Abstract
Human adenoviruses (HAdV) and human polyomavirus JCPyV have been previously proposed as indicators of fecal viral contamination in the environment. Different wastewater matrices have been analyzed by applying real-time quantitative PCR procedures for the presence, quantity, and stability of a wide diversity of excreted HAdV and JCPyV. High quantities of HAdV and JCPyV were detected in sewage, effluent wastewater, sludge, and biosolid samples. Both viruses showed high stability in urban sewage. These results confirm the suitability of both viruses as indicators of human fecal viral pollution.
Two groups of DNA viruses have been proposed as indicators of the presence of human viral pathogens in the environment: human adenoviruses (HAdV) and human polyomaviruses (2, 13, 14). PCR-based procedures have been described for the detection of these viruses in environmental samples (2, 13). The aim of this study was to analyze the presence, quantity, and stability of HAdV and human polyomavirus JCPyV in different wastewater matrices. The applied real-time quantitative PCR (QPCR) procedures showed enough sensitivity to detect not only specific serotypes but also a wide diversity of excreted strains. The presence of hepatitis E virus (HEV), considered an emergent pathogen in developed countries, was also evaluated in this study by conventional nested-PCR techniques (5).
A total of 28 samples were obtained from a wastewater treatment plant located in the south of Barcelona (Spain) which treats the domestic and industrial wastewater from a population equivalent of 400,000 with a capability of 72,000 m3 per day. The treatment includes primary sedimentation and aerobic activated sludge digestion. The samples analyzed consisted of six raw sewage samples, seven treated wastewater samples, eight sludge samples (dry weight, 3.6% to 4%) and, finally, seven biosolid samples (dry weight, 25%).
Recovery of viral particles from sewage was carried out following a previously described procedure (13) presenting an estimated recovery efficiency of 34% for JCPyV. Recovery of viral particles from effluent wastewater was carried out following EPA procedure 600/4-84/013 (N14) (www.epa.gov/microbes/chapt14.pdf) combined with a second concentration procedure (13). The recovery efficiencies observed for this combined procedure were 0.15 to 0.16% for JCPyV and 2 to 25% for HAdV. Recovery of viral particles from sludge and biosolid samples was carried out by applying a method based on EPA 600/4-84/013 (R7) (www.epa.gov/nerlcwww/chap7.pdf) with minor modifications. Viral nucleic acids were extracted by a procedure described by Boom et al. (3) which have been previously detailed (7).
Two QPCR procedures based on the use of TaqMan probes (8, 9) were evaluated for their suitability in the analysis and quantification of HAdV present in different wastewater matrices (Table 1) . After comparing the two QPCR methods by statistically analyzing the results obtained by applying two different analysis of variation (ANOVA) tests, we concluded that assay 1 (9) was significantly different from assay 2 (8) in that it detected a higher number of HAdV (P = 0.012 in the repeated measures ANOVA for sewage and effluent wastewater; P = 0.004 in the repeated measures ANOVA for sludge and biosolids; P < 0.001 for both sewage and effluent wastewater and for sludge and biosolids in the nested hierarchical ANOVA). The specificity of assay 1 for detecting the diverse HAdV serotypes had been previously evaluated, and adenoviruses belonging to all six human species have been detected. However, some strains of animal adenoviruses excreted by farm animals may also be detected with the described protocol (data not shown).
TABLE 1.
