Abstract
Chlortetracycline and the macrolide tylosin were identified as commonly used antimicrobials for growth promotion and prophylaxis in swine production. Resistance to these antimicrobials was measured throughout the waste treatment processes at five swine farms by culture-based and molecular methods. Conventional farm samples had the highest levels of resistance with both culture-based and molecular methods and had similar levels of resistance despite differences in antimicrobial usage. The levels of resistance in organic farm samples, where no antimicrobials were used, were very low by a culture-based method targeting fecal streptococci. However, when the same samples were analyzed with a molecular method detecting methylation of a specific nucleotide in the 23S rRNA that results in resistance to macrolides, lincosamides, and streptogramin B (MLSB), an unexpectedly high level of resistant rRNA (approximately 50%) was observed, suggesting that the fecal streptococci were not an appropriate target group to evaluate resistance in the overall microbial community and that background levels of MLSB resistance may be substantial. All of the feed samples tested, including those from the organic farm, contained tetracycline resistance genes. Generally, the same tetracycline resistance genes and frequency of detection were found in the manure and lagoon samples for each commercial farm. The levels of tetracycline and MLSB resistance remained high throughout the waste treatment systems, suggesting that the potential impact of land application of treated wastes and waste treatment by-products on environmental levels of resistance should be investigated further.
During animal production, antimicrobials are routinely used at subtherapeutic levels for growth promotion and prophylaxis and at therapeutic levels for treatment of infections. This agricultural use of antimicrobials could contribute to increased antimicrobial resistance in human pathogens through several routes, including human consumption of antimicrobial residues in animal products, human exposure to antimicrobials and resistant microorganisms during animal care (6), and contamination of ground and surface waters, soils, and crops by wastes containing antimicrobials and resistant microorganisms. While it is difficult to demonstrate a direct link between agricultural usage of antimicrobials and increased levels of resistance in pathogenic microorganisms, the potential consequences are serious and the World Health Organization has recommended that antimicrobials that are currently used or are under development for human therapy be phased out as animal growth promoters (36).
Animal waste handling and treatment practices vary considerably, but land application of solid and liquid side streams produced during waste handling and treatment is common, making contamination of ground and surface waters, soils, and crops with antimicrobials and antimicrobial resistance genes a realistic concern. Studies focusing on the fate of antimicrobials and the resistance levels in waste handling and treatment processes are scarce (17; L. T. Angenent, M. Mau, U. George, J. A. Zahn, and L. Raskin, submitted for publication). In studies of soil and groundwater samples, antimicrobial residues (e.g., references 22 and 33), resistant organisms (e.g., references 18, 26, and 30), and resistance genes (e.g., reference 8) have been found, but a comprehensive study is lacking. Two soil studies found only a transient increase in antimicrobial resistance following manure application (18, 30), while in another a significant difference in resistance levels was observed between farms using subtherapeutic and therapeutic antimicrobials and those at which antimicrobial use was restricted to therapeutic applications (26). The selection imposed by various culture media is an acknowledged bias of all three studies, and differences in the media used may account for the differing results. Groundwater in the vicinity of swine production facilities has also been found to contain tetracycline resistance determinants, and identical sequences were found in lagoon and groundwater samples, but the level of resistance has not been established (8).
Tetracyclines are broad-spectrum antibiotics exhibiting activity against a wide range of gram-positive and gram-negative bacteria, as well as atypical organisms such as chlamydiae, mycoplasmas and rickettsiae, and protozoan parasites (9). On the basis of their favorable antimicrobial properties and absence of major adverse side effects, tetracyclines are extensively used in the therapy of human and animal infections. Tetracycline resistance commonly results from ribosomal protection proteins (RPP) or from efflux genes coding for membrane-associated proteins that pump tetracycline from the cell (9). A recent phylogeny-based molecular ecology approach for detection of 8 tetracycline resistance genes encoding RPP and 11 tetracycline efflux pump (EFP) genes has been validated for genotyping tetracycline resistance in the environment (2, 3, 8). This approach has not been applied to investigate the impact of different antibiotic usage and waste-handling systems employed on commercial swine farms.
Macrolide antimicrobials are also used in human therapy and in swine production. Macrolides inhibit protein synthesis by binding to the 23S rRNA, and resistance mechanisms include target modification by mutation or methylation, active efflux, and structural modification of the macrolide (29, 35). Methylation of the adenine in position 2058 (A2058) of the 23S rRNA (Escherichia coli numbering) and macrolide EFPs combined account for about 99% of macrolide-resistant isolates (15), although the relative frequency of occurrence of these two mechanisms appears to vary widely (e.g., methylase genes were found in 22 and 90% of macrolide-resistant Streptococcus pneumoniae in isolates from two different countries) (15, 21). These mechanisms may be distinguished on the basis of phenotype because the methylation provides cross-resistance to lincosamides and streptogramin B antimicrobials (collectively known as macrolide-lincosamide-streptogramin B or MLSB resistance), while the EFPs are specific to 14- and 15-member ring macrolides. Lincosamides and streptogramin B antimicrobials are structurally unrelated to macrolides but bind to an overlapping region of the 50S ribosomal subunit. Because the methylation of A2058 prevents oligonucleotide binding at the sites flanked by this nucleotide, it is possible to quantify MLSB resistance by oligonucleotide probe hybridization (Angenent et al., submitted).
The focus of this work was to evaluate the effects of common conventional swine production practices on the level of antimicrobial resistance and the diversity of antimicrobial resistance genes in swine waste handling and treatment systems. As there was little information available on either antimicrobial usage or common waste handling and treatment scenarios, a survey including 11 farms was performed. Five farms were selected for analysis of the performance of the waste treatment systems and the levels of resistance to tetracyclines and MLSB antimicrobials throughout the treatment systems. Both culture-based and molecular measurements of resistance were performed to provide a detailed picture of resistance levels.
