Abstract
We used flow cytometry to examine the process of cell death in the bloom-forming alga Heterosigma akashiwo during infection by a double-stranded DNA virus (OIs1) and a single-stranded RNA virus (H. akashiwo RNA virus [HaRNAV]). These viruses were isolated from the same geographic area and infect the same strain of H. akashiwo. By use of the live/dead stains fluorescein diacetate and SYTOX green as indicators of cellular physiology, cells infected with OIs1 showed signs of infection earlier than HaRNAV-infected cultures (6 to 17 h versus 23 to 29 h). Intracellular esterase activity was lost prior to increased membrane permeability during infection with OIs1, while the opposite was seen with HaRNAV-infected cultures. In addition, OIs1-infected cells accumulated in the cultures while HaRNAV-infected cells rapidly disintegrated. Progeny OIs1 viruses consisted of large and small morphotypes with estimated latent periods of 11 and 17 h, respectively, and about 1,100 and 16,000 viruses produced per cell, respectively. In contrast, HaRNAV produced about 21,000 viruses per cell and had a latent period of 29 h. This study reveals that the characteristics of viral infection in algae are virus dependent and therefore are variable among viruses infecting the same species. This is an important consideration for ecosystem modeling exercises; calculations based on in situ measurements of algal physiology must be sensitive to the diverse responses of algae to viral infection.
Heterosigma akashiwo (Rhaphidophyceae) is a harmful bloom-forming alga that inhabits temperate coastal waters around the world (14, 35). It occurs at densities of up to 5 × 105 cells · ml−1 and can cause indiscriminate fish kills of both wild and cultured fish (35). The severity and duration of these blooms are unpredictable from year to year, largely because the factors controlling them have remained elusive. However, transmission electron microscopy (TEM) examination of H. akashiwo populations during bloom termination suggests the direct involvement of viruses (23, 24, 25). Viral infection has only recently been appreciated for its role in influencing the population dynamics of phytoplankton (1, 3, 9, 26, 31). Infections are especially important during blooms, since the rate of infection is dependent on the abundance of host cells (22). Algal-virus interactions are not well understood, and we have only a limited appreciation for the diversity of algal viruses. Among these are a diverse group of unrelated viruses that all infect H. akashiwo. These include double-stranded DNA (dsDNA) viruses (H. akashiwo virus [HaV]) (24), a single-stranded RNA (ssRNA) virus (H. akashiwo RNA virus [HaRNAV]) (17, 34), and two other uncharacterized virus systems (H. akashiwo nuclear inclusion virus and OIs1) (19; J. Lawrence, unpublished data).
Viral infections of phytoplankton populations can be difficult to detect. TEM analysis is labor-intensive and time-consuming and has very high limits of detection, as cells have to be individually examined for ultrastructural changes associated with infection. Molecular detection of infection requires the development of specific probes targeted to known viruses or groups of related viruses. Flow cytometry (FCM) has been used to detect viral infection of phytoplankton (6) and has been used to study infections of Phaeocystis pouchetii and Micromonas pusilla by two dsDNA viruses (5). FCM permits rapid and precise cell-by-cell analysis of phytoplankton in which light scattering and fluorescence are measured simultaneously. FCM in combination with fluorescent live/dead stains allows the detection of mortality due to viral infection in phytoplankton (5) by revealing changes in cellular physiology associated with cell death.
The current study uses FCM to compare the lytic cycles of two viruses, HaRNAV and OIs1, and the progression of cell death in H. akashiwo. HaRNAV was isolated from the Strait of Georgia, British Columbia, Canada. This ssRNA virus belongs to the Marnaviridae, has a genome of 9.1 kb and a particle diameter of 25 nm, and is assembled in the cytoplasm (17, 34). OIs1 is a partially characterized dsDNA virus isolated from the sediment of Malaspina Inlet, British Columbia, Canada (20). There are two cooccurring icosahedral morphotypes, differing in particle size (30 versus 80 nm) and genome size (20 kb versus 130 kb), which assemble in the cytoplasm of their host (J. Lawrence, unpublished data). In this system, both morphotypes are visible in an infected host at the same time. By comparing infections with these two viruses, we demonstrate that the characteristics of viral infection are virus dependent.
MATERIALS AND METHODS
Experimental design.