Quantification by HAdV and JCPyV QPCR and presence of HEV evaluated by nested RT-PCR in different types of wastewater matrices obtained from a wastewater treatment plant
| Sample type | Date of collection (day/mo/yr) | Genome copies of:
|
Presence of HEV | ||
|---|---|---|---|---|---|
| HAdV (assay 1) | HAdV (assay 2) | JCPyV | |||
| Sewage | 23/11/2004 | 1.16 × 105/ml | 1.40 × 104/ml | 7.99 × 103/ml | − |
| 29/11/2004 | 9.54 × 104/ml | 3.12 × 104/ml | 6.76 × 103/ml | + | |
| 21/12/2004 | 1.58 × 104/ml | 8.59 × 103/ml | 8.39 × 103/ml | + | |
| 07/02/2005 | 4.76 × 102/ml | NTa | 2.50 × 103/ml | + | |
| 07/03/2005 | 2.56 × 103/ml | NT | 8.92 × 103/ml | NT | |
| 11/04/2005 | 2.07 × 103/ml | NT | 1.15 × 103/ml | + | |
| Effluent | 23/11/2005 | 3.25 × 103/liter | 1.16 × 103/liter | 4.21 × 102/liter | − |
| 29/11/2004 | 9.03 × 103/liter | 7.80 × 103/liter | 9.69 × 102/liter | NT | |
| 21/12/2004 | 1.78 × 103/liter | 5.94 × 102/liter | 2.33 × 102/liter | − | |
| Sludgeb | 21/11/2004 | 5.06 × 103/ml | 4.33 × 103/ml | 3.95 × 104/ml | NT |
| 29/11/2004 | 1.26 × 103/ml | 3.94 × 101/ml | 6.00 × 101/ml | NT | |
| 21/12/2004 | 8.05 × 102/ml | 1.54 × 102/ml | 6.90 × 102/ml | − | |
| 25/01/2005 | 6.49 × 103/ml | 5.30 × 103/ml | 1.07 × 102/ml | NT | |
| 25/01/2005 | 1.04 × 104/ml | 2.29 × 102/ml | 2.80 × 101/ml | NT | |
| 07/02/2005 | 1.11 × 104/ml | 7.73 × 102/ml | 2.40 × 101/ml | NT | |
| 07/02/2005 | 4.97 × 102/ml | 3.67 × 101/ml | 6.02 × 103/ml | NT | |
| 01/06/2004 | 1.67 × 103/ml | 7.25 × 102/ml | 4.54 × 101/ml | NT | |
| Biosolidc | 23/11/2004 | 4.34 × 105/g | NT | NT | − |
| 29/11/2004 | 2.45 × 103/g | 2.43 × 102/g | 4.24 × 103/g | − | |
| 25/01/2005 | 1.18 × 103/gl | 1.02 × 104/g | 7.71 × 103/g | + | |
| 25/01/2005 | 2.17 × 103/g | 5.00 × 102/g | 4.39 × 103/g | NT | |
| 07/02/2005 | 5.96 × 102/g | 4.05 × 102/g | 6.20 × 102/g | NT | |
| 07/02/2005 | 2.02 × 103/g | 2.94 × 102/g | 3.42 × 102/g | + | |
| 01/06/2004 | 2.37 × 103/g | 8.09 × 102/g | 2.03 × 103/g | NT | |
NT, not tested.
Corresponding to 0.18 to 0.2 g of dry weight.
Corresponding to 0.25 g of dry weight.
HAdV assay 1 had been performed using probe AdP1. A second probe, AdP2, originally designed to better detect some adenovirus serotype B strains, did not improve the results when added to AdP1 in the analysis of environmental samples (data not shown).
The quantification of JCPyV has been evaluated using a QPCR assay (11) that showed to be specific and highly sensitive for JCPyV, which is a very stable DNA virus presenting a low level of genetic variability. To the best of our knowledge, this is the first attempt to detect JCPyV in treated urban sewage and in the sludge and/or biosolids generated in wastewater treatment plants (Table 1).
The sensitivities of both QPCR assays applied in this study were estimated to be of 1 to 10 genome copies (GC).
High concentrations of HAdV and JCPyV were found in sludge and biosolid samples (Tables 1 and 2) . HEV RNA was also detected in 6/12 analyzed samples by seminested reverse transcription-PCR (RT-PCR) with degenerated primers (6) as previously described (5) by use of a one-step RT-PCR procedure (QIAGEN OneStep RT-PCR kit) (Table 1). HEV strains were typed as genotype 3, but in two samples genotype 1 was observed. HEV has traditionally been considered nonendemic in industrialized areas, but several isolates of the virus have been recently identified in these areas. Previous studies carried out in Barcelona revealed a high frequency of HEV-positive sewage samples (43.5%) and identified clinical cases of sporadic acute hepatitis E (4, 5, 12).
TABLE 2.
