Abstract
In a lab-scale upflow anaerobic sludge blanket reactor inoculated with granular sludge from a full-scale wastewater treatment plant treating paper mill wastewater, methanethiol (MT) was degraded at 30°C to H2S, CO2, and CH4. At a hydraulic retention time of 9 h, a maximum influent concentration of 6 mM MT was applied, corresponding to a volumetric loading rate of 16.5 mmol liter−1 day−1. The archaeal community within the reactor was characterized by anaerobic culturing and denaturing gradient gel electrophoresis analysis, cloning, and sequencing of 16S rRNA genes and quantitative PCR. Initially, MT-fermenting methanogenic archaea related to members of the genus Methanolobus were enriched in the reactor. Later, they were outcompeted by Methanomethylovorans hollandica, which was detected in aggregates but not inside the granules that originated from the inoculum, the microbial composition of which remained fairly unchanged. Possibly other species within the Methanosarcinacaea also contributed to the fermentation of MT, but they were not enriched by serial dilution in liquid media. The archaeal community within the granules, which was dominated by Methanobacterium beijingense, did not change substantially during the reactor operation. Some of the species related to Methanomethylovorans hollandica were enriched by serial dilutions, but their growth rates were very low. Interestingly, the enrichments could be sustained only in the presence of MT and did not utilize any of the other typical substrates for methylotrophic methanogens, such as methanol, methyl amine, or dimethylsulfide.
Organosulfur compounds present in petroleum and other fossil fuels are receiving considerable attention because of their potential direct and indirect negative effects on the environment. Thiols and H2S, the major (organo)sulfur compounds in liquefied petroleum gas (LPG), are extracted using alkaline solutions. Currently, thiol-containing LPG (mainly methanethiol [MT]) is treated by the Merox process, in which H2S and thiols are extracted from the LPG separately. The thiols are then catalytically oxidized to water-insoluble disulfide oil (33).
Alternatively, H2S and thiols can be extracted simultaneously from LPG, which results in a solvent stream loaded with H2S, MT, and at least 1 M of Na+. The thiols present in the solvent are converted in an anaerobic bioreactor to H2S, CO2, and CH4 (26). In a second reactor, H2S is biologically oxidized to elemental sulfur, a process that has been studied in detail (7, 8) and is already being applied. After separation of the elemental sulfur, the solvent is reused for the extraction process.
Under anaerobic conditions, MT is fermented by methanogens, but oxidation by sulfate reducers has also been reported (15, 30). Kiene et al. showed that sulfate reducers compete with methanogens for MT and dimethylsulfide (DMS), but only at concentrations below 10 μM (10). Theoretically, conversion of MT to acetate or to H2, CO2, and H2S, as described for methanol (20, 21), is also possible, but this has never been found for methylated sulfur compounds. Several MT-fermenting methanogens have been isolated from marine, estuarine, and soda lake sediments, and more recently also from freshwater sediments (13, 25). These methanogens were usually isolated on DMS, methylated amines, or methanol, but never on MT. It is assumed that DMS-fermenting methanogens also ferment MT, and this was demonstrated for several species (17). Finster et al. (4) proposed a stoichiometry for the anaerobic conversion of MT, similar to the methanol metabolism of Methanosarcina barkeri (equation 1). The anaerobic degradation of DMS (equation 2) is assumed to proceed via MT as the intermediate (15).
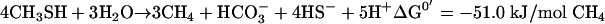 |
(1) |
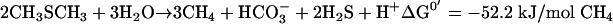 |
(2) |
MT-fermenting methanogens can be used for the treatment of waste streams rich in MT and related volatile organic sulfur compounds (VOSC). In previous studies, the anaerobic degradation of MT by granular sludge was studied in a lab-scale upflow anaerobic sludge blanket (UASB)-type reactor (26, 27). In these studies, however, the microbial diversity and activity of the anaerobic community were not assessed. Here, we present the results of reactor experiments in which we focused on the microbiology of MT degradation. We studied the metabolic properties of the reactor sludge, followed the microbial community in the reactor over time, and enriched MT-degrading microorganisms.
MATERIALS AND METHODS
Lab-scale reactor.
Two continuous-flow reactor experiments were carried out in a laboratory-scale UASB reactor with a liquid volume of 1.6 liters. The reactor was fed with an oxygen-free synthetic influent at pH 12 containing 50 to 100 mM MT (stock solution) mixed with a nutrient solution at pH 4 (see below) using peristaltic pumps (Watson Marlow, Falmouth, Cornwall, United Kingdom). The stock solutions were stored in closed glass bottles in which the volume of the consumed liquid was replaced by N2.
The nutrient solution contained the following macronutrients: 9.35 mM NH4Cl, 0.86 mM K2HPO4, 0.30 mM MgCl2, 1.61 mM KCl, 1.36 mM CaCl2, and 15 mg/liter yeast extract. Trace elements were added from a stock solution (1 ml/liter) according to the method of Paulo et al. (19). An influent containing 2 to 6 mM MT was composed by mixing the MT stock solution with the nutrient solution near the inlet of the reactor.
The superficial liquid upflow velocity in the reactor was maintained at 1.0 m/h for the first 100 days and was then increased to 1.5 m/h to improve the mixing by applying external circulation of the reactor liquid. The influent and circulation flows were 4.2 to 4.8 liters/day and 188 to 283 liters/day, respectively. The hydraulic retention time was 8 to 9 h. The reactor was operated at 30°C using a thermostat bath. The pH of the reactor liquid was measured with an H2S-resistant Flushtrode pH electrode (Hamilton Flushtrode; Hilkomij B. V., Rijswijk, The Netherlands) and controlled between 7.2 and 7.5 by adding sodium hydroxide or hydrochloric acid from 0.1 M stock solutions. Between the two reactor experiments, the reactor was stopped for 69 days. The sludge was kept in the reactor anaerobically at ambient temperature.
