Abstract
Several models (animal caliciviruses, poliovirus 1 [PV1], and F-specific RNA bacteriophages) are usually used to predict inactivation of nonculturable viruses. For the same UV fluence, viral inactivation observed in the literature varies from 0 to 5 logs according to the models and the methods (infectivity versus molecular biology). The lack of knowledge concerning the mechanisms of inactivation due to UV prevents us from selecting the best model. In this context, determining if viral genome degradation may explain the loss of infectivity under UV radiation becomes essential. Thus, four virus models (PV1 and three F-specific RNA phages: MS2, GA, and Qβ) were exposed to UV radiation from 0 to 150 mJ · cm−2. PV1 is the least-resistant virus, while MS2 and GA phages are the most resistant, with phage Qβ having an intermediate sensitivity; respectively, 6-log, 2.3-log, 2.5-log, and 4-log decreases for 50 mJ · cm−2. In parallel, analysis of RNA degradation demonstrated that this phenomenon depends on the fragment size for PV1 as well as for MS2. Long fragments (above 2,000 bases) for PV1 and MS2 fell rapidly to the background level (>1.3-log decrease) for 20 mJ · cm−2 and 60 mJ · cm− 2, respectively. Nevertheless, the size of the viral RNA is not the only factor affecting UV-induced RNA degradation, since viral RNA was more rapidly degraded in PV1 than in the MS2 phage with a similar size. Finally, extrapolation of inactivation and UV-induced RNA degradation kinetics highlights that genome degradation could fully explain UV-induced viral inactivation.
Noroviruses, including the RNA genome, are nonculturable viruses of major concern for the safety of drinking water because they are an important cause of waterborne disease (14). Therefore, viral models should be used to estimate their loss of infectivity due to UV radiation. Depending upon the model virus (animal caliciviruses, enteric viruses such as poliovirus 1 [PV1], or RNA phages such as the MS2 phage) and the methods (infectivity versus molecular biology), the UV inactivation of human noroviruses can be estimated between 0 and 5 logs for a 20-mJ · cm−2 fluence (1, 5, 6, 12, 21, 25, 34, 35). Moreover, if only one model, such as animal caliciviruses, is considered, UV inactivation studies describe a 1.5- to 5-log unit decline for 20 mJ · cm−2 for both feline and canine caliciviruses (5, 6, 25, 34). Data obtained from these studies show a high variability for the UV susceptibility of viruses. The diversity of the proposed methods also highlights the lack of knowledge concerning the effect of UV on viral particles. Clear data will have to come forward defining the main parameter (i.e., genome size or capsid structure) responsible for viral inactivation before one or another approach can be favored.
Although UV inactivates viruses by altering their genome or capsid, most inactivation studies have focused attention on the influence of radiation on DNA rather than on RNA genomes. Specific targets of UV have been demonstrated among the bases constituting the viral genome. In both DNA and RNA, pyrimidines are more sensitive than purines; thymine and uridine are the most-sensitive bases in DNA and RNA, respectively (4, 7, 10). UV radiation generates different types of photoproducts (i.e., dimers or adducts) depending on several parameters such as the wavelength or the dose delivered (4, 15, 30, 36). At 254 nm, loss of viral infectivity could thus be associated with formation of photoproducts such as cyclobutane pyrimidine dimers or pyrimidine hydrates. Regarding the impact on proteins, UV radiation could affect protein integrity or conformation by breaking disulfur (S-S) bonds or creating cross-links between proteins and the genome, for example (4, 10). In the literature, a recent study hypothesized that viral inactivation by UV is mainly due to alterations in the genome (18). Therefore, these authors proposed using only the type and the size of the viral genome to predict viral inactivation. On the contrary, UV radiation also modifies the viral capsid (26).