The raphidophycean alga Heterosigma akashiwo (NEPCC strain 522, nonaxenic), which was originally isolated from English Bay, Vancouver, British Columbia, Canada, was used throughout this study. Cultures were grown in 29-practical-salinity-unit, f/2-enriched seawater media (11) supplemented with 10 nM sodium selenite at 20°C, while providing 260 μmol photons m−2 s−1 during a 14-h:10-h light:dark regimen. Cultures were grown in 1-liter polycarbonate culture flasks (Nalgene) and were monitored for growth by measuring in situ chlorophyll a (chl a) fluorescence using a Turner Designs fluorometer. Inocula of HaRNAV and OIs1 were produced by serially filtering lysates obtained from clonal isolates through GC50 glass fiber filters (1.2-μm nominal pore size; Micro Filtration Systems) and 0.45-μm-pore-size polyvinylidene difluoride Durapore filters (Millipore) to remove cell debris while retaining the large morphotype of OIs1 and stored at 4°C. The clonal isolate of HaRNAV was isolated from Strait of Georgia waters (British Columbia, Canada) as described by Tai et al. (34), and OIs1 was isolated from sediments collected in the Strait of Georgia, British Columbia, Canada.
Exponentially growing cultures of H. akashiwo (600 ml) were inoculated with 1% (vol/vol) HaRNAV or OIs1 in duplicate. Noninfected cultures served as a control. Samples for in vivo fluorescence and FCM were taken every 6 h for 59 h, after which a final sample was taken at 71 h for those cultures that still contained live cells.
Analyses.
Algal cell counts and two viability assays were performed immediately after sampling on unfixed material by use of a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) equipped with a 15-mW, 488-nm, air-cooled argon ion laser and a standard filter setup (5, 21). The trigger was set on red autofluorescence, and samples were run for 1 min at a delivery rate of 150 μl min−1. Sheath fluid for both assays consisted of filtered seawater (0.2-μm-pore-size filter). Sphero Nile Red high-intensity particles (1.7 to 2.2 μm; Becton Dickinson, San Jose, CA) were added as an internal reference to all algal samples. Fluorescence data were normalized to the bead internal standard. Chl a autofluorescence per cell was examined using FCM. A window (window A) was set around cells with relatively high red autofluorescence in the control culture (t = 0) (ca. 100% of the cells), and cells within this window were analyzed.
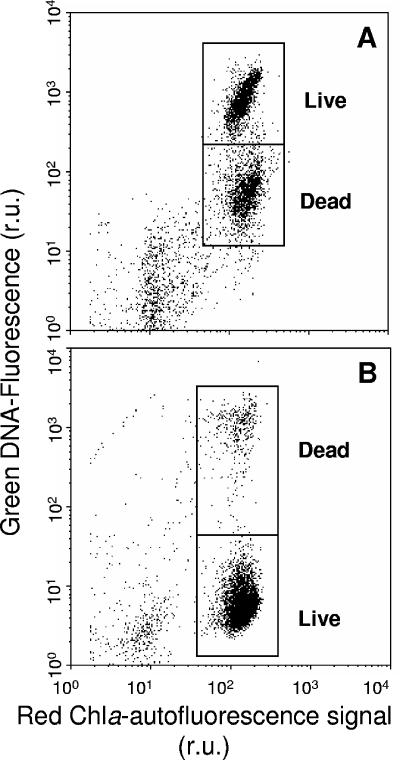
Metabolically active (“live”) cells were detected using the dye fluorescein diacetate (FDA) (Sigma-Aldrich), which readily diffuses into cells and becomes fluorescent upon hydrolysis by intracellular esterases (16). FDA was used instead of calcein-AM (5), as the latter gave unsatisfactory results with H. akashiwo. Prior to experiments, the FDA assay was optimized for dye concentration and incubation time. Cells were stained with FDA to a final concentration of 2 nM FDA (using a 2 mM working stock of FDA in pure acetone stored at −20°C) and incubated in the dark at in situ temperature (20°C) for 30 min. Incubations of 20 to 40 min gave optimal green fluorescence of live cells without significant increases in signal from fixed dead cells. No difference was observed for incubations in the dark or in situ light. Figure 1A shows a typical example of live and dead subpopulations of virally infected H. akashiwo cells after staining with FDA.