Sludge characteristics.
The reactor was inoculated with 96 g volatile suspended solids (VSS) of fresh anaerobic granular sludge (dry weight) from a full-scale anaerobic bioreactor treating paper mill wastewater (Eerbeek, The Netherlands). The VSS content of the sludge was 66% of the dry weight. The granule strength was measured with a tension-and-compression test apparatus (Overload Dynamics S900; Overload Dynamics BV, Schiedam, The Netherlands) according to the method of Hulshoff-Pol (6). Microscopic analysis of sludge and batch samples was performed on an Olympus BH2 epifluorescence microscope. To measure the sludge washout, the concentrations of VSS and total suspended solids (TSS) were determined in effluent samples.
Batch incubations.
To enrich for MT-fermenting methanogens and to study the activity of the reactor sludge, reactor sludge samples were incubated in 120-ml serum flasks filled with 50 ml bicarbonate-buffered medium with 0.1 g/liter yeast extract and a headspace composed of 1.7 bar N2-CO2 (80:20) (2). To localize specific methanogenic activities, the sludge was separated into a suspended and a granular fraction by decanting the suspended fraction of the granules. The granules were washed three times with freshly prepared anaerobic medium. Bromoethane sulfonate was used to inhibit methanogenesis. Substrates and bromoethane sulfonate were added from sterile stock solutions, except for H2, which was added to the batches by flushing the bottles with H2-CO2 (80:20). Methanogenic activities were quantified by measuring the CH4 produced over time. Sterile controls were prepared by sterilizing the batches for 20 min at 120°C after addition of the sludge. The batches were incubated in duplicate at 30°C in the dark.
DNA extraction and 16S rRNA gene amplification.
Samples withdrawn from the reactor were fixed in 60% (vol/vol) ethanol containing 25 mM NaCl. Cells from enrichment cultures were collected by centrifugation at 17,500 × g. DNA was extracted using a FastDNA SPIN kit (for soil) (Q Biogene, Cambridge, United Kingdom) and quantified with a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, Del.). The 16S rRNA genes were amplified from genomic DNA using Taq DNA polymerase (Invitrogen, Breda, The Netherlands) with primers targeting conserved domains (see Table S1 in the supplemental material). The primers were purchased from Eurogentec (San Diego, CA). 16S rRNA genes were amplified using the following thermocycling program; predenaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 40 s, and elongation at 72°C for 90 s; and postelongation at 72°C for 5 min. To amplify bacterial (V6 to V8) and archaeal (V2 to V3) 16S rRNA gene fragments for denaturing gradient gel electrophoresis (DGGE) analysis, the annealing temperature was 56°C and the elongation step was 60 s. The PCR product size was checked by electrophoresis in 1.2% (wt/vol) agarose gels stained with ethidium bromide using a 100-bp DNA ladder (MBI Fermentas, Vilnius, Lithuania). Quantitative real-time PCR amplification was performed with universal primers for bacteria and archaea (28, 35) (see Table S1 in the supplemental material) using the Bio-Rad iQ SYBR green supermix (Bio-Rad Laboratories) according to the manufacturer's instructions. Each of the primer sets was optimized with respect to the annealing temperature and time required for extension. Real-time PCR amplification was performed in a Bio-Rad iCycler programmed for 10 min at 95°C for initial heat activation, followed by 45 cycles of denaturation for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. For archaea, the annealing and extension steps were combined (30 s at 60°C). DNA samples for standard curves were prepared by amplifying group-specific cloned 16S rRNA genes using vector-targeted primers and purifying the products with the Bioké PCR purification kit. DNA standards were quantified with a Nanodrop spectrophotometer.
DGGE.
Denaturing gradient gel electrophoresis was performed in 8% (wt/vol) polyacrylamide (37.5:1 acrylamide-bisacrylamide) gels containing a 30 to 50% or 30 to 60% denaturing gradient for archaeal and bacterial 16S rRNA gene amplicons, respectively. A 100% denaturing solution contained 7 M urea and 40% (vol/vol) formamide. Gelbond (Amersham Biosciences, Little Chalfont, United Kingdom) was used as a physical support. Electrophoresis was performed in 0.5× TAE buffer (0.02 M Tris, 0.01 M acetic acid, and 0.5 mM EDTA, pH 8) at 85 V and 60°C for 16 h using a DCode System (Bio-Rad, Hercules, CA). Silver staining and development of the gels were performed according to the method of Sanguinetti et al. (24).
Cloning and sequencing.
For cloning and sequencing, 16S rRNA gene amplicons were purified with a Bioké PCR purification kit (Leiden, The Netherlands) and cloned into Escherichia coli XL1 blue (Stratagene, Amsterdam, The Netherlands). The pGEM-T Easy vector system (Promega, Madison, WI) was used to transform the 16S rRNA gene amplicons into E. coli and to select for positive clones using ampicillin selection and blue/white screening. The clones were screened by amplified rRNA gene restriction analysis, using the restriction enzymes MspI, CfoI, and AluI (Promega, Madison, WI). The restriction fragments were analyzed by electrophoresis in 12% (wt/vol) agarose gels (Elchrom, Cham, Switzerland) and ethidium bromide staining. Sequence analysis was performed using the pGEM-T vector-targeted sequencing primers Sp6 and T7 and the 16S rRNA gene-targeted internal primers Uni-519f and Arch-915r (see Table S1 in the supplemental material). Phylogenetic trees were constructed in ARB, using the neighbor-joining method (E. coli positions 125 to 1469) and the Felsenstein correction (16, 23). Sequences were aligned with FastAligner, followed by manual alignment according to secondary-structure models.
Analytical techniques.