In the present work, the objective was to determine if viral genome degradation might explain the loss of infectivity under UV radiation. UV photoproducts correspond to entities which deform or change the configuration, and sometimes the integrity, of RNA. Theoretically, this would block the primary function of the genome, which is to serve as a template during replication and translation to produce a new virion. We monitored genome degradation using reverse transcription (RT)-PCR, because with this technique the viral genome must undergo enzymatic transcription before detection. For the considered viruses, detection of viral RNA would thus decline with increasing fluence whatever the targeted fragment, a rationale previously used for DNA of irradiated phage M13 (16). Hence, the decrease of viral infectivity due to UV was monitored simultaneously with the effect of UV radiation on nucleic acids. The following four nonenveloped, culturable, single-stranded RNA viruses measuring 20 to 30 nm were used: poliovirus 1, phage MS2, phage Qβ, and phage GA. These models belong to two different families (Picornaviridae and Leviviridae) and two different genera within the Leviviridae family (Levivirus and Allolevivirus). The genome size varies somewhat (3,500 to 7,500 bases), as does capsid structure (Table 1).
TABLE 1.
Characteristics of the four virus models useda
| Family | Genus | Virus | Size in nm (avg) | No. of capsid proteins (%)b | Length of RNA in bases | No. of genes (%)c |
|---|---|---|---|---|---|---|
| Leviviridae | Levivirus | MS2 | 22.4-28.8 (26.3) | 1 (100) | 3,569 | 4 (100) |
| GA | 22.7-28.9 (26.3) | 1 (62) | 3,466 | 3 (56.3) | ||
| Allolevivirus | Qβ | 21.3-29.4 (27.0) | 1 (20) | 4,160 | 4 (NCS) | |
| Picornaviridae | Enterovirus | PV1 | 26.3-32.6 (30) | 4 (ND) | 7,440 | Polyprotein (ND) |
MATERIALS AND METHODS
Virus propagation and assay. (i) Vaccine poliovirus.
The Poliovirus 1 Sabin Lsc 2ab strain was propagated and assayed in Buffalo green monkey cell lines. Growing medium (Sigma; catalog no. M-5650) (minimum essential medium containing 5% fetal calf serum [Gibco; catalog no. 10500-056] and completed with 1% l-glutamine [Sigma; catalog no. G-7513] and 1% penicillin-streptomycin [Sigma; catalog no. P-4333] solutions) was decanted from 175-cm2 tissue culture flasks containing complete monolayers. Two milliliters of an approximate 106-most probable number of cytopathic units (MPNCU)-per-ml suspension was inoculated onto the monolayer. The cells were incubated at 37°C for 2 h, where the flasks were rocked every 30 min throughout the 2-h incubation. After allowing 2 h for virus attachment, the remaining volume was removed and 30 ml of maintenance medium (minimum essential medium with 1% fetal calf serum instead of 5%) was inoculated and incubated at 37°C for 24 to 48 h until viral replication produced a cytopathogenic effect (CPE) visualized by the destruction of the continuous monolayer. The cells were then exposed to three successive freeze-thaw cycles, and the supernatant was centrifuged at 4°C at 10,000 × g for 60 min in order to remove cell debris. Further purification of PV1 was accomplished by performing ultrafiltration through a 100,000-Da membrane (UFVABHK00; Millipore). The retentate recovered from the membrane was resuspended in phosphate-buffered saline. This solution consisting of the viral stock has a titer of 9 × 108 MPNCU/ml and was stored at −80°C. In the context of poliomyelitis eradication, such strains should be used by taking into account the WHO recommendations.
(ii) Bacteriophages.
MS2 (ATCC 15597-B1) and GA and Qβ (kindly provided by J. Jofre, University of Barcelona) phages were replicated according to the standard procedure (ISO 10705-1, 1997) without a CHCl3 lysis step using Escherichia coli Hfr K12 (ATCC 23631) as the bacteria host. The sample was centrifuged at 27,000 × g for 60 min at 4°C (Beckman; model J2-22), and the supernatant was filtered through a 0.22-μm-pore-size filter (Millipore; catalog no. SLGP033RS). Finally, the viral suspension was stored as stock suspension at 4°C and the final viral concentration was 1011 PFU/ml.
Cell culture quantification of infectious virus. (i) Poliovirus 1.