FIG. 1.
Representative biparametric plots of green fluorescence versus chlorophyll red autofluorescence of virally infected cultures of H. akashiwo after staining with green fluorescent live/dead stains (A) FDA and (B) SYTOX green. r.u., relative units.
Physiologically “dead” cells were enumerated after staining with SYTOX green (Invitrogen, Carlsbad, CA), a nucleic acid-specific stain that penetrates cells with compromised membranes but does not cross membranes of live cells. Dye concentrations (0.25 to 1.0 μM) and incubation times (5 to 30 min) were tested to determine the optimal staining conditions of a 10-min incubation with 0.5 μM SYTOX green (final concentration) in the dark at room temperature. Figure 1B shows the enhanced green fluorescence upon staining a dead subpopulation of virally infected H. akashiwo cells with SYTOX green. The low-level fluorescence detected from the live subpopulation is due to chlorophyll autofluorescence.
Viruses and bacteria were enumerated with FCM using SYBR green I stain (Invitrogen) after fixation with glutaraldehyde (0.5% final concentration, EM grade) and freezing in liquid nitrogen according to Brussaard (2). Since tests with other ssRNA viruses done using SYBR green II did not provide better results (4), we used SYBR green I for all analyses. All samples were zeroed with a buffer blank (Tris-EDTA buffer with sterile filtered seawater [0.2-μm-pore-size filter]), and the infected cultures were corrected for background counts using uninfected control cultures. Unfortunately, several virus samples of the HaRNAV-infected cultures were lost prior to analysis.
Flow cytometric parameters were collected as logarithmic signals and analyzed with CYTOWIN (freely available at http://www.sb-roscoff.fr/Phyto/index.php). For analyses of the collected data, the discrimination between various subpopulations was set for an example typical of the uninfected controls and kept constant for all samples.
RESULTS
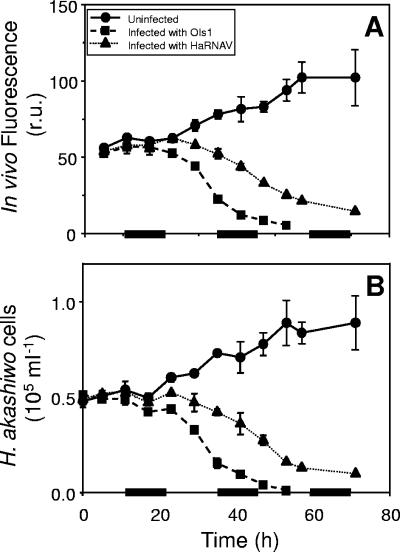
Total in vivo chl a fluorescence of the control and infected Heterosigma akashiwo cultures tracked very well with the total cell numbers for each of the cultures. Chl a fluorescence and cell number in the control cultures increased until the experiment was terminated (Fig. 2). Fluorescence and cell number of HaRNAV-infected cultures departed from levels for the control cultures after 23 h and decreased slowly over the remainder of the 71 h (Fig. 2). Natural fluorescence and cell abundance declined faster for OIs1-infected cultures than for HaRNAV-infected cultures (Fig. 2). Fluorescence of OIs1-infected cultures started to deviate from that of the control cultures after 17 h, and complete lysis of the infected cultures was reached 53 h after infection.
FIG. 2.
Biomass of H. akashiwo cultures measured by (A) in vivo natural fluorescence and (B) cell abundance of cultures infected with either HaRNAV or OIs1 compared to uninfected control cultures. Bars on the x axis indicate the dark periods. Experiments were performed in duplicate; mean values are presented, with standard errors of the means indicated by bars (if not visible, the error is smaller than the symbol). r.u., relative units.