MT, DMS, dimethyldisulfide (DMDS), and ethanethiol in the reactor effluent were analyzed by high-pressure liquid chromatography (Separations, Hendrik Ido Ambracht, The Netherlands) using a Chrompack (Bergen op Zoom, The Netherlands) C18 column with a length of 20 cm. The oven temperature was 30°C. The composition of the eluent was 35% acetonitrile and 65% water, and the flow rate was 0.6 ml/min. The injection volume of the samples was 20 μl, and a UV detector (Gynotek Germering, Germany) was used to monitor the VOSC at a wavelength of 210 nm. Total and volatile suspended solids were analyzed according to standard methods (1). The reactor biogas composition (CH4, CO2, N2, and H2S) was analyzed on a Packard Becker gas chromatograph, model 433 (Delft, The Netherlands), equipped with two columns connected in parallel (split 1:1): 1.5 m by 1/8 in, Teflon packed with Chromosorb 108 (60 to 80 mesh), and 1.2 m by 1/8 in, stainless steel packed with a molecular sieve of 5 Å (60 to 80 mesh). Helium was used as a carrier gas (45 ml/min). The temperatures were 40°C for the column and 100°C for the injection port and hot-wire detector. The injection volume was 100 μl. The VOSC contents of the headspaces of batch incubations were measured on a Hewlett-Packard CP9000 gas chromatograph equipped with a CP-Porabond Q column (25 m by 0.53 mm) and a flame ionization detector. Volatile compounds were separated on the column in a 1-min linear gradient from 150 to 190°C. For quantitative experiments, 150 μl propane was included in the batches as an internal standard. The total dissolved sulfide was measured photometrically according to the methylene blue method described by Trüper and Schlegel (31). CH4 in the headspaces of batches was measured gas chromatographically with a Shimadzu GC-14B gas chromatograph equipped with a thermal-conductivity detector and molecular sieve 13× (60/80 mesh). The column temperature was 50°C, and the carrier gas was argon at a flow rate of 30 ml/min.
Chemicals.
All chemicals used were of analytical grade. A sodium mercaptide solution was supplied by the Arkema Group (Rotterdam, The Netherlands).
Calculations.
The removal efficiency of the reactor was calculated in two ways (based on the VOSC or H2S concentration in the effluent) according to the following equations:
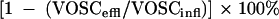 |
(3) |
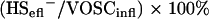 |
(4) |
in which VOSCinfl is the VOSC concentration in the influent (MT and some DMDS produced by auto-oxidation of MT), VOSCeffl is the sum of VOSC in the effluent (MT, ethanethiol [ET], DMS, and DMDS), and HS−effl is the H2S concentration in the effluent. The sludge retention time (τ) is calculated by equation 5:
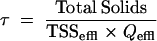 |
(5) |
where Total Solids is the amount of dry sludge in the reactor (g), TSSeffl is the measured total suspended solids in the effluent (g/liter), and Qeffl is the effluent flow (liters/day).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been deposited in GenBank (accession no. DQ631884 to DQ631889).
RESULTS
Degradation of methanethiol in a lab-scale reactor.
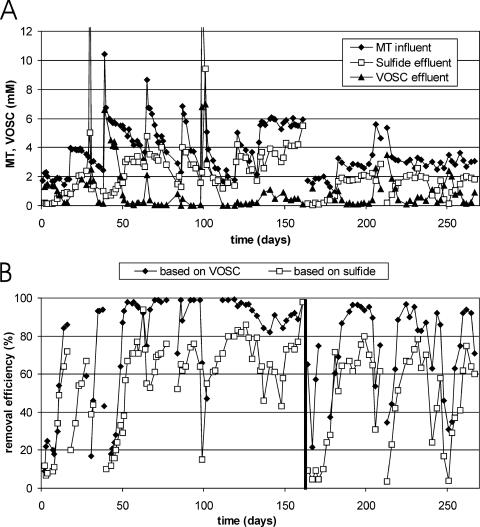
Eight days after startup of the reactor at an influent MT concentration of 2 mM, H2S production started (Fig. 1A). The influent MT concentration was increased to 4 mM at day 16 and to 6 mM at day 39. This corresponded to maximum volumetric and specific loading rates (at 6 mM MT in the influent) of 16.5 mmol MT · liter−1 · day−1 and 0.33 mmol MT · g VSS−1 · day−1. Besides MT, DMDS, DMS (<0.1 mM), and ET (<0.05 mM) were also detected in the effluent, and for this reason the total VOSC concentration was used to determine removal efficiencies. During the first reactor run (0 to 163 days), it was difficult to maintain a constant MT concentration in the influent. MT shock loads disturbed the reactor performance substantially (Fig. 1). For instance, at day 30, due to failure of one of the pumps, the influent MT concentration increased to 20 mM for 20 h. The effluent H2S concentration increased to 5 mM, indicating that degradation of MT continued. The reactor performance recovered within a few days after the influent MT concentration was reduced back to 3 mM. In general, MT shock loads resulted in increased VOSC (mainly MT) and H2S concentrations in the effluent, but the effluent VOSC concentration was always restored (to <0.5 mM) within 10 days after the influent concentration was lowered to 6 mM MT or less. After 163 days of operation, the reactor was stopped and started up again 69 days thereafter. During the second part of the reactor experiment, in which the influent MT concentration was kept more constant, the reactor performance was sometimes strongly affected by exposure to air. At day 209, the reactor had to be stopped and opened for a short time due to clogging problems. After that, it took 10 days before the removal efficiency was restored to the previous level.
FIG. 1.
Performance of the MT-degrading UASB reactor. (A) MT influent, VOSC total (the sum of MT, DMS, and DMDS), and H2S in the effluent. (B) MT removal efficiency. The reactor was stopped after 163 days of operation and restarted 69 days later.
Biogas production and composition.