The most probable number quantification method was used for poliovirus 1. A 200-μl cell suspension containing 7 × 104 cells/ml in growing medium was mixed with 50 μl of the sample to be analyzed, or its dilution, and was introduced into 96-well microplates. Quantification was performed on three successive dilutions, each dilution being inoculated in 40 wells. A single microplate thus held two dilutions (80 wells) and 16 controls (cells without viral inoculum). The microplates were held at 37°C under 5% CO2 for six days. The number of wells of each dilution presenting a CPE was noted at this time. This provided three CPE readings for each sample. Results were expressed in MPNCU/ml.
(ii) Bacteriophages.
The double layer agar procedure (Norm ISO 10705-1, 1997) was used to quantify bacteriophage infectious units of MS2, GA, and Qβ in one milliliter. Briefly, a pure or diluted sample was mixed with 2.5 ml of molten agar (semisolid nutrient medium). One milliliter of the culture of the host strain E. coli Hfr K12 was added to the previous mixture and plated on a solid nutrient medium (tryptone-yeast-glucose agar). After an incubation time of 12 to 18 h at 37°C, the plaques were counted. Results were expressed in PFU/ml.
Viral genome characterization. (i) Viral RNA extraction.
Viral RNA was extracted by using a QIAamp viral RNA minikit (QIAGEN; catalog no. 52906). Extraction was performed on 140-μl samples, yielding the RNA extract in a 60-μl volume. All steps were performed in compliance with the manufacturer's instructions.
(ii) Probes and primers.
For poliovirus 1, the various primers and probes used for RT and PCR have been described in a previous report (28). Concerning MS2, the primers were designed with Primer Express software 1.2 (Applied Biosystems) applied to the MS2 genome sequence (GenBank accession no. NC001417). The resulting primers are described in Table 2.
TABLE 2.
Primers used for each amplified region of the MS2 genome during real-time RT-PCR
| Localization | Primer(s) | Sequence (5′ to 3′) | Position | Size (in bases) |
|---|---|---|---|---|
| 5′ NCa | 5′NC-f | 5′TGCTCAACTTCCTGTCGAGCT3′ | 24-44 | 111 |
| 5′NC-r | 5′CGCACAGGTCAAACCTCCTAG3′ | 114-134 | ||
| Capsid gene | Cap-f | 5′AAGTGGCAACCCAGACTGTTG3′ | 1534-1554 | 111 |
| Cap-r | 5′GCTCGCAGTCGGAATTCGT3′ | 1626-1644 | ||
| Replicase gene | Rep-f | 5′TTCTCCAACGGTGCTCCTATG3′ | 2145-2165 | 81 |
| Rep-r | 5′GGTAACGGTTGCTTGTTCAGC3′ | 2205-2225 | ||
| 3′ NC | 3′NC-r | 5′TCTTTCGAGCACACCCACC3′ | 3424-3442 | —b |
NC, noncoding region.
—, only for RT.
(iii) cDNA synthesis.
cDNA was synthesized from the extracted viral RNA following the protocol developed in our laboratory (28).
(iv) Real-time PCR.
Concerning poliovirus 1, real-time PCR was performed using the method originally developed in our laboratory (28) with minor modifications. DNase/RNase-free water (25 μl) and 14.5 μl of TaqMan universal master mix were used, instead of 14.5 μl and 25 μl, respectively. For bacteriophage MS2, 5 μl of cDNA was mixed with 25 μl of 2× TaqMan SYBR green master mix (Applera; catalog no. 4309155), 15 μl of nuclease-free water, and 2.5 μl of forward and 2.5 μl of reverse primers to obtain a final concentration of 0.5 μM, 0.75 μM, or 1 μM in the function of the primers used. Amplification and detection were performed for the two viruses with an ABI Prism 7700 sequence detection system (Perkin Elmer Inc.). The amplification ramp was carried out as follows: (i) 2 min at 50°C to activate the uracil N-glycosylase, and subsequently (ii) 10 min at 95°C to release the activity of the hot-start DNA polymerase, and finally (iii) 50 cycles of 15 s at 95°C and 60 s at 60°C. For MS2, the amplification stage was then followed by three temperature cycles, in order to dissociate double strands and obtain a typical fusion curve of the targeted fragment. The temperature cycles are as follows: 30 s at 95°C, 30 s at 60°C, slow ramping temperature from 60°C to 95°C performed in 19 min 59 s, and finally 1 min at 95°C. Real-time fluorescence measurements were obtained and directly analyzed using ABI Prism 7700 sequence detection system software. Concerning SYBR green, Dissociation Curve software was used to assess the specificity of the amplification of the different fragments only for MS2. Electrophoresis gels have also been made. In addition, several controls with and without primers and with and without RNA template were included to eliminate fluorescence from the nonspecific amplification.