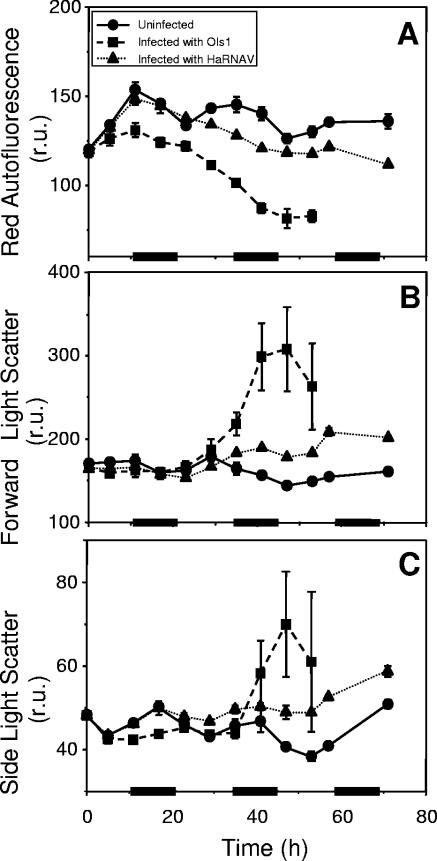
Cells in the control cultures (window A) exhibited a diel rhythm of chl a autofluorescence, with the maximum level corresponding to the end of the light phase (Fig. 3A). For the virus-infected cultures, the number of cells in window A decreased due to cell lysis, and the average autofluorescence of these cells declined with respect to that of the control cultures throughout the experiment (Fig. 3A). Cellular chl a autofluorescence in OIs1-infected cells deviated from that of the uninfected cells after 6 h, whereas the HaRNAV-infected cells showed cellular fluorescence comparable to that of the controls until 23 h after infection (Fig. 3A).
FIG. 3.
Cellular characteristics (normalized to fluorescent 1-μm beads) of H. akashiwo cultures as obtained by FCM. Presented are the natural parameters red autofluorescence, forward light scatter, and side-angle light scatter. Bars on the x axis indicate the dark periods. Experiments were performed in duplicate; mean values are presented, with standard errors of the means indicated by bars (if not visible, the error is smaller than the symbol). r.u., relative units.
Forward scatter, which is affected by cell size (28), increased considerably in OIs1-infected cells after 29 h (Fig. 3B). Forward scatter of HaRNAV-infected cells was only slightly enhanced compared to that of the uninfected control cells. The side scatter signal, which is thought to be related to internal structure and morphology (30), showed a similar but less distinct response to viral infection (Fig. 3C).
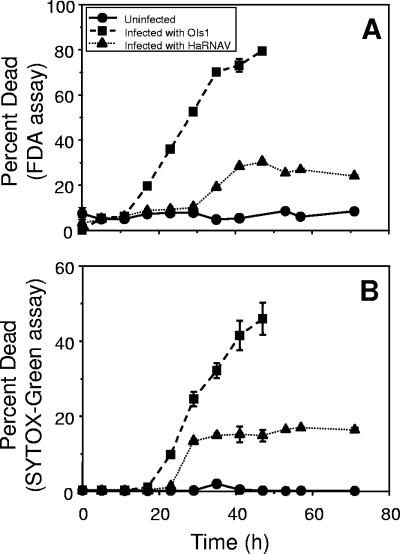
By use of the live/dead stains FDA and SYTOX green, H. akashiwo cells in the uninfected control cultures were found to be in good physiological condition throughout the experiment (Fig. 4). Independent of the type of live/dead stain, the percentage of dead OIs1-infected cells started to increase earlier than did the percentage of HaRNAV-infected cultures (17 to 23 h versus 29 to 35 h) and continued to increase until complete lysis 48 h postinfection. In contrast, the percentage of dead HaRNAV cells leveled off at a maximum of 30% according to the FDA assay. For OIs1-infected cultures, both assays gave comparable data, except that the maximum percentage of dead cells was higher by use of the FDA stain. This was not the case for HaRNAV-infected cultures, where there was a steep increase in the percentage of SYTOX-dead cells 29 h after infection with HaRNAV, although the cells still stained with FDA. Six hours later, however, the percentages of dead HaRNAV-infected cells were equal by use of the FDA and SYTOX green assays.
FIG. 4.
Percentages of dead cells of H. akashiwo cultures infected with either HaRNAV or OIs1 compared to percentages of dead cells of uninfected cultures. Live/dead stains used were FDA and SYTOX green. The percentage of dead cells was obtained directly from the percentage of fluorescently labeled dead cells when using the SYTOX green assay or indirectly as the inverse of the percentage of fluorescently labeled live cells when using the FDA assay. Experiments were performed in duplicate; mean values are presented, with standard errors of the means indicated by bars (if not visible, the error is smaller than the symbol).