The CH4 content of the biogas was approximately 60% and was fairly constant throughout both reactor experiments (data not shown). The remaining 40% was N2, as the influents were kept anaerobically under an N2 atmosphere. The CO2 content was around 0.3%, and no H2S was detected in the biogas.
Part of the CH4 dissolved in the reactor liquid and left the reactor via the effluent. This was verified qualitatively by gas chromatography, in which a soluble CH4 peak was identified in effluent samples. The distribution coefficient (m = Cg/Cl [dimensionless], where Cg is the concentration in the gas phase and Cl is the concentration in the liquid phase) of CH4 at 30°C was calculated from different sources (3, 9, 12, 34). The average m value from these sources was 32. Using this distribution coefficient, the ideal gas law, and a CH4 concentration of 60% in the biogas, the estimated CH4 concentration in the effluent was 0.7 mM. For an average effluent flow of 4.4 liters/day, this was equivalent to a CH4 flow of 81 ml/day at 30°C.
A complete conversion of MT to CH4, CO2, and H2S (equation 1) at an MT load of 16.5 mmol MT liter−1 day−1 (MT influent, 6 mM; 4.4 liters/day; reactor volume = 1.6 liters) provides a theoretical CH4 production of 490 ml/day. The average daily CH4 production recorded over the period between days 120 and 160 was 316 ml/day (biogas, 235 ml/day; effluent, 81 ml/day), which accounts for only 65% of the theoretical maximum. The amount of H2S recovered during the period between days 120 and 160 was also between 60% and 80%.
Sludge washout and sludge characteristics.
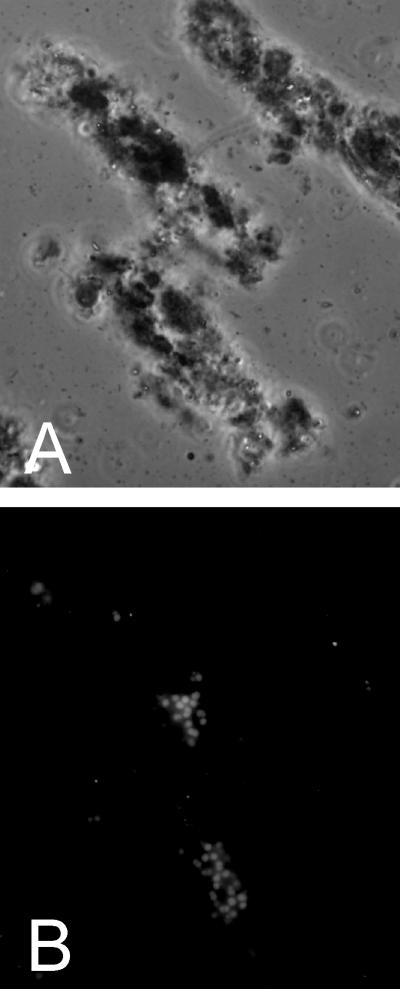
During the second part of the reactor experiment, sludge washout was monitored. It varied between 27 and 165 mg TSS/liter. The average amount of sludge in the reactor was around 110 g total solids. The sludge retention time was estimated at between 153 and 787 days, using the minimal and maximal sludge washouts in equation 5. At the start of the reactor experiment, the granules were black and varied in diameter between 1 and 4 mm. The granule strength of the starting sludge was 3.9 ± 0.1 · 105 N/m2, which decreased by 36% to 2.5 ± 0.1 · 105 N/m2. Microscopic examination of the reactor sludge showed that it was composed of granules and a “suspended” fraction containing aggregates varying in size between 10 and 100 μm. Epifluorescence microscopy revealed mainly long (Methanobacterium-like) rod-shaped methanogens within the granules, while Methanosarcina-shaped cells were detected in the suspended fraction of the sludge embedded in aggregates (Fig. 2). At the end of the reactor experiment, the suspended sludge fraction comprised approximately 5% of the total reactor sludge.
FIG. 2.
Phase-contrast images of the suspended sludge fraction (A) and localization of methanogenic cells in the same preparation by epifluorescence (B).
Sludge activity tests.
To localize the MT-degrading activity in the sludge (immediately after sampling at the end of the reactor experiment), a sludge sample was divided into three fractions: total sludge, a granular sludge fraction, and a suspended sludge fraction. Of each fraction, 0.5 g wet sludge was incubated in 120-ml serum bottles with methanogenic medium and either 1 mM MT, 20 mM acetate, or 1.7 · 105 Pa H2-CO2 (80:20) as a substrate. The batches with the suspended fractions and those with the total sludge clearly degraded MT faster than the batches with the granular sludge fraction only (Table 1). In contrast, the specific CH4 production rate with acetate was significantly lower with the suspended sludge fraction than with the granular fraction, whereas CH4 production levels from H2-CO2 were comparable for all fractions (Table 1).
TABLE 1.
Specific activities of the different sludge fractions from the reactor at the end of the experimenta
| Sludge fraction | mmol MT degraded g VSS−1 day−1 on MT substrate | mmol CH4 produced g VSS−1 day−1 on substrate:
|
|
|---|---|---|---|
| H2-CO2 | Acetate | ||
| Total | 1.34 | 1.68 | 1.35 |
| Granules | 0.134 | 1.35 | 1.41 |
| Suspended | 2.91 | 1.40 | 0.70 |
Measured at 30°C in batch with MT, H2-CO2, or acetate as a substrate. The values are averages of duplicates (sludge).
Enrichment of MT-fermenting methanogenic archaea.
