 from the Biogeochemical Research Initiative for Education.
We thank Paul Babitzke and Carol Baker for cell and plasmid stocks and advice and Mary Ann Bruns for helpful discussions.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Adler, B., and S. Faine. 2002. The genus Leptospira. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.9. Springer-Verlag, New York, N.Y.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Christgau, S., T. Sandal, L. V. Kofod, and H. Dalboge. 1995. Expression cloning, purification and characterization of a beta-1,4-galactanase from Aspergillus aculeatus. Curr. Genet. 27:135-141. [DOI] [PubMed] [Google Scholar]
- 6.Coker, J. A., P. P. Sheridan, J. Loveland-Curtze, K. R. Gutshall, A. J. Auman, and J. E. Brenchley. 2003. Biochemical characterization of a beta-galactosidase with a low temperature optimum obtained from an Antarctic Arthrobacter isolate. J. Bacteriol. 185:5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coombs, J., and J. E. Brenchley. 2001. Characterization of two new glycosyl hydrolases from the lactic acid bacterium Carnobacterium piscicola strain BA. Appl. Environ. Microbiol. 67:5094-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 9.Daniel, R. A., J. Haiech, F. Denizot, and J. Errington. 1997. Isolation and characterization of the lacA gene encoding beta-galactosidase in Bacillus subtilis and a regulator gene, lacR. J. Bacteriol. 179:5636-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degnan, B. A., and G. T. Macfarlane. 1995. Arabinogalactan utilization in continuous cultures of Bifidobacterium longum: effect of co-culture with Bacteroides thetaiotamicron. Anaerobe 1:103-112. [DOI] [PubMed] [Google Scholar]
- 11.De Vries, R. P., L. Parenicova, S. W. Hinz, H. C. Kester, G. Beldman, J. A. Benen, and J. Visser. 2002. The beta-1,4-endogalactanase A gene from Aspergillus niger is specifically induced on arabinose and galacturonic acid and plays an important role in the degradation of pectic hairy regions. Eur. J. Biochem. 269:4985-4993. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau, E. J., K. Cabane, and I. Smith. 1987. Regulation of spo0H, an early sporulation gene in bacilli. J. Bacteriol. 169:1182-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrlich, S. D. 1977. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 74:1680-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errington, J., and J. Mandelstam. 1986. Use of a lacZ gene fusion to determine the dependence pattern and the spore compartment expression of sporulation operon spoVA in spo mutants of Bacillus subtilis. J. Gen. Microbiol. 132:2977-2985. [DOI] [PubMed] [Google Scholar]
- 15.Errington, J., and C. H. Vogt. 1990. Isolation and characterization of mutations in the gene encoding an endogenous Bacillus subtilis beta-galactosidase and its regulator. J. Bacteriol. 172:488-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutshall, K. R., D. E. Trimbur, J. J. Kasmir, and J. E. Brenchley. 1995. Analysis of a novel gene and β-galactosidase isozyme from a psychrotrophic Arthrobacter isolate. J. Bacteriol. 177:1981-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. 2001. BioEdit Sequence Alignment Editor, version 5.0.9. North Carolina State University, Raleigh, N.C.
- 18.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons, New York, N.Y.