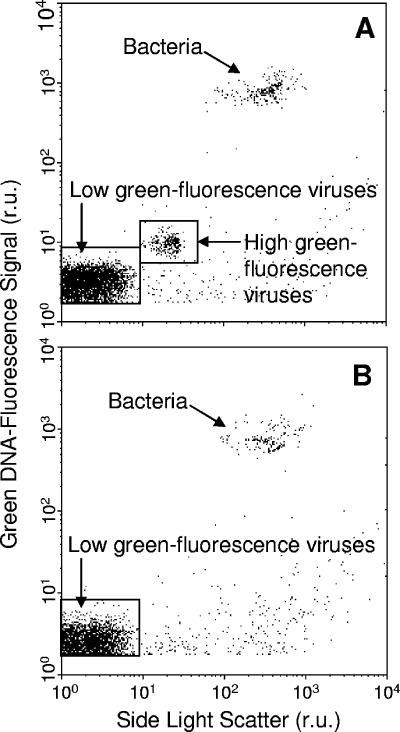
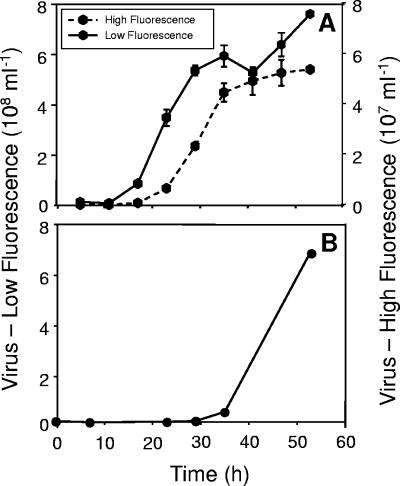
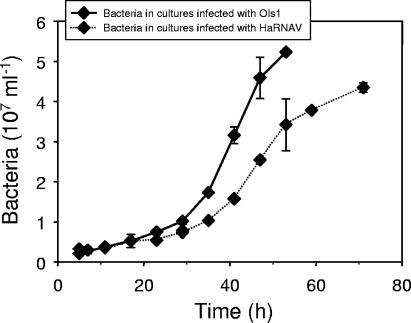
FCM allowed clear discrimination of two virus populations in OIs1-infected cultures, while only one population of viruses, which exhibited low fluorescence, was present in the HaRNAV-infected cultures (Fig. 5A). The high-fluorescing population of viruses produced in the OIs1-infected cultures began to appear after 17 h, while the low-fluorescing population appeared after 11 h (Fig. 6A). Viruses did not appear in HaRNAV-infected cultures until 28 h postinfection (Fig. 6B). Bacteria were also detected in all of the cultures (Fig. 5A); however, their abundance did not increase substantially until the onset of cell lysis at ∼20 h (Fig. 7).
FIG. 5.
Biparametric dot plots of green fluorescence versus side scatter representing virus systems (A) OIs1 and (B) HaRNAV. Viruses were stained with the nucleic acid-specific green fluorescent dye SYBR green I. The low-green-fluorescence virus population also included bacteriophages. r.u., relative units.
FIG. 6.
Flow cytometric enumeration of H. akashiwo virus systems (A) HaV-OIs1 and (B) HaRNAV. The HaNAV-OIs1 system consisted of two virus morphotypes, one with a high green fluorescence and the other with a low green fluorescence after staining with a nucleic acid-specific dye. The HaRNAV system consisted of only one morphotype with a very low green fluorescence. Data were corrected for the blank and the uninfected controls. Experiments were performed in duplicate; mean values are presented, with standard errors of the means indicated by bars (if not visible, the error is smaller than the symbol).
FIG. 7.
Variation in bacterial abundance in H. akashiwo cultures infected with OIs1 or HaRNAV. Data were corrected for the blank and the uninfected controls. Experiments were performed in duplicate; mean values are presented, with standard errors of the means indicated by bars (if not visible, the error is smaller than the symbol).
DISCUSSION
FCM permits rapid, single-cell analysis of phytoplankton populations. Here we have used FCM to examine the lytic cycles of the viruses HaRNAV and OIs1 in Heterosigma akashiwo and shown that the characteristics of viral infection were virus dependent.