To enrich for the MT-fermenting methanogens, several liquid dilution series were prepared from sludge from the reactor and from batches used for the sludge activity tests. For dilutions from suspended sludge, degradation of MT coupled to CH4 formation was observed up to a dilution of 10−7 (equal to 10−5% inoculum), and for some even up to a dilution of 10−8. Subculturing from these dilutions was possible only with MT concentrations below 2 mM, and the lag phases were up to 1 month, while complete fermentation of MT required up to 2 months. When fresh medium was inoculated from lower positive dilutions, MT was usually completely fermented within 2 weeks. For a dilution series from washed and crushed granules, fermentation of MT was not observed beyond a dilution of 10−6. The enrichment did not metabolize DMS, methanol, acetate, trimethylamine, methylamine, H2, or formate, and it required either yeast extract, peptones, or Casamino Acids for growth. No CH4 was produced in any of the controls or in batches to which no MT was added (endogenous controls). Further attempts to isolate the MT-fermenting microorganisms were not successful.
Community analysis of the sludge samples and enrichment cultures.
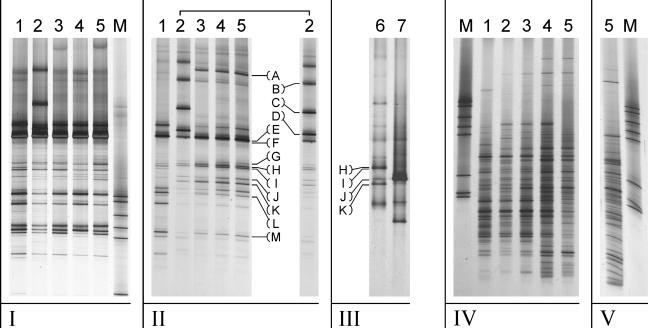
Four sludge samples were taken from the reactor over time and were used to characterize the composition and dynamics of the microbial community. Sludge from the full-scale wastewater treatment reactor that was used to inoculate the lab-scale reactor, and for which the microbial community was characterized in detail in a previous study (22), was also included in the analyses. Since MT was converted to CH4 and H2S in the reactor and batch tests, we mainly focused on the archaeal community of the reactor. Amplification of the V2-V3 region of the archaeal 16S rRNA genes, followed by DGGE analysis of the DNA isolated from each of the samples, did not reveal any substantial changes in the archaeal community. For the sample from day 126, however, some additional amplicons were clearly visible. Exclusion of granules from the DNA isolation procedure revealed that some archaeal phylotypes had become more abundant over time in the suspended sludge fraction (Fig. 3). Some of these amplicons were not visible in the DGGE fingerprint of the original sludge (Fig. 3II, A and G to K), while some others did seem to represent species from the original granular sludge (E, L, and M). Some amplicons were detected only in the sample from day 126 and not in the samples taken thereafter (B, C, and D).
FIG. 3.
DGGE fingerprints showing changes in the microbial community in sludge from the lab-scale UASB reactor fed with MT. (I) Archaeal community profiles of the total reactor sludge (granular plus suspended fractions). (II) Archaeal community profiles of the suspended fraction of the sludge (granules excluded). (III) Archaeal community profiles of enrichment cultures. (IV) Bacterial community profiles of the total reactor sludge. (V) Bacterial community profile of the suspended fraction of the reactor sludge. Lanes: 1, sludge from the full-scale UASB reactor treating paper mill wastewater at Eerbeek, The Netherlands, sampled on 16 January 2003; 2 to 5, samples from the lab-scale reactor fed with MT sampled at 126 days (lane 2), 187 days (lane 3), 248 days (lane 4), and 269 days (lane 5); 6, enrichment culture on MT; 7, enrichment culture on MT plus DMS; M, marker. (A to M) Amplicons for which the identities of the corresponding populations were determined by sequence analysis of the corresponding cloned 16S rRNA genes or of amplicons excised from the gels.
To determine the phylogenetic affiliations of the archaeal populations detected by DGGE analysis, DNA from the suspended sludge fraction on day 269 was used to construct a clone library of archaeal 16S rRNA gene fragments (V2 to V8; primers Arch-109f and Uni-1492r). Sequence analyses of 16S rRNA gene inserts of clones that were selected by DGGE analysis revealed that the majority of archaeal populations detected in the suspended sludge fraction of the reactor over time were most closely related to members of the genus Methanomethylovorans that were closely related to Methanomethylovorans hollandica (Table 2) . Nevertheless, not all amplicons that appeared in the archaeal 16S rRNA gene fingerprints were represented in the clone library. The sequence of the amplicon (Fig. 3 II, band E) that was assumed to represent Methanosaeta concilli based on a previous study (22) appeared to be most closely related to Methanobacterium beijingense, suggesting that the band representing this species might have overlapped with that of Methanosaeta in the fingerprint. Therefore, five additional clones representing this band were selected and analyzed, but those clones also contained Methanobacterium beijingense 16S rRNA gene fragments. To determine the 16S rRNA gene sequences of species that appeared in the reactor but were not present in the clone library, the corresponding amplicons were excised from DGGE gels, reamplified, and subjected to sequence analysis. Using this strategy, we were able to confirm that the dominant amplicon in the DGGE fingerprints indeed represented Methanobacterium beijingense. The amplicons (C and D) representing the archaeal phylotypes that were visible only in the DGGE fingerprint of the sample from day 126 (Fig. 3 II) were both more closely related to Methanolobus taylorii (Table 2). To determine the full 16S rRNA gene sequences of these species, a second clone library (V2 to V8; Arch-109f and Uni-1492r) from the day 126 (suspended sludge) sample was prepared. DGGE analysis showed that both populations of interest were represented by 7 out of 19 clones (Table 2). DGGE analysis of the enrichment cultures revealed that archaeal species that were enriched in the reactor also dominated the enrichments (Fig. 3II and 3III). Moreover, significant differences in the intensities of the dominant bands observed for the enrichments in the absence or presence of DMS suggested that the relative abundances of the different dominant populations differed substantially (Fig. 3II and 3III).
TABLE 2.