Mean values and sigma for HAdV and JCPyV GC in the different wastewater matrices analyzeda
| Type of sample analyzed | HAdV assay 1
|
HAdV assay 2
|
JCPyV
|
|||
|---|---|---|---|---|---|---|
| Mean value | Sigma | Mean value | Sigma | Mean value | Sigma | |
| Sewage (ml) | 3.87 × 104 | 3.78 × 103 | 1.79 × 104 | 4.58 × 103 | 6.11 × 103 | 2.58 × 103 |
| Effluent (ml)b | 4.69 | 4.69 | 3.19 × 100 | 0.95 | 0.63 | 0.28 |
| Sludge (g [dry wt]) | 1.83 × 102 | 8.64 × 101 | 5.50 × 101 | 2.36 × 101 | 2.35 × 102 | 1.02 × 102 |
| Biosolid (g [dry wt]) | 1.59 × 104 | 2.24 × 104 | 5.19 × 102 | 1.89 × 102 | 8.05 × 102 | 5.94 × 102 |
Sigma is the repeatability error for any individual measure and consists of the square root of the mean square error of the one-way ANOVA. Results are expressed as GC/ml for wastewater or GC/g of dry weight for sludge and biosolids.
Samples were concentrated using electropositive filters and organic flocculation.
The recovery efficiency of the concentration method used for sewage is much higher than that for the method applied to treated water samples, which are processed using filtration-elution methods. By applying the procedure used for sewage to recover viruses from 40 ml of treated effluent wastewater, higher values for HAdV were obtained (four samples; mean value = 8.08 × 101 GC/ml).
Enzymatic inhibition has been observed by other authors when applying this methodology to environmental samples (10). In this study, enzymatic inhibition was also observed when external controls, known quantities of target, were added to nucleic acid extraction from the environmental samples without previous dilution of the extractions. However, the high level of viral contamination in this kind of sample allows the preparation of dilutions, thus avoiding inhibitory conditions and producing reliable information on viral concentration; this reliability has been proven by the statistical analysis of two different 10-fold dilutions (1:10 and 1:100) of the sample tested (P = 0.281 and 0.430 for sewage/effluent wastewater and sludge/biosolids, respectively).
In order to study the stability of HAdV and JCPyV in sewage samples, raw sewage was kept in nonhermetic glass containers in a 20°C thermostatic room, and aliquots of 42 ml were collected and concentrated at days 0, 1, 4, 7, 14, 21, 28, 42, 84, 105, 135, 168, 225, and 300 as described above. Viral genomes present in the samples were evaluated by the described QPCR assays. Sterile spiked phosphate-buffered saline (PBS) was kept as a control for the experiment. Samples were tested in two parallel assays with and without previous treatment with DNase before the nucleic acid extraction in order to identify potential free DNA in the samples. The analysis applied, using simple linear regression, had previously been described when the stability of human polyomaviruses in sewage samples was studied by use of limiting-dilution nested-PCR assays (1).
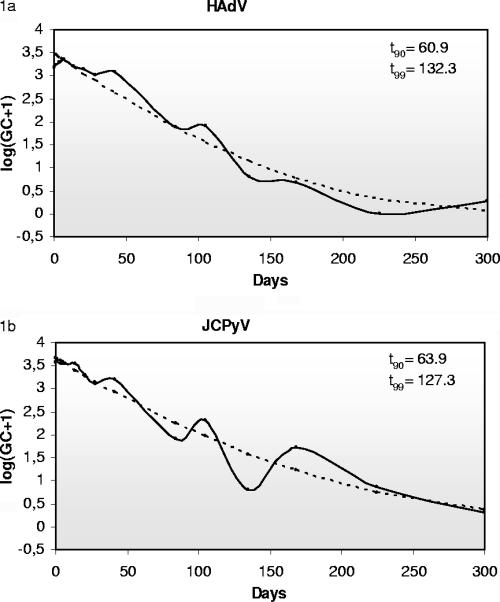
The t90 (time required to observe a reduction of 90% in the initial viral concentration) and t99 values have been calculated according to the regression curve obtained. A t90 of 60.9 days and a t99 of 132.3 days for HAdV (Fig. 1a) and a t90 of 63.9 days and t99 of 127.3 days for JCPyV (Fig. 1b) were estimated. When DNase was added, we estimated a t90 of 55.8 days and a t99 of 126.1 days for HAdV and a t90 of 59.3 days and t99 of 121.4 days for JCPyV. As expected, the studied viruses showed higher stability in PBS than in sewage. The estimated t90 values for HAdV and JCPyV in PBS were 236.8 and 110.8 days, respectively, and were 140.7 and 80.8 days when samples were pretreated with DNase. The high stability of the HAdV observed is in agreement with what has been previously described (15). No remarkable differences have been noticed when DNase was added to the experiment, and the genomes detected presumably represent full viral particles. The infectivities of viruses present in the analyzed samples have not been tested due to the difficulty of culturing low concentrations of these environmental strains, especially JCPyV strains, which typically present archetypal regulatory regions and are very difficult to grow in cell culture.