MATERIALS AND METHODS
Farm survey.
Questionnaires were collected from 10 commercial swine farms in Central Illinois and from the University of Illinois swine research farm. The questionnaires requested information about current and past therapeutic and subtherapeutic antimicrobial usage patterns, manure handling and treatment schemes, and manure and waste disposal practices.
Sample collection and transport.
After evaluation of the information collected from the survey, four commercial farms and the University of Illinois research farm were selected for sampling and analysis. The first set of samples was collected in December 2001 and January 2002 (phase 1); the second set of samples was obtained in April, May, and June 2002 (phase 2). Samples were collected from the swine feed, each stage of waste collection and treatment, nearby ground and/or surface water, and manure-amended soils. Waste samples were collected from each of the finisher buildings (the buildings in which the animals are housed after the grower stage until slaughter, typically from 10 to 16 weeks to 26 to 28 weeks of age), solids separators (when present), and the lagoons or holding ponds. To evaluate the level of mixing within the finisher buildings, where possible, three different locations inside a single building were sampled; one sample was obtained from each subsequent building on the same farm. For the University of Illinois research farm, the combined waste from all finisher buildings was sampled. For the solid separator units and the lagoons, samples were obtained from two to three locations within the units and at two different depths whenever possible, about 14 cm from the bottom and top surfaces of the units. Waste samples were collected with a PVC pipe fitted with a toilet plunger and steel rod; the sampling apparatus was washed with well water between samples. One sampler was dedicated for each farm. During phase 1, the lagoons at farm M were frozen and could not be sampled.
Soil samples were obtained from the soil surface or from a depth of about 15 to 20 cm, depending upon whether manure and waste treatment by-products were applied to the surface or injected into the soil, respectively. Five soil samples were typically taken, 25 m in from the four corners and the center of the field. For each sample, soil cores were obtained from the chosen site and from locations about 1 m away from the center in each direction and these soil cores were pooled. The latitude and longitude of the sampling locations were noted so that the same place could be sampled again during the second phase. The soil sampler was sterilized by washing with ethanol and then flaming it between samples.
Soil cores were stored in soil-sampling bags (Bageroft Packaging L.L.C., Chicago, IL), and all other samples were stored in new high-density polyethylene bottles (Cole-Parmer, Vernon Hills, IL), which were washed with an HCl solution when phosphate analyses were to be performed (1). All samples were transported to the lab on ice and analyzed or processed for storage as described below within 24 h of collection.
Chemical analyses.
Samples from phase 1 were analyzed for pH, alkalinity, solids (total, suspended, and volatile suspended solids [TS, SS, and VSS, respectively]), soluble and total chemical oxygen demand (SCOD and TCOD, respectively), 5-day biochemical oxygen demand (BOD5), ammonia, nitrate, nitrite, total nitrogen, total phosphorus, and orthophosphate. The pH was measured within 4 h of sample collection. For other analyses, samples were processed for storage according to standard methods (1). Samples for nitrate, nitrite, SCOD, and orthophosphate analyses were filtered through 0.45-μm-pore-size filters before storage at 4°C. For ammonia, TCOD, and total-phosphorus analyses, 2-N or 5.25-N H2SO4 was added to the samples to reduce the pH to less than 2 and samples were stored at 4°C. When prompt analysis was not possible, samples were stored at −20°C.
Alkalinity, solids, COD, and BOD5 analyses were performed according to standard methods (1). The ammonia analysis was also performed by standard methods (1), except that the reaction volume was scaled down and a microplate reader (340 ATTC; SLT Lab Instruments, Grödig Salzburg, Austria) was used. Nitrate, nitrite, orthophosphate, and total phosphorus were analyzed by Hach protocols (methods 8192, 8153, 8114, and 10127, respectively) and with a DR 4000 spectrophotometer (model 4000U; Hach Company, Loveland, CO). Total nitrogen analysis was performed by a combustion (Dumas) method (4).
Culture-based analyses.
Samples from phase 2 were stored in 30% glycerol stock at −80°C for culture-based analysis. A selective medium, mEnterococcus Agar (Becton & Dickinson, Sparks, MD), was used to cultivate fecal streptococci by incubation at 35.0 ± 0.5°C for 48 h, according to standard methods (1). Four sets of plates were incubated for each sample at each dilution: (i) control with no antimicrobials, (ii) with 20 μg/ml of tylosin (from tylosin tartrate; product no. T 6271; Sigma-Aldrich, St. Louis, MO), (iii) with 20 μg/ml tetracycline (from tetracycline hydrochloride; product no. T 7660; Sigma-Aldrich, St. Louis, MO), and (iv) with both antimicrobials at 20 μg/ml each. Dark red colonies were counted as fecal streptococcal colonies. A negative catalase test (1, 31) was considered a confirmation of the correct identity of fecal streptococci. The 20-μg/ml concentration of tetracycline is higher than the relevant Clinical and Laboratory Standards Institute breakpoint of 8 μg/ml but provides a conservative estimate of resistance and allows comparison with previous work.
RNA and DNA extractions.
Samples for RNA and DNA extractions were stored at −80°C. RNA extractions were performed by a low pH-hot phenol method (31) with the following slight modifications. Immediately prior to separating the aqueous phase each time, the samples were centrifuged at 16,000 to 18,000 × g for a few seconds; this was found to substantially improve the quality of the RNA as seen by the ratio of absorbances of the extracted RNA at 260 and 280 nm. In addition, for some samples (soil and swine building samples) not all of the material precipitated in the final recovery step could be dissolved in water. In such cases, the solution was heated twice at 60°C for 1 min and vortexed for a few seconds, followed by centrifugation at 16,000 × g to remove the remaining insoluble material.