OIs1-infected cultures showed initial signs of infection 6 h earlier than HaRNAV-infected cultures (17 versus 23 h) as indicated by in vivo fluorescence and cell abundance and 17 h earlier as indicated by the relative fluorescence levels of individual cells (6 versus 23 h). The results from the live/dead assays supported this finding; the percentage of dead cells increased after 11 to 17 h for OIs1-infected cultures but only after 23 to 29 h for the HaRNAV-infected cultures. This provides us with estimates for the latent periods of 11 to 17 h for OIs1 and 23 to 29 h for HaRNAV.
The live/dead stains provided insight into specific physiological changes occurring during infection. OIs1-infected cells lost intracellular esterase activity (FDA assay) more rapidly than membrane permeability increased (SYTOX green assay), whereas the opposite occurred in HaRNAV-infected cultures. In a previous study, TEM also showed that OIs1 produced viruses earlier than HaRNAV, as only 10% of HaRNAV-infected cells showed visible signs of infection 24 h postinfection, compared to 93% of OIs1-infected cells (18). While the maximum percentage of dead HaRNAV-infected cells was low by either of the live/dead assays, in vivo fluorescence and cell numbers indicated that lysis occurred in most HaRNAV-infected cells. This suggests that dying cells undergo rapid lysis, thus preventing a standing stock of dead cells, as reported previously for Phaeocystis pouchetii (5). In contrast, dead and dying cells infected with OIs1 accumulate in the culture prior to lysis.
In contrast, changes in light scattering were similar in H. akashiwo cultures infected with either OIs1 or HaRNAV. In either case, the light scattering signals were higher in infected cells than in uninfected control cells. Enhanced forward scatter has also been reported previously for infected Micromonas pusilla cells and was related to swelling of the cells (6). However, the scatter increased more and for a longer time in cells infected with OIs1 than in cells infected with HaRNAV, possibly because of the proliferation of the two virus morphotypes.
High (OIs1HI)- and low (OIs1LO)-fluorescence populations of virus-sized particles were observed for OIs1-infected cultures. As the OIs1 virus strain consists of two different morphotypes infecting and replicating inside individual cells at the same time (J. Lawrence, unpublished data), OIs1HI most likely represents the larger-diameter virus with the larger genome size (80 nm and 130 kb), while OIs1LO is likely the smaller virus (30 nm and 20 kb). Although bacteriophages are also included in OIs1LO, this low-fluorescence population grew rapidly after 10 h (Fig. 6A) while there was no change in the bacterial population at this time that could be attributable to phage release (Fig. 7). Additionally, the control cultures contained, at maximum, 6.4 × 105 ml−1 viruses throughout the experiment, or about 2% of total virus counts in the infected cultures.
The small-genome ssRNA HaRNAV (8.5 kb) could not be separated from the phages (Fig. 5B), nor did it increase in abundance earlier than the bacteria (Fig. 6B and 7). While we cannot be certain we were detecting HaRNAV, other ssRNA viruses have been stained successfully with fluorescent dyes, such as SYBR green I and II, OliGreen, and PicoGreen (4, 27). Bacteria rapidly accumulate and respond to the products of viral lysis of phytoplankton (7, 10), and the delayed increase in bacterial abundance of HaRNAV-infected cultures was likely due to the delayed lysis of HaRNAV-infected compared to OIs1-infected cultures.
The latent period of OIs1HI was approximately 6 h longer than that of OIs1LO (17 h instead of 11 h) (Fig. 6A). Therefore, the appearance of OIs1LO precedes lysis of the host cell, while release of OIs1HI occurs concomitantly with cellular lysis. The latent period of either particle is noticeably shorter than the 30 to 33 h for HaV, a large dsDNA virus infecting H. akashiwo (26). The latent period of HaRNAV was at least three times longer than that of the OIs1 model system.
Based on the FCM virus counts, we estimate the virus-to-alga host ratio at the start of the experiment for OIs1HI to be ∼7:1, which should result in near-synchronous infection of the cells. The virus-to-host ratio at the start of the experiment for OIs1LO and HaRNAV could not be determined, because the number of phages introduced with the algal viruses was unknown. From the net production of OIs1HI and net loss of H. akashiwo cells, we estimate that 1,100 large OIs1 particles were produced per cell. In contrast, if most of the low-fluorescence virus population prior to 35 h is attributed to OIs1LO then ∼16,000 small OIs1 particles were produced per cell. Estimates of viral production for HaRNAV are even less certain, but if most of the particles produced prior to 53 h were HaRNAV, then ∼21,000 viruses cell−1 were produced.