The cycle threshold (CT) was calculated by determining the point at which fluorescence exceeded a threshold limit. This value is directly linked to viral genome quantity by following this equation: log10 (Qn/Qo) = −{[CT(n) − CT(o)]/3.33}, in which Qn/Qo represents the ratio of the concentration of viral genomes after irradiation at fluence n to the concentration of nonirradiated genomes, and CT(n) and CT(o) are, respectively, the CT obtained by the real-time RT-PCR after irradiation (n) and before irradiation (o). The 3.33 value represents a 100% efficiency of PCRs. Even if this is an approximation, this value should be a constant inside each system used. Changing this value did not change the value of the slope of the linear model applied to compare the different genome degradation kinetics. RT-specific efficiency was not defined for each system but should also be constant and should not change the previous slope value.
Low-pressure UV radiation system and radiometry.
UV inactivation and RNA degradation of both virus models spiked in phosphate buffer were conducted using a low-pressure mercury vapor lamp emitting monochromatic (253.7-nm) UV light. The collimated beam apparatus consisted of a single 10-W Slimline germicidal lamp (ozone-free Ster-L-Ray G12T6L; Atlantic UV Corporation) that was suspended horizontally in a metal box and emitted UV irradiation directed through a circular opening controlled by an automatic shutter according to the exposure time. UV radiance was measured with a radiometer according to the standard method (3). The UV intensity of each experiment was determined with a calibrated UV 254-nm detector (catalog no. IL-1800A, photodetector SED 240/NS254/W; International Light, Newburyport, Mass.) by placing the radiometer at the same location and elevation as the water surface of the irradiated sample. The average irradiance in the mixed suspension was determined by the UV absorbance of the test suspension at the wavelength of 254 nm, the sample depth, and the incident average irradiance. Required exposure times were calculated by dividing the desired UV fluence by the average UV irradiance. For uniform lamp output, the lamp was warmed up for at least 30 min before each experiment. A stir plate was placed directly under the collimated beam for slow stirring of the viral suspensions. Radiation experiments were performed at room temperature with continuous stirring. Linear regression analyses were carried out for comparisons of virus inactivation rates.
Degradation of viral genome and viral inactivation by UV.
Viral stock was used to study viral inactivation and viral genome degradation. Viral stock was introduced into a 250-ml glass vessel containing 158.4 ml of sterile phosphate buffer (constituted by mixing salts K2HPO4 and KH2PO4) at pH 7.2 to give a final concentration between 106 and 107 MPNCU/ml or PFU/ml. After homogenizing, the 160-ml sample was distributed into 10 petri dishes (60 by 15 mm) with 15 ml per dish, 1 ml being transferred to a disposable UV transparent cuvette (trUView cuvettes, catalog no. 170-2510; Bio-Rad, Hercules, CA) to measure absorption of the test suspension at 254 nm. Viral inactivation and genome degradation were noted as a function of the UV fluence delivered and the exposure time. Measurements were made at 0 (control), 20, 40, 50, 60, 75, 90, 100, 125, and 150 mJ · cm−2.