Total genomic DNA was extracted by lysozyme and freeze-thaw treatments, followed by phenol chloroform extraction, as developed by Tsai and Olson for environmental samples (34). Creek and groundwater samples were centrifuged at 17,700 × g, and the extraction was performed on the pellets. When necessary, isolated DNA was purified to remove substances inhibitory to the PCR (such as humic acids) with polyvinylpolypyrrolidone (Sigma, St. Louis, MO) and Sepharose 2B or 4B as described by Zhou et al. and Miller (23, 38).
Molecular quantitation of MLSB resistance rRNA.
The specific ribosomal methylation resulting in MLSB resistance was quantified through membrane hybridizations as originally proposed by Angenent et al. (L. T. Angenent, M. Mau, A. Jindal, U. George, J. A. Zahn, and L. Raskin, 74th Annu. Conf. Water Environ. Fed., 2001; Angenent et al., submitted). Oligonucleotide probes (Table 1) were synthesized and purified by the University of Illinois W. M. Keck Center for Comparative and Functional Genomics (Urbana, IL) or by MegaBases Inc. (Evanston, IL). Membrane hybridizations were performed as described elsewhere (28), with minor modifications. Membranes were prehybridized at 29°C for the L-*-Bact-2053-a-A-13 (MLSB sensitivity) and L-*-Bact-2053-b-A-13 (MLSB sensitivity and resistance) probes or at 40°C for the universal probe for 9 to 12 h with 30 ml of hybridization buffer (Perfect Hyb Plus Hybridization Buffer, Sigma-Aldrich, St. Louis, Missouri). Hybridizations were performed with 32P-labeled probes at 29°C (MLSB probes) or 40°C (universal probe) for 12 to 18 h. Subsequently, two washing steps of 1 h each were carried out at 29°C for the MLSB probes and at 40°C for the universal probe, and one wash was performed at the previously determined (see below) temperature of dissociation (Td) of each probe (45.1°C for the MLSB sensitivity probe, 35.5°C for the MLSB resistance and sensitivity probe, and 44°C for the universal probe). For the MLSB probes, a wash buffer consisting of 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate was used. The amount of radioactivity was quantified with an Instant Imager (Packard Instruments, Meriden, CT). Hybridization results were quantified by comparison to known quantities of RNA from reference strains.
TABLE 1.
Oligonucleotide probes used in this study
| Target group | Probe | Sequence (3′ to 5′) | Reference organism | Reference |
|---|---|---|---|---|
| All organisms | S-*-Univ-1390-a-A-18 | AACATGTGTGGCGGGCAG | E. faecalis JH2-2/pAMβ1 | 37 |
| MLSB-sensitive bacteria | L-*-Bact-2053-a-A-13 | CTGCCTTTCTGGG | E. faecalis JH2-2 | Angenent et al., submitted |
| MLSB-resistant and -sensitive bacteria | L-*-Bact-2053-b-A-13 | CTGCC5TTCTGGGa | E. faecalis JH2-2/pAMβ1 | Angenent et al., submitted |
5 stands for the synthetic nucleotide 5-nitroindole.
The Td for the MLSB probes was determined through a Td study (37) with resistant and sensitive strains of Enterococcus faecalis (JH2-2 pAMβ1 and JH2-2, respectively) and an organism with a G-T mismatch at position 2057 (Rhodococcus coprophilus). Because of the short length of the probes, prehybridization and hybridization were performed at 29°C and a modified wash buffer (4× SSC and 1% sodium dodecyl sulfate) was used. The Td study was performed in duplicate (data not shown).
PCR detection of tetracycline resistance genes.
PCR was performed on duplicate extractions of phase 1 samples to monitor eight tetracycline resistance genes encoding RPP [tet(M), tet(O), tet(Q), tet(W)] and EFP proteins (tet(A), tet(C), tet(H) and tet(Z) with the class-specific primer sets designed and validated by Aminov et al. (2, 3). These eight genes were chosen because they were the most frequently detected tetracycline resistance genes in previous studies on swine farms (2, 3, 8). The presence of bacterial DNA was confirmed by positive amplification of the V3 region of the bacterial 16S rRNA gene with primers 341F and 534R (24). A typical reaction mixture contained 25 pmol of each primer, 100 μM each deoxyribonucleotide triphosphate, 1× ExTaq reaction buffer (Pan Vera Corporation, Madison, WI), 0.5 U of ExTaq DNA polymerase (Pan Vera Corporation), and 50 to 100 ng DNA extracted from environmental samples in a total volume of 25 μl. PCR amplification was carried out with a GeneAmp PCR System 2400 Thermocycler (Applied Biosystems, Foster City, CA) for 30 cycles as described by Aminov et al. (2, 3). Sterile water was used as the negative control template, and strains containing the target genes were used as templates in positive control reactions (2, 3, 8). PCR product aliquots (5 μl) were separated by electrophoresis in 2.0% agarose gels and viewed after staining with ethidium bromide. Analytical sensitivity of the PCR method is 100 to 1,000 copies of the resistance genes. Environmental detection limits are an order of magnitude less sensitive than this (104 copies). The frequency of detection of each resistance gene was calculated as the number of positive signals as a percentage of the total number of samples for each sample type on each farm.
Statistical analysis.
Pairwise comparisons were performed in SAS v8.2 by the least significant difference method.
RESULTS AND DISCUSSION
Swine farms.