- 20.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidaka, M., S. Fushinobu, N. Ohtsu, H. Motoshima, H. Matsuzawa, H. Shoun, and T. Wakagi. 2002. Trimeric crystal structure of the glycosyl hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 322:79-91. [DOI] [PubMed] [Google Scholar]
- 23.Hinz, S. W., L. van den Brock, G. Beldman, J. P. Vincken, and A. G. Voragen. 2004. β-Galactosidase from Bifidobacterium adolescentis DSM20083 prefers β(1,4)-galactosides over lactose. Appl. Microbiol. Biotechnol. 66:276-284. [DOI] [PubMed] [Google Scholar]
- 24.Hinz, S. W. A., M. I. Pastink, L. A. M. van den Broek, J.-P. Vincken, and A. G. J. Voragen. 2005. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 71:5501-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirata, H., S. Negoro, and H. Okada. 1984. Molecular basis of isozyme formation of beta-galactosidases in Bacillus stearothermophilus: isolation of two beta-galactosidase genes, bgaA and bgaB. J. Bacteriol. 160:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, M. L., and M. L. Dyall-Smith. 2000. Sequence and expression of a halobacterial beta-galactosidase gene. Mol. Microbiol. 36:114-122. [DOI] [PubMed] [Google Scholar]
- 27.Holmes, M. L., R. K. Scopes, R. L. Moritz, R. J. Simpson, C. Englert, F. Pfeifer, and M. L. Dyall-Smith. 1997. Purification and analysis of an extremely halophilic beta-galactosidase from Haloferax alicantei. Biochim. Biophys. Acta 1337:276-286. [DOI] [PubMed] [Google Scholar]
- 28.Hung, M. N., Z. Xia, N. T. Hu, and B. H. Lee. 2001. Molecular and biochemical analysis of two beta-galactosidases from Bifidobacterium infantis HL96. Appl. Environ. Microbiol. 67:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, M.-N., and B. H. Lee. 1998. Cloning and expression of beta-galactosidase gene from Bifidobacterium infantis into Escherichia coli. Biotechnol. Lett. 20:659-662. [Google Scholar]
- 30.Hung, M.-N., and B. H. Lee. 2002. Purification and characterization of a recombinant beta-galactosidase with transgalactosylation activity from Bifidobacterium infantis HL96. Appl. Microbiol. Biotechnol. 58:439-445. [DOI] [PubMed] [Google Scholar]
- 31.Ito, Y., and T. Sasaki. 1997. Cloning and characterization of the gene encoding a novel β-galactosidase from Bacillus circulans. Biosci. Biotechnol. Biochem. 61:1270-1276. [DOI] [PubMed] [Google Scholar]
- 32.Kamionka, A., and M. K. Dahl. 2001. Bacillus subtilis contains a cyclodextrin-binding protein which is part of a putative ABC-transporter. FEMS Microbiol. Lett. 204:55-60. [DOI] [PubMed] [Google Scholar]
- 33.Kang, S. K., K. K. Cho, J. K. Ahn, J. D. Bok, S. H. Kang, J. H. Woo, H. G. Lee, S. K. You, and Y. J. Choi. 2005. Three forms of thermostable lactose-hydrolase from Thermus sp. IB-21: cloning, expression, and enzyme characterization. J. Biotechnol. 116:337-346. [DOI] [PubMed] [Google Scholar]
- 34.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotake, T., S. Dina, T. Konishi, S. Kaneko, K. Igarashi, M. Samejima, Y. Watanabe, K. Kimura, and Y. Tsumuraya. 2005. Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1→3)- and β-(1→6)-galactosyl residues of arabinogalactan protein. Plant Physiol. 138:1563-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krispin, O., and R. Allmansberger. 1998. The Bacillus subtilis AraE protein displays a broad substrate specificity for several different sugars. J. Bacteriol. 180:3250-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 38.Labavitch, J. M., L. E. Freeman, and P. Albersheim. 1976. Structure of plant cell walls. Purification and characterization of a beta-1,4-galactanase which degrades a structural component of the primary cell walls of dicots. J. Biol. Chem. 251:5904-5910. [PubMed] [Google Scholar]
- 39.Luonteri, E., C. Laine, S. Uusitalo, A. Teleman, M. Siika-aho, and M. Tenkanen. 2003. Purification and characterization of Aspergillus beta-d-galactanases acting on beta-1,4- and beta-1,3/6-linked arabinogalactans. Carbohydr. Polym. 53:155-168. [Google Scholar]
- 40.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Megazyme International. 2002. Galactan (Lupin) (Lot 01001). Megazyme International, Wicklow, Ireland. [Online.] http://www.megazyme.com/downloads/en/data/P-GALLU.pdf.