The degradation of genome fragments of different sizes was measured by associating different primers for RT and PCR. Degradation of the following fragments was monitored for PV1: a 76-base fragment in the 3C region (3C primers for RT and PCR); a 145-base fragment in the 5′ untranslated (UTR) region (5′ UTR primers for RT and PCR); a 1,869-base fragment (oligodT15 primer for RT and 3C primers for PCR); a 5,429-base fragment (3C reverse primer for RT and 5′ UTR primers for PCR); and a 6,989-base fragment representing nearly the entire viral genome (oligodT15 primer for RT and 5′ UTR primers for PCR). Degradation of the following fragments was monitored for MS2: an 81-base fragment in the replicase region (replicase primers for RT and PCR); a 111-base fragment in the capsid region (capsid primers for RT and PCR); a 111-base fragment in the 5′ UTR region (5′ UTR primers for RT and PCR); a 692-base fragment (replicase reverse primer for RT and capsid primers for PCR); a 1,298-base fragment (3′ UTR reverse primer for RT and replicase primers for PCR); and finally a 1,909-base fragment (3′ UTR reverse primer for RT and Cap region primers for PCR).
All the RT-PCR systems were different in terms of sensitivity, fluorescence (SYBR green versus TaqMan style probe), and reaction efficiency. Nevertheless, a decrease in fluorescence was defined for each system and modeled by a linear regression. The comparison between each system is then based on the slope. Differences for each system therefore should not influence the comparison.
All experiments and measurements of viral inactivation and genome degradation were taken in triplicate.
Statistical analysis.
A t test (Student test) was performed by using EXCEL (Microsoft Office 2003) for the inactivation kinetics for each virus as well as for the RNA degradation kinetics for each fragment of the virus. Linear regression analysis was also performed in order to verify the relationship between the viral inactivation and the fluence (dose dependence). The P values were computed and compared at the confidence level of 95%, or 0.05, in both cases.
RESULTS
Inactivation by UV radiation.
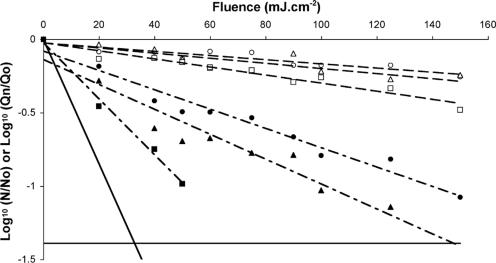
UV inactivation curves for PV1, MS2, GA, and Qβ are shown in Fig. 1. A dose-dependent relationship was first noted between inactivation and fluence as assessed by the linear regression test; the number of infectious units decreased with increasing fluence. The second observation concerned the shape of the curve. Bacteriophages MS2 (slope, −0.0389; r2 = 99.0%), GA (slope, −0.0375; r2 = 97.0%), and Qβ (slope, −0.0595; r2 = 96.2%) exhibited typical first-order inactivation kinetics. Tailing was not observed for any of these inactivation curves. However, a common sensitivity pattern to UV was not observed for the same family. Phages MS2 and GA (Levivirus) displayed equivalent sensitivity to UV (P > 0.05), whereas phage Qβ was inactivated significantly more quickly than the two others (P < 0.05). Unlike the phages, poliovirus 1 exhibited first a linear curve up to 60 mJ · cm−2 (slope, −0.1106; r2 = 97.9%), which then tailed off approaching the detection threshold. Residual infectious particles are detected until 125 mJ · cm−2. MS2 and GA thus appeared to be the most resistant to UV radiation, while PV1 was the most sensitive (P < 0.05). The inactivation curve for Qβ, which is from the Leviviridae family but belongs to the Allolevivirus genus, was intermediate (P < 0.05). No infectious virus particles were detected for a fluence delivered higher than 150 mJ · cm−2 regardless of the virus studied.
FIG. 1.
UV fluence response for bacteriophages MS2 (○), GA (⋄), and Qβ (▵) and for poliovirus 1 (□) in phosphate buffer. The dashed lines are regression lines for bacteriophages MS2 (— —), GA (- -), and Qβ (— - - —); the solid line is the regression line for poliovirus 1. Standard deviations are not plotted to avoid too many marks.
Impact of UV on viral RNA.