To focus measurements of antimicrobial resistance on the appropriate antimicrobials, a survey of antimicrobial usage practices at 10 commercial swine farms and 1 research farm in central Illinois was performed (19). One of these farms was an organic farm, and antimicrobials had not been used at this farm since 1996. The herd sizes varied from 1,100 to 8,500 swine. Nine of the 11 farms were all in-all out facilities, which means that all of the animals in a building were brought in at the same time and shipped out at the same time. Most farms cleaned barns by a power wash with a disinfectant. The frequency of cleaning varied from once every 2 weeks to about once every 6 months and most often occurred between groups of animals (because of the all in-all out mode of operation).
On the basis of the survey results, five farms were selected for further analyses. As detailed below, these farms were chosen to represent both a range of antimicrobial usage patterns and a variety of waste handling and treatment processes. Farm M included operations at two sites, designated M1 and M2. Because the two sites had different antimicrobial usage and waste handling and treatment processes, samples from these two sites were analyzed and reported separately.
Antimicrobial usage.
Chlortetracycline (most commonly used at 400 g/ton feed) and bacitracin (30 g/ton feed) were the two most commonly used antimicrobials for growth promotion and prophylaxis and were in use at nine and eight farms, respectively. Five farms reported regular subtherapeutic use of tiamulin, while tylosin and roxarsone were each in use at three farms. If historical usage was also considered, tylosin usage was more prevalent. Specifically, of the eight farms not currently using tylosin, all four of the farms that reported historical usage patterns had discontinued the use of tylosin only 1 to 6 years prior to sampling. Various other antimicrobials were in use at subtherapeutic levels at one or two farms, including a mixture of chlortetracycline, sulfathiazole, and penicillin (CSP 250); lincomycin; virginiamycin; carbadox; and tilmicosin. These results are consistent with results from a national survey performed by the National Animal Health Monitoring Survey in which tylosin, chlortetracycline, and bacitracin were the most commonly used antimicrobials in grower-finisher animals (25). These results were also consistent with those of a slightly older survey of antimicrobial use at Canadian swine farms in which tylosin was the most common antimicrobial used in finisher animals (12, 13). On the basis of these results and on the importance of various antimicrobials in human treatment, resistance to macrolides (including tylosin) and tetracyclines was examined further. Bacitracin resistance was not examined because bacitracin is currently used only topically in human medicine and exhibits no known cross-resistance with other antimicrobials (27).
Eight of the farms reported therapeutic usage of antimicrobials in the last 5 years. The types of antimicrobials used included tetracyclines (six farms), penicillins and related cephalosporins (five farms), the lincosamide lincomycin (three farms), the macrolide tylosin (three farms), sulfa drugs (three farms), the arsenical roxarsone (two farms), and the polypeptides polymyxin B (two farms) and bacitracin (one farm). Therapeutic antimicrobials were frequently used at high concentrations by injection or through addition to the water supply, while subtherapeutic antimicrobials were routinely included at low levels in the feed.
Antimicrobial usage practices at the five farms selected for detailed analyses are described in Table 2. Except for the organic farm (farm O), all of the farms used multiple antimicrobials at subtherapeutic levels, sometimes simultaneously and sometimes sequentially during the production cycle, and the treatments varied in number, dosage, and duration. Additional important considerations include therapeutic doses, which were applied as needed, and the historical usage patterns for each site. Farm L began using tilmicosin and CSP250 in 2000 and 2001, respectively; no antimicrobials were reported to have been discontinued recently at this farm. The subtherapeutic use of tylosin was discontinued in 1995 at farm H, in 2000 at farm M1, and in 1999 at farm M2. Lincomycin also had been used subtherapeutically at farm M1, and this use was discontinued in 1998. Finally, although for purposes of resistance measurements the antimicrobials were considered by class, it is also important to remember that antimicrobials within a class vary in characteristics such as degradation and sorption potentials, causing differences in environmental fate.
TABLE 2.
Antimicrobial usage at selected farmsc
| Farm and subtherapeutic drug used | Age | Therapeutic drug(s) used |
|---|---|---|
| H | ||
| Tiamulin | 2-3 wk | Penicillin |
| Chlortetracycline | 2-15, 21 wk | |
| Bacitracin | 16-25 wk | |
| M1 | ||
| Chlortetracycline | 1 wk/mo | Penicillin, tylosin, roxarsone, bacitracin, chlortetracycline |
| Bacitracin | 3 wk/mo | |
| M2 | ||
| Chlortetracycline | 2 wk/mo nursery | Penicillin, tylosin, roxarsone, bacitracin, chlortetracycline |
| Bacitracin | 2 wk/mo nursery | |
| Chlortetracycline | 1 wk/mo | |
| Bacitracin | Continuous | |
| Roxarsone | Continuous | |
| La | ||
| Carbadox | 3-7 wk | Lincomycin, sulfatrimethoprim, oxytetracycline, polymyxin B |
| Tilmicosin | 8-10 wk | |
| Tylosin | 11-13 wk | |
| CSP 250b | 14-15 wk | |
| Virginiamycin | 16-28 wk | |
| R | ||
| CSP 250 | 3-7 wk | None |
| Tylosin | 8-22 wk | |
| O, none | None |
Antimicrobial usage at farm L was previously reported (39).
CSP 250 contains chlortetracycline, sulfathiazole, and penicillin.
Historical usage of antimicrobials is described in the text.
Waste handling and treatment procedures.
Survey results reflected the wide variety of systems currently used for the removal of waste from buildings, including scrapers, recharge, pull plugs, and flushing, and often multiple systems were used in different buildings at the same farm. The frequency of removal also varied from daily to yearly, depending on the capacity of the system. Following removal of wastes from the buildings, most of the conventional farms used a solids-settling basin, followed by one or more lagoons. The remainder used deep pits beneath the buildings, and waste from these pits was typically removed once or twice yearly and land applied without further treatment. The organic farm had a bedded pack of straw and hay to collect waste. Manure from deep pits and solids from solids-settling basins were land applied one to four times yearly, either by injection or without incorporation in the soil. In most cases, there was no removal of lagoon effluent or solids and the farmers thus relied on evaporation and solids build-up.