Identities of archaeal 16S rRNA genes retrieved from a lab-scale reactor fed with MT and percent similarities to the closest related sequence in the NCBI database
| Positiona | Cloneb | Abundance | Closest related sequences in database (NCBI) | Accession no. | Size (bp) | % Similarity |
|---|---|---|---|---|---|---|
| A | 0/44c | Uncultured archaeon WCHD3-30 (Methanosarcinaceae) | AF050612 | 385 | 87.8 | |
| B | 0/44c | Artifact/hybrid | ||||
| C | VII-A11 | 7/19 | Uncultured archaeon clone PL-7A3 | AY570661 | 1,355 | 98.1 |
| Methanolobus taylorii | U20154 | 1,327 | 96.4 | |||
| D | VII-A7 | 7/19 | Uncultured archaeon clone PL-7A3 | AY570661 | 1,358 | 98.2 |
| Methanolobus taylorii | U20154 | 1,330 | 96.3 | |||
| E | 10/44 | Methanobacterium beijingense strain 8-2 | AY350742 | 384 | 99.7 | |
| F | 1/44 | Methanosaeta concilli | X51423 | 714 | 99.7 | |
| G | 1/44 | Uncultured archaeon clone MP104-1109-a25 | DQ88782 | 560 | 99.6 | |
| Methanobacterium beijingense strain 4-1 | AY552778 | 560 | 98.4 | |||
| H/I | I-B1 | 12/44 | Methanomethylovorans hollandica strain ZB | AY260433 | 1,360 | 98.9 |
| J | I-G4 | 6/44 | Methanomethylovorans hollandica strain ZB | AY260433 | 1,360 | 98.8 |
| K | I-A10 | 3/44 | Methanomethylovorans hollandica strain ZB | AY260433 | 1,361 | 98.5 |
| L | 0/44c | Heterogeneous sequence | ||||
| M | 1/44 | Methanosaeta concilli | X51423 | 719 | 99.9 | |
| N | I-D9 | 1/44 | Methanomethylovorans hollandica strain ZB | 1,363 | 98.4 | |
| N | 6/44d | Methanomethylovorans hollandica | >700 | >98 | ||
| N | 2/44d | Methanobacterium beijingense | >500 | >97 | ||
| N | 1/44 | Uncultured crenarchaeote | AJ576209 | 700 | 97.6 |
A to K, positions of the amplicons in the DGGE gel (Fig. 3); N, not identified in the DGGE gel or position does not correspond to any of the bands in the fingerprint of the reactor sludge.
Codes for sequences that were deposited in GenBank.
Not represented in clone library; 16S rRNA gene fragments were retrieved directly from DGGE gels.
None of these sequences were 100% similar.
DGGE analysis of the bacterial population in the reactor, based on the V6-V8 region amplified from bacterial 16S rRNA genes present in the samples (Bac-968f and Bac-1401r), showed that the majority of the amplicons detected in the inoculum (granules) remained constant during reactor operation (Fig. 3 IV). Some additional amplicons did appear in the DGGE fingerprints, and most of these were also detected in the suspended sludge fraction from the day 269 sample (Fig. 3V), the fingerprint of which was very similar to the corresponding fingerprint of the total sludge.
Quantification of archaea and bacteria in the reactor samples revealed that bacteria dominated the reactor sludge, as well as the suspended fraction of the sludge, even if the differences in the average numbers of rRNA gene operons between bacteria and archaea were taken into account (4.1 for bacteria and 1.5 for archaea). The ratio of both groups in the granules did not change, with 13% (±2.3%) archaea in the inoculum sludge and 15% (±1%) in the total sludge after 269 days. The ratio in the suspended sludge fraction changed in favor of the bacteria, from 33% (±3.6%) archaea after 126 days to 11% (±0.7%) after 269 days.
DISCUSSION
Granular sludge from a full-scale UASB reactor treating paper mill wastewater can be used as the inoculum for a lab-scale UASB reactor to treat MT-containing waste streams, which is in agreement with previously published data (26, 27). During initial experiments in which the reactor was fed with 6 mM MT during startup, no MT was degraded, most likely due to the toxicity of MT (data not shown). MT was shown to inhibit methanogens at concentrations between 6 and 10 mM (32). This also might explain why incidental increases in the influent MT concentration seriously affected reactor performance, requiring up to 10 days for full recovery (e.g., day 209) (Fig. 1). However, sulfide may also have contributed to toxicity effects after shock loads, since the sulfide concentration in the effluent usually increased initially with increased influent MT concentrations (Fig. 1A).
The small amounts of DMS and ET detected in the reactor effluent suggest that methylation of MT, and possibly also other reactions, occurred in the reactor. Sipma et al. have reported DMS formation from MT in an anaerobic bioreactor treating MT (26), but it has also been found in freshwater sediments and chemostat cultures of Methanomethylovorans hollandica (13, 14, 29). Possibly, the MT-fermenting archaea produce DMS from MT themselves by methylation, since MT is also an intermediate of DMS fermentation. However, other microorganisms may also be involved in DMS formation from MT (15, 29). More difficult to explain is the formation of ET, which, unlike MT and DMS, is not a known intermediate of MT degradation. Since ET is coextracted from LPG with MT and H2S, ideally it would be treated in the same step as MT to produce H2S, which then can be oxidized to elemental sulfur in the ultimate step of the process (27). However, batch experiments showed that both ET and PT are not degraded under methanogenic or sulfate-reducing conditions (32).