The two virus systems examined in this study were isolated from nearby areas. Their coexistence presents a paradox reminiscent of the paradox of the plankton, referring to species that “coexist in a relatively isotropic or unstructured environment all competing for the same sorts of materials” (15) rather than competitively excluding each other (12). In our case, two unrelated viruses, and likely many others, coexist in the same environment and compete for the same host when we might expect one virus to outcompete the other. Harris' (13) explanation for the paradox of the plankton is that the ocean is not homogeneous. Equilibrium is rarely attained; the frequency and intensity of environmental fluctuations are high enough that competitive exclusion is disrupted (8). The paradox of the viruses can likewise be explained by heterogeneity, but in this case it may be biological variability and the inherent dynamics of phytoplankton blooms that allow two virus systems infecting the same host to coexist. The latent period of OIs1 is shorter than that of HaRNAV. The rapid release of progeny viruses following infection and the subsequent swift propagation of OIs1 infection lend this virus a competitive advantage over HaRNAV when the host density is high. Although the lytic cycle of HaRNAV is longer, HaRNAV also displays high viral production rates. Thus, HaRNAV has an advantage when host density is low. In this manner, latent period and burst size interact with the rapidly fluctuating host densities inherent in phytoplankton blooms and may lead to one virus being temporarily favored over another on short time scales. If this variability is overlaid against complex patterns of host range (28, 29, 32) and decay rates of viral infectivity (9, 33), it seems apparent that the biological variability is great enough to allow the coexistence of viruses infecting the same host.
We are only beginning to appreciate the diversity of viruses that infect marine phytoplankton. As we continue to discover novel algal viruses and gain insight into their biology, we will be able to better comprehend their undoubtedly complex role in shaping marine microbial ecosystems.
Acknowledgments
This research was supported by NSERC Discovery and RTI grants to C. A. Suttle, a grant to C. P. D. Brussaard from the European Commission Research Programs Environment and Sustainable Development under contract EVK3-CT-1999-00015 BIOHAB, and an NSERC PDF to J. E. Lawrence.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Bratbak, G., J. K. Egge, and M. Heldal. 1993. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93:39-48. [Google Scholar]
- 2.Brussaard, C. P. D. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70:1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussaard, C. P. D., B. Kuipers, and M. J. W. Veldhuis. 2005. A mesocosm study of Phaeocystis globosa population dynamics. I. Regulatory role of viruses in bloom control. Harmful Algae 4:859-874. [Google Scholar]
- 4.Brussaard, C. P. D., D. Marie, and G. Bratbak. 2000. Flow cytometric detection of viruses. J. Virol. Methods 85:175-183. [DOI] [PubMed] [Google Scholar]
- 5.Brussaard, C. P. D., D. Marie, R. Thyrhaug, and G. Bratbak. 2001. Flow cytometric analysis of phytoplankton viability following viral infection. Aquat. Microb. Ecol. 26:157-166. [Google Scholar]
- 6.Brussaard, C. P. D., R. Thyrhaug, D. Marie, and G. Bratbak. 1999. Flow cytometric analyses of viral infection in two marine phytoplankton species, Micromonas pusilla (Prasinophyceae) and Phaeocystis pouchetii (Prymnesiophyceae). J. Phycol. 35:941-948. [Google Scholar]
- 7.Cole, J. J., M. L. Pace, and S. Findlay. 1988. Bacterial production in fresh and saltwater ecosystems: a cross system overview. Mar. Ecol. Prog. Ser. 43:1-10. [Google Scholar]
- 8.Connell, J. H. 1978. Diversity in tropical rain forests and coral reefs. Science 199:1302-1310. [DOI] [PubMed] [Google Scholar]
- 9.Cottrell, M. T., and C. A. Suttle. 1995. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 40:730-739. [Google Scholar]
- 10.Gobler, C. J., D. A. Hutchins, N. S. Fisher, E. M. Cosper, and S. A. Sanudo-Wilhelmy. 1997. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 42:1492-1504. [Google Scholar]
- 11.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 12.Hardin, B. 1960. The competitive exclusion principle. Science 131:1292-1297. [DOI] [PubMed] [Google Scholar]
- 13.Harris, G. P. 1986. Phytoplankton ecology: structure, function and fluctuation. Chapman and Hall, New York, N.Y.