The three conventional, commercial farms chosen for further study all made use of the most common treatment system described above, i.e., a solids-settling basin followed by one or two lagoons. However, several differences were evident in their manure-removal systems and retention times. Farm H had shallow pits underneath the building floors which were emptied monthly with a plug pull system. The contents of the solids-settling basin were land applied twice yearly. Farm M1 had three buildings with deep pits whose contents were emptied yearly and land applied without further treatment. The manure from these buildings was also applied on a separate stretch of land which had not received manure from any other location. Farm M2 consisted of several buildings utilizing different manure-handling systems, four of which provided influent to the same solids-settling basin. In one building, waste was scraped to an external underground storage tank three times daily. The contents of this storage tank were pumped to the solids-settling basin monthly. A second building had a shallow pit which was flushed with recycle water from the lagoon every 2 months. This waste was transferred to another underground storage tank and then to the solids-settling basin. Two other buildings had deep pits, from which waste was pumped to the solids-settling basin about every 4 months, typically shortly before land application of solids from the solids-settling basin. At farm L, buildings were flushed weekly with water recycled from the lagoon. Farm L was also unusual in that the contents of the lagoon, as well as the solids-settling basin, were land applied yearly (i.e., the lagoon was operated as a holding pond). At farm R, waste was scraped out of the buildings daily, collected in a pit, and pulled into a lagoon weekly. Effluent from the lagoon was either discharged into the municipal sewer line or land applied. The organic farm used a bedded pack of straw and hay for waste collection, and this mixture was removed and land applied without further treatment.
Chemical characterization of samples collected from buildings, solids-settling basins, and lagoons provided information on the degree of treatment taking place in the different unit processes (Table 3). In general, the strength of swine waste in terms of VSS, SCOD, and nutrient content (nitrogen and phosphorus) was comparable to previous analyses of swine manure samples (11) and high, with values often over 100-fold higher than a typical domestic wastewater (32). A comparison of the results for the building samples from the four conventional farms indicated that samples from farm R were of lower strength, those from farms L and H were similar and of intermediate strength, and those from farm M were substantially stronger (Table 3). Strength did not correlate directly with the number of animals at each farm since it reflects the combined effects of number and density of animals and the different dilutions involved with various manure removal processes. For the lagoon samples, VSS, SCOD, and nutrient levels were lowest in the samples from farm R, and farm H samples generally had lower levels than farm L. This could reflect the higher ratio of lagoon volume to number of animals at farm H versus farm L (9 and 2 m3/head, respectively).
TABLE 3.
Chemical analysis of swine waste treatment samples
| Farm and locationb | VSSc | VSS/SS ratio | SCODc | Total Nc | NH3c | Total Pc | Orthophosphatec |
|---|---|---|---|---|---|---|---|
| H | |||||||
| Bld | 58 | 0.83 | 19.8 | 8.27 | 3.48 | 3.9 | 0.82 |
| SSB | 28 | 0.76 | 13.6 | 3.48 | 2.64 | 2.5 | 0.71 |
| Lag | 2.6 | 0.47 | 2.9 | 0.61 | 0.54 | 0.46 | 0.34 |
| M2 | |||||||
| Bld | 105 | 0.74 | 41.5 | 10.3 | NDd | 4.6 | 0.87 |
| SSB | 36 | 0.68 | 32.2 | 6.2 | ND | 2 | 0.67 |
| La | |||||||
| Bld | 53 | 0.82 | 17.5 | 6.7 | 4.14 | 2.1 | 0.47 |
| SSB | 36 | 0.61 | 5.5 | 4.9 | 3.57 | 3.5 | 0.35 |
| Lag | 13 | 0.48 | 1.5 | 2.0 | 0.82 | 1.4 | 0.14 |
| R | |||||||
| Bld | 16 | 0.84 | 18 | 3.9 | 3.13 | 1.4 | 0.19 |
| Lag | 1.5 | 0.75 | 0.76 | 0.85 | 0.37 | 0.09 | 0.03 |
Chemical analysis of farm L samples was reported previously (39).
Bld, building, SSB, solids-settling basin, Lag, lagoon.
Measurements are presented in grams per liter.
ND, not determined.
Substantial improvements were observed in the quality of the wastewater from the building to the lagoon, as indicated by the decreases in the levels of VSS, SCOD, and nutrients, with SCOD removal efficiencies ranging from 85 to 96%. It is important to note that samples collected include the biomass present in the lagoon and therefore underestimate the treatment performance with respect to VSS and nutrient removal. For land application, farm L used the contents of the solids-settling basin and the lagoon while farms H and M2 applied only the contents of the solids-settling basin. Considering the land-applied stages, a much broader range of SCOD removal was observed, from 23 to 86%.
Culture-based resistance measurements.