Granules can be considered spherical biofilms consisting of densely packed anaerobic microbial consortia. Due to their high settling velocities and the ability to withstand hydraulic shear, they are an essential feature of the UASB process (5, 11). In previous studies, it was assumed that the microorganisms responsible for MT fermentation were located in the granules (26, 27). Our results showed that the MT-fermenting archaea were embedded in small aggregates, which varied in size between approximately 10 and 100 μm, but not in the granules (Fig. 2). Initially, a population related to Methanolobus dominated the archaeal community of the suspended sludge fraction (Fig. 3), but they were eventually outcompeted by Methanomethylovorans hollandica, which was represented by five different 16S rRNA gene sequences. Methanomethylovorans hollandica is the only methanogen known to ferment MT and DMS in freshwater environments (13). The original type strain of this genus (Methanomethylovorans hollandica strain DMS1) was isolated on DMS, but like most other DMS-fermenting methanogens, it has also been reported to ferment MT. To our knowledge, methylotrophic methanogens have never been isolated on MT, so our experiments revealed that microorganisms enriched on MT do not represent different genera or species than DMS-fermenting species. However, the MT-fermenting species related to Methanolobus that were initially detected in the reactor represent a novel species, and possibly even a new genus, of freshwater methanogens degrading MT. In contrast, the Methanomethylovorans enrichment obtained by using MT as the sole substrate fermented only MT and not DMS, methylamine, or methanol, compounds which are fermented by all other DMS-fermenting species described so far (15). Our culture, however, produced traces of DMS, which may explain why the reactor was not dominated by one archaeal species. DNA samples of enrichments on MT were dominated by two of the four Methanomethylovorans species found in the reactor. When both MT and DMS were used as substrates in dilution series, one of the other species, which must have been growing on DMS in the reactor as well, dominated. Unfortunately, none of the strains was obtained in pure culture, which would have allowed a more detailed characterization.
Remarkably, the dominant species in the granular sludge was Methanobacterium beijingense, a hydrogenotrophic methanogen, while previous studies reported that species of the genus Methanosaeta dominated that sludge (18, 22). One explanation may be that the microbial community in the Eerbeek full-scale reactor changed between our study and previous studies. However, such a dramatic community change in a full-scale reactor would be rather unusual, especially as the composition of the waste stream did not change. Furthermore, the archaeal 16S rRNA gene-based DGGE fingerprint of the original sludge sample used in our study was remarkably similar to the fingerprint presented by Roest et al. (22), who used the same primer sets for DGGE analyses. A possible explanation for this unexpected discrepancy could be that the procedure previously used for the selection of clones for sequencing by amplified rRNA gene restriction analysis was not able to distinguish any of the Methanomethylovorans species found in the reactor and, in addition, was not suitable for discriminating Methanosaeta and Methanobacterium from Methanomethylovorans (data not shown).
At the end of the reactor experiment, it was estimated that 95% of the biomass was still present in granules. Since the granules were not involved in the degradation of MT, we expect that much higher specific loading rates should be possible, provided that the biomass is retained in the reactor. For instance, the specific activity of suspended sludge in batch (Table 1) was ninefold higher than the maximum specific loading rate calculated for the reactor. Future studies, therefore, should also focus on other types of sludge retention or on stimulating the MT-degrading microorganisms to form granules.
Supplementary Material
Acknowledgments
This work was supported by the Technology Foundation STW, the Applied Science Division of NWO, and the technology program of the Ministry of Economic Affairs. We are thankful to Paques B.V. and Universal Oil Products for their financial support.
We are grateful to Ad Konijnendijk from Arkema Group (Rotterdam, The Netherlands) for providing sodium methyl mercaptide solutions and to Walter Hulshof (Industriewater Eerbeek, The Netherlands) for providing anaerobic granular sludge. We also thank Zhuna for her assistance with the reactor experiments; Wim Roelofsen, Wim van Doesburg, Mahmut Altinbas, and Hans Heilig for technical support; Mark Sturme for assisting with real-time PCR; Diana Sousa and Stan Brouns for providing archaeal genomic-DNA samples; and Kees Roest for critical reading of the manuscript.
Footnotes
Published ahead of print on 29 September 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.American Public Health Association. 1985. Standard methods for the examination of water and wastewater, 16th ed. American Public Health Association, Washington, D.C.
- 2.de Bok, F. A. M., E. H. A. Roze, and A. J. M. Stams. 2002. Hydrogenases and formate dehydrogenases of Syntrophobacter fumaroxidans. Antonie Leeuwenhoek 81:283-291. [DOI] [PubMed] [Google Scholar]
- 3.Dean, J. A. 1992. Lange's handbook of chemistry. Mc Graw-Hill, Inc., New York, N.Y.
- 4.Finster, K., Y. Tanimoto, and F. Bak. 1992. Fermentation of methanethiol and dimethylsulfide by a newly isolated methanogenic bacterium. Arch. Microbiol. 157:425-430. [Google Scholar]
- 5.Forster, C. F., and J. Quarmby. 1995. The physical characteristics of anaerobic granular sludges in relation to their internal architecture. Antonie Leeuwenhoek 67:103-110. [DOI] [PubMed] [Google Scholar]
- 6.Hulshoff-Pol, L. W. 1989. The phenomenon of granulation of anaerobic sludge. Ph.D. thesis. Wageningen Agricultural University, Wageningen, The Netherlands.
- 7.Janssen, A. J. H., S. C. Ma, P. Lens, and G. Lettinga. 1997. Performance of a sulfide-oxidizing expanded-bed reactor supplied with dissolved oxygen. Biotechnol. Bioeng. 53:32-40. [DOI] [PubMed] [Google Scholar]
- 8.Janssen, A. J. H., R. Sleyster, C. van der Kaa, A. Jochemsen, J. Bontsema, and G. Lettinga. 1995. Biological sulfide oxidation in a fed-batch reactor. Biotechnol. Bioeng. 47:327-333. [DOI] [PubMed] [Google Scholar]
- 9.Kavanaugh, M. C., and R. R. Trussell. 1980. Design of aeration towers to strip volatile contaminants from drinking-water. J. Am. Water Works Assoc. 72:684-692. [Google Scholar]
- 10.Kiene, R. P., R. S. Oremland, A. Catena, L. G. Miller, and D. G. Capone. 1986. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl. Environ. Microbiol. 52:1037-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lettinga, G. 1995. Anaerobic digestion and waste-water treatment systems. Antonie Leeuwenhoek 67:3-28. [DOI] [PubMed] [Google Scholar]
- 12.Lide, D. R., and H. P. R. Frederikse. 1995. CRC handbook of chemistry and physics, 76th ed. CRC Press, Inc., Boca Raton, Fla.