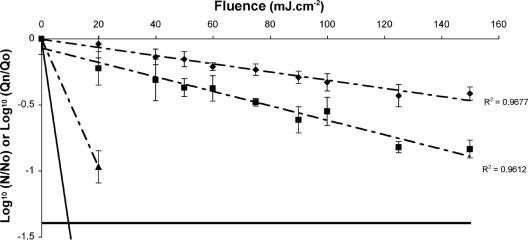
The effect of UV was investigated on five fragments of PV1 RNA: 76, 145, 1,869, 5,429, and 6,989 bases representing 1%, 2%, 25%, 73%, and 94%, respectively, of the genome. Six fragments were studied for MS2: 81, 111, 111, 692, 1,298, and 1,909 bases representing 2%, 3%, 3%, 19%, 36%, and 53%, respectively, of the genome. Since the RT and PCR steps involved different locations for long fragments (>400 bases), a positive signal was proof that a segment having a length equal to the distance between the two considered locations was present. All negative controls (with or without primers) were negative. When viral suspension was included, PCR produced specific amplification even if there were no primers during the RT step. This was considered to occur because of the presence of small oligonucleotides able to act as random primers during the RT step. Thus the amplification signal came from a specific region selected for PCR but unrelated to the distance between the locations selected for RT and PCR. Consequently, analyses were performed on signals obtained with and without primers during RT step. The signal obtained without primers was taken as a background level under which any signal observed was considered not to be a reliable measure of the size of the selected region. This background level was about 1 to 2 log units less than for RT-PCR with the specific primers. Results are given in Fig. 2 and 3 for MS2 and PV1, respectively.
FIG. 2.
Influence of the size of the targeted fragment on the kinetics of UV-induced degradation of MS2: 81 bases (○), 111 bases in the capsid region (▵), 111 bases in the 5′ UTR region (□), 692 bases (•), 1,298 bases (▴), and 1,909 bases (▪). The dashed lines are regression lines for small fragments (— —) and long fragments (— - —); the solid line is the regression line for infectivity and is given for reference. Standard deviations are not plotted to avoid too many marks. The horizontal black line designates the highest value for the background.
FIG. 3.
Influence of the size of the targeted fragment on the kinetics of UV-induced degradation of poliovirus 1 RNA: 76 bases (⧫), 145 bases (▪), and 1,869 bases (▴). The dashed lines are regression lines for genome fragments, the solid line is the regression line for infectivity and is given for reference. The horizontal black line designates the highest value for the background.
The first observation was a dose-dependent relationship between genome degradation and fluence delivered as assessed by the linear regression test, with the quantity of targeted fragment decreasing with increasing fluence. Loss of the detected fragment under UV radiation produced a first-order relationship for each fragment (81 bases [slope, −0.0014; r2 = 84.7%], 111 bases coding the capsid gene [slope, −0.002; r2 = 75.1%], 111 bases from 5′ UTR [slope, −0.0028; r2 = 94.2%], 692 bases [slope, −0.0066; r2 = 96.3%], 1,298 bases [slope, −0.0085; r2 = 92.6%], and 1,909 bases [slope, −0.0191; r2 = 99.1%] for MS2 and 76 bases [slope, −0.0031; r2 = 96.8%] and 145 bases [slope, −0.0055; r2 = 96.1%] for PV1). For MS2 and PV1, viral RNA degradation was proportional to the size of the targeted fragment, with more rapid degradation affecting longer fragments. For MS2 RNA fragments, no significant difference in the degradation pattern (P > 0.05) was observed between the fragment of 81 bases and the fragment of 111 bases coding the capsid gene and also between the fragment of 111 bases coding the capsid gene and the fragment of 111 bases from 5′ UTR. Comparisons between all of the other fragments reveal a significant difference in the degradation kinetic (P < 0.05). Concerning PV1, a significant difference in the degradation pattern (P < 0.05) was obtained for all the fragments studied. Degradation of the viral RNA for small fragments (<200 bases) was very low, less than 0.5 log, except for the 145-base PV1 (0.9 log for the maximal fluence of 150 mJ · cm−2). Fragments longer than 500 bases degraded very quickly, even declining to the background level for fragments around 2,000 bases. Moreover, the two longest PV1 fragments, 5,429 and 6,989 bases, could not be analyzed because the signal fell to the background level after a single UV fluence delivery.