Antimicrobial resistance was measured with a culture-based resistance assay targeting fecal streptococci, and results are reported in Fig. 1. As expected, the levels of tetracycline, tylosin, and dual resistance were lowest at the organic farm, where antimicrobials were not used. The organic farm had significantly lower resistance than farms M2, L, and R for tetracycline and farms H, M1, and M2 for tylosin (P value < 0.05). However, despite differences in antimicrobial usage, no clear difference in tetracycline or tylosin resistance was observed among the other farms. This result was particularly striking given that farms H, M1, and M2 had discontinued subtherapeutic tylosin usage 6 years, 1 year, and 2 years, respectively, before this study. Tylosin was still used therapeutically at farm M, but recent results from Denmark suggest that this would not provide sufficient selective pressure to maintain such high levels of resistance (F. M. Aarestrup, Int. Invitational Symp. Beyond Antimicrob. Growth Promoters Food Anim. Prod., 2002). Continued selective pressure from the accumulation and persistence of tylosin in the waste treatment process is also unlikely, because tylosin is reported to be rapidly degraded in these systems (10, 14, 20; Angenent et al., submitted). The most likely explanations, therefore, appear to be either the persistence of other MLSB antimicrobials or coselection via the continued subtherapeutic use of chlortetracycline at the farms, since linkage between macrolide and tetracycline resistance genes has been observed (9, 16). These results emphasize the complexity of the system, as the simple case of discontinuing antimicrobial use followed by a decrease in resistance levels was not observed. Soil samples could not be analyzed by this method because of the low numbers of fecal streptococci. With respect to waste treatment procedures, a decrease in the resistance through the waste treatment train was observed at farms M2 (P < 0.05) and R. Although the decrease at farm R could not be confirmed statistically because there were insufficient samples across the waste treatment train, a similar trend was observed when farm R samples were analyzed by the molecular method (below).
FIG. 1.
Percentages of antimicrobial-resistant fecal streptococci. Values shown are the average percent resistance from all samples from the buildings (bld; black), solids-settling basin(s) (ssb; stripes), or lagoon(s) (lag; white) of a given farm. For triplicates of the same sample, the standard deviations averaged 16% of the total counts. When results from each treatment stage at a particular farm were averaged as shown here, standard deviations were typically 20 to 30%, reflecting the heterogeneity of these unmixed systems. The total number of samples for a treatment stage was three to eight, except for farm L buildings (two samples), farm R (buildings, one sample; lagoon, two samples), and farm O (buildings, one sample).
Molecular measurements of MLSB-resistant microbial rRNA.
To better evaluate antimicrobial resistance in the overall microbial community, oligonucleotide probe hybridization was used to quantify MLSB resistance due to target modification (Angenent et al., submitted). The level of MLSB-sensitive microbial rRNA was quantified and compared to the total level of rRNA. The probe targeting the 23S rRNA of sensitive and resistant organisms resulted in a low signal even after optimization of hybridization conditions, and in several cases the hybridization signals obtained with this probe could not be used for normalization. Therefore, results obtained with the probe targeting the 23S rRNA of sensitive organisms were normalized to results obtained with a universal probe targeting the 16S rRNA (Table 1). There are a small number of organisms that have a mismatch in the region of their 23S rRNA targeted by the probes used in this study, and since these sequences, if present, would not bind to the probe targeting the sensitive organisms but would be quantified with the 16S rRNA-targeted universal probe, there is a possibility of overestimation of resistance. However, in those cases where the probe targeting the 23S rRNA of sensitive and resistant strains had a signal above the background, the two methods of normalization resulted in similar resistance levels (data not reported), so this possible overestimation appears not to be substantial.
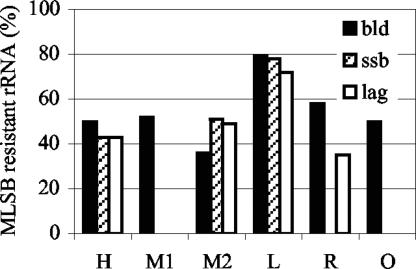
The highest level of MLSB-resistant microbial rRNA was observed at farm L, which used the highest level of MLSB antimicrobials (Fig. 2). However, there was no significant difference among the other farms (P value < 0.05), and the fraction of rRNA that was resistant at the organic farm, where no antimicrobials were used, was unexpectedly high (50%). It remains unclear whether the 50% resistance observed at the organic farm is a typical level of MLSB resistance in the absence of anthropogenic selective pressures. Other explanations include contamination of the feed (below and Table 4) or persistence from usage of the site prior to 1996. Although samples of soil amended with manure were taken at each farm, sufficient RNA was not retrieved from the soil samples for quantification of resistance by this method.
FIG. 2.
Percentage of MLSB-resistant microbial rRNA. Values shown are the average percentages from all samples from the buildings (bld; black), solids-settling basin(s) (ssb; stripes), or lagoon(s) (lag; white) of a given farm. Resistant microbial rRNA was calculated as 1 − the ratio of sensitive to universal hybridization signals. Samples were hybridized in duplicate, and the percent half range, calculated as 100 × |Signal1 − SignalAve|SignalAve, was typically below 20%.
TABLE 4.
Pattern of tetracycline resistance genes in swine waste treatment samples and in soil and water samples
| Farm | Sequence of sample from:
|
||||
|---|---|---|---|---|---|
| Feed | Building | Lagoon | Soil | Water | |
| H | -OQW----a | MOQW-CHZ | MOQW-CHZ | ---W---- | -O------ |
| M1 | MOQW--HZ | MOQW--HZ | NAb | MOQW--HZ | NA |
| M2 | -OQW-CHZ | MOQWACHZ | MOQW--HZc | --QW---Z | -OQW---- |
| L | -OQW---- | MOQW--HZ | MOQW--HZ | MOQW--HZ | NA |
| R | -OQW---- | MOQW--HZ | MOQW--HZ | NA | NA |
| O | MOQW--HZ | MOQW--HZ | NA | -------Z | --QW--H- |
The letters represent a positive amplification with primers specific to the class of tet genes represented by the letter shown [e.g., M refers to tet(M) genes]. A dash indicates that no PCR product was detected; the detection limit was approximately 104 copies.
NA, not applicable.
Because the lagoon was frozen and could not be sampled during phase 1, data from the solids-settling basin are included here as well as in Fig. 3.