- 14.Honjo, T. 1993. Overview on bloom dynamics and physiological ecology of Heterosigma akashiwo, p. 33-42. In T. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier Science Publishers B.V., New York, N.Y.
- 15.Hutchinson, G. E. 1961. The paradox of the plankton. Am. Nat. 882:137-145. [Google Scholar]
- 16.Jochem, F. J. 2000. Probing the physiological state of phytoplankton at the single-cell level. Scientia Marina 64:183-195. [Google Scholar]
- 17.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, J. E., and C. A. Suttle. 2004. Effect of viral infection on sinking rates of Heterosigma akashiwo and its implications for bloom termination. Aquat. Microb. Ecol. 37:1-7. [Google Scholar]
- 19.Lawrence, J. E., A. M. Chan, and C. A. Suttle. 2001. A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae). J. Phycol. 37:216-222. [Google Scholar]
- 20.Lawrence, J. E., A. M. Chan, and C. A. Suttle. 2002. Viruses causing lysis of the toxic bloom forming alga Heterosigma akashiwo (Raphidophyceae) are widespread in coastal sediments of British Columbia, Canada. Limnol. Oceanogr. 47:545-550. [Google Scholar]
- 21.Marie, D., F. Partensky, D. Vaulot, and C. P. D. Brussaard. 1999. Enumeration of phytoplankton, bacteria, and viruses in marine samples, p. 11.11.11-11.11.15. In J. Robinson (ed.), Current protocols in cytometry, vol. supplement 10. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 22.Murray, A. G., and G. A. Jackson. 1992. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms on other particles. Mar. Ecol. Prog. Ser. 89:103-116. [Google Scholar]
- 23.Nagasaki, K., and M. Yamaguchi. 1997. Isolation of a virus infectious to the harmful bloom causing microalga Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 13:135-140. [Google Scholar]
- 24.Nagasaki, K., M. Ando, I. Imai, S. Itakura, and Y. Ishida. 1994. Virus-like particles in Heterosigma akashiwo (Raphidophyceae): a possible red tide disintegration mechanism. Mar. Biol. 119:307-312. [Google Scholar]
- 25.Nagasaki, K., M. Ando, S. Itakura, I. Imai, and Y. Ishida. 1994. Viral mortality in the final stage of Heterosigma akashiwo (Raphidophyceae) red tide. J. Plankton Res. 16:1595-1599. [Google Scholar]
- 26.Nagasaki, K., K. Tarutani, and M. Yamaguchi. 1999. Growth characteristics of Heterosigma akashiwo virus and its possible use as a microbiological agent for red tide control. Appl. Environ. Microbiol. 65:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagasaki, K., Y. Tomaru, N. Katanozaka, U. Sirai, K. Nishida, S. Itakura, and M. Yamaguchi. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagasaki, K., and M. Yamaguchi. 1998. Intra-species host specificity of HaV (Heterosigma akashiwo virus) clones. Aquat. Microb. Ecol. 14:109-112. [Google Scholar]
- 29.Sahlsten, E. 1998. Seasonal abundance in Skagerrak-Kattegat coastal waters and host specificity of viruses infecting the marine photosynthetic flagellate Micromonas pusilla. Aquat. Microb. Ecol. 16:103-108. [Google Scholar]
- 30.Shapiro, H. M. 1988. Practical flow cytometry, 2nd ed. Wiley-Liss, New York, N.Y.
- 31.Suttle, C. A. 2000. The ecological, evolutionary and geochemical consequences of viral infection of cyanobacteria and eukaryotic algae, p. 248-286. In C. J. Hurst (ed.), Viral ecology. Academic Press, London, United Kingdom.
- 32.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar. Ecol. Prog. Ser. 92:99-109. [Google Scholar]
- 33.Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 35.Taylor, F. J. R., and R. Haigh. 1993. The ecology of fish-killing blooms of the chloromonad flagellate Heterosigma in the Strait of Georgia and adjacent waters, p. 705-771. In T. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier Science Publishers, B.V., New York, N.Y.