FIG. 1.
Stability of HAdV (1a) and JCPyV (1b) in sewage samples. The regression (discontinuous line) and transformed (solid line) values of the averages of GC detected by QPCR in the three samples (sewage) are represented.
Adenoviruses present in urban sewage samples kept at 20°C after 4, 42, and 105 days were amplified by nested PCR (1), and the amplicons obtained were cloned and sequenced in order to study the variability of the serotypes present in the samples through time. Three clones of two different nucleic acid extractions per day were analyzed; thus, six clones per day were obtained in the end. Nucleotide sequences corresponding to serotypes 40, 41, 31, 34, 35, 11, and 12 were detected over the 105 analyzed days, showing no significant differences in stability between the different serotypes present during the experiment.
The specific detection and quantification by QPCR of HAdV and JCPyV in different wastewater matrices might serve as a useful tool for indicating the presence of viral pathogens and for evaluating the efficiency of virus removal in wastewater treatment plants and the risk associated to the applications of biosolids in agricultural practices, as well as for controlling the virological quality of recycled water.
Acknowledgments
During the development of this study, Nestor Albinana-Gimenez was a fellow of the Generalitat de Catalunya. This study was partially founded by Centre de Referència D'R+D en Biotecnologia de Catalunya (CERBA) and Grup de Recerca Consolidat de la Generalitat de Catalunya “Grup de Microbiologia d'aigües relacionades amb la salut.”
We thank Serveis Científico Tècnics of the University of Barcelona for the sequencing of PCR products.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Bofill-Mas, S., M. Formiga-Cruz, P. Clemente-Casares, F. Calafell, and R. Girones. 2001. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J. Virol. 75:10290-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bofill-Mas, S., S. Pina, and R. Girones. 2000. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 66:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buti, M., P. Clemente-Casares, R. Jardi, M. Formiga-Cruz, M. Schaper, A. Valdes, F. Rodriguez-Frias, R. Esteban, and R. Girones. 2004. Sporadic cases of acute autochthonous hepatitis E in Spain. J. Hepatol. 41:126-131. [DOI] [PubMed] [Google Scholar]
- 5.Clemente-Casares, P., S. Pina, M. Buti, R. Jardi, and R. Gironés. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erker, J. C., S. M. Desai, and I. K. Mushahwar. 1999. Rapid detection of hepatitis E virus RNA by reverse transcription-polymerase chain reaction using universal oligonucleotide primers. J. Virol. Methods 81:109-113. [DOI] [PubMed] [Google Scholar]
- 7.Formiga-Cruz, M., A. Hundesa, P. Clemente-Casares, N. Albiñana-Gimenez, A. Allard, and R. Girones. 2005. Nested multiplex PCR assay for detection of human enteric viruses in shellfish and sewage. J. Virol. Methods 125:111-118. [DOI] [PubMed] [Google Scholar]
- 8.Heim, A., C. Ebnet, G. Harste, and P. Pring-Akerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 9.Hernroth, B. E., A. C. Conden-Hansson, A. S. Rehnstam-Holm, R. Girones, and A. K. Allard. 2002. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 68:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, S., H. Dezfulian, and W. Chu. 2005. Real-time quantitative PCR for enteric adenovirus serotype 40 in environmental waters. Can. J. Microbiol. 51:393-398. [DOI] [PubMed] [Google Scholar]
- 11.Pal, A., L. Sirota, T. Maudru, K. Peden, and A. M. Lewis, Jr. 2006. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods 135:32-42. [DOI] [PubMed] [Google Scholar]
- 12.Pina, S., M. Buti, M. Cotrina, J. Piella, and R. Girones. 2000. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J. Hepatol. 33:826-833. [DOI] [PubMed] [Google Scholar]
- 13.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puig, M., J. Jofre, F. Lucena, A. Allard, W. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson, S. S., J. L. Jackson, M. Suva-Castillo, W. A. Yanko, Z. El Jack, J. Kuo, C. L. Chen, F. P. Williams, and D. P. Schnurr. 2003. Detection of infectious human adenoviruses in tertiary-treated and ultraviolet-disinfected wastewater. Water Environ. Res. 75:163-170. [DOI] [PubMed] [Google Scholar]