Because the culture-based and molecular measurements of MLSB resistance target different groups of microorganisms (cultured fecal streptococci versus the overall microbial community), comparison of these results provides insight into the distribution of resistance within a community. The agreement between the methods used for most of the samples is consistent with the idea that horizontal gene transfer is common and supports the use of culture-based methods as surrogates for overall resistance measurements. This agreement further suggests that macrolide resistance via efflux is not the dominant resistance mechanism in these samples, because the efflux resistance mechanism is detected by the culture-based method but not the molecular method and would therefore have resulted in higher resistance measurements from the culture-based versus the molecular method. The reverse discrepancy between these two methods in the organic farm samples (molecular higher than culture based) may indicate that the level of resistance in the fecal streptococci is substantially different from that in the overall community. This possibility raises serious concerns because current monitoring efforts for antimicrobial resistance focus on specific fecal organisms (5, 7) and would likely not have detected the relatively high resistance at the organic farm.
Distribution of tetracycline resistance genes.
The presence of tetracycline resistance genes throughout the production and waste-handling process was evaluated, and the patterns observed in different sample types are shown in Table 4. For sample types for which multiple samples were taken from the same farm, the frequency of detection is reported in Fig. 3. These genes were chosen on the basis of their detection in previous studies, and as expected, they were found in most samples, except for the EFP gene tet(A), which was only detected once. Two points are clear from this analysis. First, several tetracycline resistance determinants were present in the feed at all of the farms, including the organic farm. Invariably, the feed samples contained at least three tetracycline resistance genes [tet(O), tet(Q), and tet(W)] encoding RPP. On the basis of these results, animal feed deserves consideration as a source of antimicrobial resistance genes, and the origin and fate of these genes should be examined further.
FIG. 3.
Frequency of detection of tetracycline resistance genes. The detection of RPP [tet(M), tet(O), tet(Q) and tet(W)] and EFP protein [tet(C), tet(H) and tet(Z)] gene families in building, lagoon, and soil samples is presented for each of four commercial swine operations (farms H, M1, M2, and L). Frequency was calculated as the number of positive detections in the total number of samples from a particular farm and treatment stage. The total number of samples was five or more, except for the building and lagoon samples from farm L, which were duplicates. Results from the other farms and additional sample types are not included because the number of samples was too low. The tet(A) gene family was not included because there was only one positive response for this gene among all of the samples tested.
Second, although the tetracycline resistance genes detected and their frequency of detection did not differ substantially between the building and lagoon samples, the soil and water samples typically had fewer positive responses (Table 4 and Fig. 3). These results suggest a decrease in both the richness and the abundance of resistance genes following land application, although the magnitude and implications of that decrease are difficult to assess without a quantitative analysis of tetracycline resistance gene levels in all samples, which was beyond the scope of the present study. The exception to this general trend were the soil samples from farms M1 and L, which showed the presence of all four of the RPP genes tested [tet(M), tet(O), tet(Q), and tet(W)], as well as the EFP genes tet(H) and tet(Z). When frequency of detection is also considered, only the soil samples from farm L exhibited results comparable to those obtained with the corresponding building and lagoon samples. This farm was the only one to use a spray gun to apply lagoon effluent and slurry across the field surface. While these data could result from an increased persistence of resistance genes associated with this application method, it could also reflect differences in affected area and heterogeneity, resulting in a sampling bias that is specific to the application method. All creek and groundwater samples from the farms also contained tetracycline resistance genes, although again fewer genes were detected in these samples than in the building and lagoon samples. This result suggests that drinking water could also be a source of antimicrobial resistance genes, for both animals and humans. However, since multiple tetracycline resistance genes were also observed in groundwater from the organic farm, it is unclear whether these concerns are associated with the use of antimicrobials in animal production.
Concluding comments.
This study clearly demonstrates the presence of high levels of antimicrobial resistance not only in swine manure but throughout the waste treatment process, raising concerns that the land application of the treated manure contributes to an environmental reservoir of resistance. However, it is important to note that the link between this type of environmental resistance and public health concerns is currently speculative and itself worthy of investigation. This work further illustrates the complex relationship between antimicrobial usage and resistance, as high levels of tylosin resistance persisted for years after usage ceased, and indicates the need for microbiologists, swine nutritionists, and veterinarians to work together in identifying treatment regimens that are both effective and prudent. Finally, the organic farm results are striking in two respects. First, the high level of resistance observed in the absence of known selective pressure again demonstrates that our understanding of the microbial ecology of resistance is incomplete. Second, the fact that this resistance was not detected by traditional culture-based methods highlights a potentially serious problem with current antimicrobial resistance monitoring efforts.
Acknowledgments
This research was partially supported by U.S. Department of Agriculture section 224 funds (L.R. and R.I.M.), USDA-NRI 26.0 funding (AG 2001-35102-10774 to R.I.M.), and the Agricultural Experiment Station, College of Agricultural, Consumer and Environmental Sciences, University of Illinois at Urbana-Champaign.
We thank the farmers for their cooperation and participation, Toshio Shimada for assistance with statistical analysis, Ka Wai Suzanne Huang and Matt Wagoner for assistance with culture-based assays, Jim Royer for the BOD measurements, Ron Belyea for the total nitrogen assays, and Nadia Shoemaker and Abigail Salyers for providing Enterococcus faecalis strains. We also thank Rustam Aminov, Lars Angenent, Joanne Chee-Sanford, Mike Cotta, Ted Funk, Gilbert Hollis, Mike Tumbleson, Terry Whitehead, and the anonymous reviewers for helpful comments.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 2.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Association of Official Analytical Chemists. 1984. Official methods of analysis, 14th ed. Association of Official Analytical Chemists, Arlington, Va.