- 13.Lomans, B. P., R. Maas, R. Luderer, H. J. M. op den Camp, A. Pol, C. van der Drift, and G. D. Vogels. 1999. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 65:3641-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomans, B. P., H. J. M. Op den Camp, A. Pol, C. van der Drift, and G. D. Vogels. 1999. Role of methanogens and other bacteria in degradation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl. Environ. Microbiol. 65:2116-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomans, B. P., C. van der Drift, A. Pol, and H. J. M. op den Camp. 2002. Microbial cycling of volatile organic sulfur compounds. Cell. Mol. Life Sci. 59:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni, S., and D. R. Boone. 1993. Catabolism of dimethyl sulfide and methanethiol by methylotrophic methanogens, p. 796-810. In R. S. Oremland (ed.), Biogeochemistry of global change. Chapman & Hall, New York, N.Y.
- 18.Oude Elferink, S. J. W. H., W. J. C. Vorstman, A. Sopjes, and A. J. M. Stams. 1998. Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol. Ecol. 27:185-194. [Google Scholar]
- 19.Paulo, P. L., B. Jiang, S. Rebac, L. H. Pol, and G. Lettinga. 2001. Thermophilic anaerobic digestion of methanol in UASB reactor. Water Sci. Technol. 44:129-136. [PubMed] [Google Scholar]
- 20.Paulo, P. L., M. V. G. Vallero, R. H. M. Trevino, G. Lettinga, and P. N. L. Lens. 2004. Thermophilic (55°C) conversion of methanol in methanogenic-UASB reactors: influence of sulphate on methanol degradation and competition. J. Biotechnol. 111:79-88. [DOI] [PubMed] [Google Scholar]
- 21.Roest, K., M. Altinbas, P. L. Paulo, H. G. H. J. Heilig, A. D. L. Akkermans, H. Smidt, W. M. de Vos, and A. J. M. Stams. 2005. Enrichment and detection of microorganisms involved in direct and indirect methanogenesis from methanol in an anaerobic thermophilic bioreactor. Microb. Ecol. 50:440-446. [DOI] [PubMed] [Google Scholar]
- 22.Roest, K., H. G. H. J. Heilig, H. Smidt, W. M. de Vos, A. J. M. Stams, and A. D. L. Akkermans. 2005. Community analysis of a full-scale anaerobic bioreactor treating paper mill wastewater. Syst. Appl. Microbiol. 28:175-185. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Sanguinetti, C. J., E. Dias Neto, and A. J. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:914-921. [PubMed] [Google Scholar]
- 25.Simankova, M. V., O. R. Kotsyurbenko, T. Lueders, A. N. Nozhevnikova, B. Wagner, R. Conrad, and M. W. Friedrich. 2003. Isolation and characterization of new strains of methanogens from cold terrestrial habitats. Syst. Appl. Microbiol. 26:312-318. [DOI] [PubMed] [Google Scholar]
- 26.Sipma, J., A. J. H. Janssen, L. W. Hulshoff-Pol, and G. Lettinga. 2003. Development of a novel process for the biological conversion of H2S and methanethiol to elemental sulfur. Biotechnol. Bioeng. 82:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Sipma, J., R. van Bree, A. J. H. Janssen, B. Arena, L. W. H. Hulshoff-Pol, and G. Lettinga. 2002. Degradation of methanethiol in a continuously operated upflow anaerobic sludge-blanket reactor. Water Environ. Res. 74:264-271. [DOI] [PubMed] [Google Scholar]
- 28.Smits, T. H. M., C. Devenoges, K. Szynalski, J. Maillard, and C. Holliger. 2004. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57:369-378. [DOI] [PubMed] [Google Scholar]
- 29.Stets, E. G., M. E. Hines, and R. P. Kiene. 2004. Thiol methylation potential in anoxic, low-pH wetland sediments and its relationship with dimethylsulfide production and organic carbon cycling. FEMS Microbiol. Ecol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Tanimoto, Y., and F. Bak. 1994. Anaerobic degradation of methylmercaptan and dimethyl sulfide by newly isolated thermophilic sulfate-reducing bacteria. Appl. Environ. Microbiol. 60:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trüper, H. G., and H. G. Schlegel. 1964. Sulphur metabolism in Thiorhodaceae-I, quantitative measurements on growing cells of Chromatium okenii. Antoni Leeuwenhoek J. Microbiol. Serol. 30:225-238. [DOI] [PubMed] [Google Scholar]
- 32.van Leerdam, R. C., F. A. M. de Bok, A. J. M. Stams, P. N. L. Lens, and A. J. H. Janssen. Volatile sulfur compounds in anaerobic sludge and sediment: biodegradation and toxicity. Environ. Toxicol. Chem., in press. [DOI] [PubMed]
- 33.Verachtert, T. A., R. T. Cassidy, and E. S. Holmes. 1990. Presented at the Laurance Reid Gas Conditioning Conference, Norman, Oklahoma, 5 to 7 March 1990.
- 34.Wilhelm, E., R. Battino, and R. J. Wilcock. 1977. Low-pressure solubility of gases in liquid water. Chem. Rev. 77:219-262. [Google Scholar]
- 35.Yu, Y., C. Lee, J. Kim, and S. Hwang. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89:670-679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.