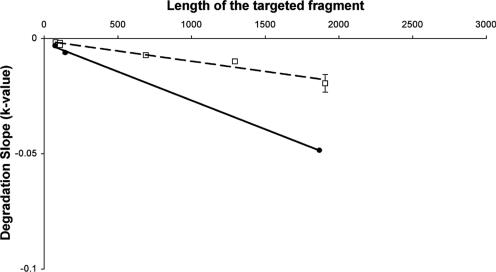
Since loss of viral RNA displayed first-order kinetics for all fragments studied, we presented the degradation slope as a function of fragment size for PV1 and MS2 (Fig. 4). For both viruses, UV radiation-induced degradation was directly proportional to fragment size, but with significantly different degradation slopes. PV1 RNA degraded more rapidly than MS2 RNA, as seen with the 1,869-base fragment of PV1 and the 1,909-base fragment of MS2. A similar degradation slope was observed for the 145-base fragment of PV1 and the 692-base fragment of MS2, which was much more rapid than for the 111-base fragment of MS2. According to the degradation slopes, the relative sensitivity of PV1 RNA to UV was twice that of MS2 RNA for a similar size.
FIG. 4.
Influence of the size of the targeted fragment versus the degradation slope of UV-induced degradation of MS2 RNA (□) and poliovirus 1 RNA (•). The dashed line and solid line are, respectively, regression lines for bacteriophage MS2 and for poliovirus 1.
DISCUSSION
Simultaneous monitoring of inactivation processes and genome degradation in four models—poliovirus 1 and three F-specific RNA phages (MS2, GA, and Qβ)—provided, for the first time to our knowledge, complementary data on the effect of UV light on RNA viruses.
UV inactivation of MS2 and PV1 has been widely studied, and our findings are very similar to previous reports (1, 12, 21, 23, 25, 29, 31, 34). As expected, the MS2 phage was significantly more resistant than PV1. This phenomenon would be related more to difference in genome size (3,569 versus 7,440 bases) than capsid structure (one versus four proteins). Others have also observed the tailing effect for PV1 (20, 32). Here, the explanation is less straightforward but could involve viral aggregation or population heterogeneity. As for MS2, a first-order kinetics model fits well with GA and Qβ inactivation. GA belongs to the same genus as MS2 and therefore has the same genome size (3,466 versus 3,569 bases) and 62% amino acid similarity for the capsid protein. UV inactivation curves were strictly identical. Therefore, a 38% difference in amino acids constituting the capsid protein would not be enough to change the UV sensitivity. Qβ belongs to another genus of the same family as MS2 (Leviviridae). Its genome is longer (4,160 bases), and the amino acid similarity of its capsid protein is only 20% with MS2; the inactivation curves are different. Noting that PV1 exhibits the greatest sensitivity, it is reasonable to hypothesize that the difference in genome size is the cause of the difference in inactivation (18). The significant difference in capsid structure (>38%) cannot, however, be ignored (26).
We therefore chose to monitor the genome degradation of both MS2 and PV1. Results show a moderate decline (≤0.9 log) of small fragments (<145 bases). Resistance of small fragments has also been previously highlighted. A 196-base fragment in the 5′ noncoding region of PV1 and PV2 and a 149-base fragment in the same region of PV1 were always detected by qualitative PCR even for high fluences (17, 22, 29). However, different findings were observed since no loss of the considered fragment before a fluence equal to 150 mJ · cm−2 could be obtained while a one-log decrease in RT-PCR signal for fluence starting from 2.5 mJ · cm−2 could also be estimated followed by no change up to 30 mJ · cm−2 (19, 29). Similarly, a 437-base fragment has even been detected up to 350 mJ · cm−2 (24).