- 5.Bager, F. 2000. DANMAP: monitoring antimicrobial resistance in Denmark. Int. J. Antimicrob. Agents 14:271-274. [DOI] [PubMed] [Google Scholar]
- 6.Borgen, K., G. S. Simonsen, A. Sundsfjord, Y. Wasteson, O. Olsvik, and H. Kruse. 2000. Continuing high prevalence of Van-A type vancomycin resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J. Appl. Microbiol. 89:478-485. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2003. National antimicrobial resistance monitoring system for enteric bacteria (NARMS): 2001 annual report. Centers for Disease Control and Prevention, Atlanta, Ga.
- 8.Chee-Sanford, J. C., R. I. Aminov, I. G. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Liguoro, M., V. Cibin, F. Capolongo, B. Halling-Sorensen, and C. Montesissa. 2003. Use of oxytetracycline and tylosin in intensive calf farming: evaluation of transfer to manure and soil. Chemosphere 52:203-212. [DOI] [PubMed] [Google Scholar]
- 11.DeRouchey, J. M., R. D. Goodband, J. L. Nelssen, M. D. Tokach, S. S. Dritz, and J. P. Murphy. 2002. Nutrient composition of Kansas swine lagoons and hoop barn manure. J. Anim. Sci. 80:2051-2061. [DOI] [PubMed] [Google Scholar]
- 12.Dunlop, R. H., S. A. McEwen, A. H. Meek, W. D. Black, R. C. Clarke, and R. M. Friendship. 1998. Individual and group antimicrobial usage rates on 34 farrow-to-finish swine farms in Ontario, Canada. Prev. Vet. Med. 34:247-264. [DOI] [PubMed] [Google Scholar]
- 13.Dunlop, R. H., S. A. McEwen, A. H. Meek, R. A. Friendship, R. C. Clarke, and W. D. Black. 1998. Antimicrobial drug use and related management practices among Ontario swine producers. Can. Vet. J. 39:87-96. [PMC free article] [PubMed] [Google Scholar]
- 14.Gavalchin, J., and S. E. Katz. 1994. The persistence of fecal-borne antibiotics in soil. J. AOAC Int. 77:481-485. [Google Scholar]
- 15.Gay, K., W. Baughman, Y. Miller, D. Jackson, C. G. Whitney, A. Schuchat, M. M. Farley, F. Tenover, and D. S. Stephens. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417-1424. [DOI] [PubMed] [Google Scholar]
- 16.Giovanetti, E., A. Brenciani, R. Lupidi, M. C. Roberts, and P. E. Varaldo. 2003. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 47:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanzawa, Y., C. Oka, N. Ishiguro, and G. Sato. 1984. Antibiotic-resistant coliforms in the waste of piggeries and dairy farms. Jpn. J. Vet. Sci. 46:363-372. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, L. B., S. Baloda, M. Boye, and F. M. Aarestrup. 2001. Antimicrobial resistance among Pseudomonas spp. and the Bacillus cereus group isolated from Danish agricultural soil. Environ. Int. 26:581-587. [DOI] [PubMed] [Google Scholar]
- 19.Jindal, A. 2002. Antimicrobial resistance in swine waste treatment processes. M.S. thesis. University of Illinois, Urbana.
- 20.Loke, M.-L., F. Ingerslev, B. Halling-Sorensen, and J. Tjornelund. 2000. Stability of tylosin A in manure containing test systems determined by high performance liquid chromatography. Chemosphere 40:759-765. [DOI] [PubMed] [Google Scholar]
- 21.Marchese, A., E. Tonoli, E. A. Debbia, and G. C. Schito. 1999. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J. Antimicrob. Chemother. 44:461-464. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, M. T., J. E. Bumgarner, J. L. Varns, J. V. Daughtridge, E. M. Thurman, and K. A. Hostetler. 2000. Use of radioimmunoassay as a screen for antibiotics in confined animal feeding operations and confirmation by liquid chromatography/mass spectrometry. Sci. Total Environ. 248:181-187. [DOI] [PubMed] [Google Scholar]
- 23.Miller, D. N. 2001. Evaluation of gel filtration resins for the removal of PCR-inhibitory substances from soils and sediments. J. Microbiol. Methods 44:49-58. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Animal Health Monitoring System. 2002. Highlights of NAHMS Swine 2000, part II. U.S. Department of Agriculture, Washington, D.C.
- 26.Onan, L. J., and T. M. LaPara. 2003. Tylosin-resistant bacteria cultivated from agricultural soil. FEMS Microbiol. Lett. 220:15-20. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, I. 1999. The use of bacitracin as a growth promoter in animals produces no risk to human health. J. Antimicrob. Chemother. 44:725-728. [DOI] [PubMed] [Google Scholar]
- 28.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittman, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengelov, G., Y. Agerso, B. Halling-Sorensen, S. B. Baloda, J. S. Andersen, and L. B. Jensen. 2003. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 28:587-595. [DOI] [PubMed] [Google Scholar]
- 31.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchobanoglous, G., and F. L. Burton. 1991. Wastewater engineering: treatment, disposal, and reuse, third ed. Irwin/McGraw-Hill, Boston, Mass.
- 33.Thiele-Bruhn, S. 2003. Pharmaceutical antibiotic compounds in soils—a review. J. Plant Nutr. Soil Sci. 166:145-167. [Google Scholar]
- 34.Tsai, Y. L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. 2000. WHO global principles for the containment of antimicrobial resistance in animals intended for food. World Health Organization, Geneva, Switzerland.
- 37.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zilles, J., T. Shimada, A. Jindal, M. Robert, and L. Raskin. 2005. Presence of macrolide-lincosamide-streptogramin B and tetracycline antimicrobials in swine waste treatment processes and amended soil. Water Environ. Res. 77:57-62. [DOI] [PubMed] [Google Scholar]