- 42.Merino, E., P. Babitzke, and C. Yanofsky. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177:6362-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Møller, P. L., F. Jørgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano, H., S. Takenishi, and Y. Watanabe. 1985. Purification and properties of two galactanases from Penicillium citrinum. Agric. Biol. Chem. 49:3445-3454. [Google Scholar]
- 46.Nakano, H., S. Takenishi, and Y. Watanabe. 1987. Substrate specificity of several beta-galactosidases towards a series of beta-1,4-linked galactooligosaccharides. Agric. Biol. Chem. 51:2267-2269. [Google Scholar]
- 47.Ohtsu, N., H. Motoshima, K. Goto, F. Tsukasaki, and H. Matsuzawa. 1998. Thermostable β-galactosidase from an extreme thermophile, Thermus sp. A4: enzyme purification and characterization, and gene cloning and sequencing. Biosci. Biotechnol. Biochem. 62:1539-1545. [DOI] [PubMed] [Google Scholar]
- 48.Phan Trân, L. S., L. Szabo, L. Fulop, L. Orosz, T. Sik, and A. Holczinger. 1998. Isolation of a beta-galactosidase-encoding gene from Bacillus licheniformis: purification and characterization of the recombinant enzyme expressed in Escherichia coli. Curr. Microbiol. 37:39-43. [DOI] [PubMed] [Google Scholar]
- 49.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 50.Russell, R. R. B., A.-O. Joseph, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631-4637. [PubMed] [Google Scholar]
- 51.Ryttersgaard, C., J. Le Nours, L. Lo Leggio, C. T. Jorgensen, L. L. Christensen, M. Bjornvad, and S. Larsen. 2004. The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. J. Mol. Biol. 341:107-117. [DOI] [PubMed] [Google Scholar]
- 52.Ryttersgaard, C., L. Lo Leggio, P. M. Coutinho, B. Henrissat, and S. Larsen. 2002. Aspergillus aculeatus beta-1,4-galactanase: substrate recognition and relations to other glycoside hydrolases in clan GH-A. Biochemistry 41:15135-15143. [DOI] [PubMed] [Google Scholar]
- 53.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheridan, P. S., and J. E. Brenchley. 2000. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic Antarctic Planococcus isolate. Appl. Environ. Microbiol. 66:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, D. L., D. A. Starrett, and K. C. Gross. 1998. A gene coding for tomato fruit beta-galactosidase II is expressed during fruit ripening. Cloning, characterization, and expression pattern. Plant Physiol. 117:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Characterization of an exo-β-d-glucosaminidase involved in a novel chitinolytic pathway from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:5175-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taron, C. H., J. S. Benner, L. J. Hornstra, and E. P. Guthrie. 1995. A novel beta-galactosidase gene isolated from the bacterium Xanthomonas manihotis exhibits strong homology to several eukaryotic beta-galactosidases. Glycobiology 5:603-610. [DOI] [PubMed] [Google Scholar]
- 58.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Focus 9:12. [Google Scholar]
- 59.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trimbur, D. E., K. R. Gutshall, P. Prema, and J. E. Brenchley. 1994. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl. Environ. Microbiol. 60:4544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsumura, K., Y. Hashimoto, T. Akiba, and K. Horikoshi. 1991. Purifications and properties of galactanases from alkalophilic Bacillus sp. S-2 and S-39. Agric. Biol. Chem. 55:1265-1271. [Google Scholar]
- 64.Van Laere, K. M. J., T. Abee, H. A. Schols, G. Beldman, and A. G. J. Voragen. 2000. Characterization of a novel beta-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl. Environ. Microbiol. 66:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vian, A., A. V. Carrascosa, J. L. Garcia, and E. Cortes. 1998. Structure of the beta-galactosidase gene from Thermus sp. strain T2: expression in Escherichia coli and purification in a single step of an active fusion protein. Appl. Environ. Microbiol. 64:2137-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong-Madden, S. T., and D. Landry. 1995. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 5:19-28. [DOI] [PubMed] [Google Scholar]
- 67.Yakhnin, H., H. Zhang, A. V. Yakhnin, and P. Babitzke. 2004. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 186:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamaguchi, F., S. Inoue, and C. Hatanaka. 1995. Purification and properties of endo-beta-1,4-d-galactanase from Aspergillus niger. Biosci. Biotechnol. Biochem. 59:1742-1744. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto, T., and S. Emi. 1988. Arabinogalactanase of Bacillus subtilis var. amylosacchariticus. Methods Enzymol. 160:719-728. [Google Scholar]
- 70.Zagorec, M., and M. Steinmetz. 1991. Construction of a derivative of Tn917 containing an outward-directed promoter and its use in Bacillus subtilis. J. Gen. Microbiol. 137:107-112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.