Several studies have attempted to correlate the degradation of the viral genome to the size of the detected fragment (8, 11, 23, 27, 28, 29). All published reports on encapsidated poliovirus 1 RNA show that longer fragments (>800 bases) are at least equal to or more sensitive to UV or oxidation treatment (i.e., ozone, chlorine, chloramines, and chlorine dioxide) than are shorter fragments (<145 bases) (23, 27, 28, 29). Earlier results in our laboratory have also shown that size has a significant impact on viral RNA degradation by chlorine dioxide but that the principal parameter is the localization of the detected fragment rather than size (28). For the moment, none of these studies have shown a similarity between genome degradation and viral inactivation. In the present study, viral RNA degradation increased as a function of fragment size. This phenomenon was clear for MS2 but also apparent for PV1. The rate of RNA degradation increased linearly with increasing fragment size. Extrapolating the two linear models to the whole genome (3,569 bases for MS2 and 7,440 bases for PV1) yielded degradation slope constants (−0.0332 and −0.1507, respectively) similar to those obtained for inactivation (−0.0389 and −0.1127, respectively). This implies that genome degradation (loss of retrotranscription capacity) may fully explain UV-induced viral inactivation for each virus. For RNA viruses, genome size is therefore a very important parameter for UV resistance (18). Nevertheless, our results also show that genome size is not the only parameter involved since similarly sized fragments degraded more rapidly for PV1 than for MS2. A 76-base PV1 RNA fragment is degraded faster than an 81-base MS2 RNA fragment. This difference cannot be explained by a different percentage of pyrimidines, known to be much more sensitive bases, because the percentage in the MS2 fragment is 49% versus 38% for the PV1 fragment. The same holds for the 1,869-base PV1 fragment (46% pyrimidine), which was much more rapidly degraded than the 1,909-base MS2 fragment (51% pyrimidine). Consequently, UV viral inactivation cannot be explained on a structural basis without an understanding of why PV1 RNA is more sensitive than MS2 RNA. A requirement for greater compactness for PV1 (7,440 bases in a 20- to 30-nm particle) than for MS2 (3,569 bases in a 20- to 30-nm particle) could be a possible explanation.
In conclusion, our study has demonstrated that genome degradation seems to be the main phenomenon explaining UV inactivation of RNA viruses. Moreover, degradation of the genome depends on fragment size. Nevertheless, another virus-dependent factor is apparently involved, since PV1 RNA is more sensitive than MS2 RNA, even for similar fragment sizes. Factors like compactness or secondary structure of RNA should be investigated before predicting viral inactivation by UV light on a structural basis. Besides, from our results, detection of the fragment of the genome was not reliable for the infectivity and may not yet be the critical parameter to determine the risk of viruses from water treated with UV. Finally, phages can be used to estimate UV treatment efficiency for enteric viruses since they displayed a high resistance to UV irradiation.
Acknowledgments
This work was supported by grants from Danone, Nestlé Waters, and Veolia Water.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Battigelli, D. A., M. D. Sobsey, and D. C. Lobe. 1993. The inactivation of hepatitis A virus and other model viruses by UV irradiation. Water Sci. Technol. 27:339-342. [Google Scholar]
- 2.Bollback, J. P., and J. P. Huelsenbeck. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J. Mol. Evol. 52:117-128. [DOI] [PubMed] [Google Scholar]
- 3.Bolton, J. R., and K. G. Linden. 2003. Standardization of methods for fluence (UV dose) determination in bench-scale UV experiments. ASCE J. Environ. Eng. 3:209-215. [Google Scholar]
- 4.Cadet, J., and P. Vigny. 1991. The photochemistry of nucleic acids, p. 3-272. In H. Morrison (ed.), Bioorganic photochemistry, vol. 1. Wiley Interscience, Hoboken, N.J. [Google Scholar]
- 5.De Roda Husman, A.-M., P. Bijkerk, W. Lodder, H. Van Den Berg, W. Pribil, A. Cabaj, P. Gehringer, R. Sommer, and E. Duizer. 2004. Calicivirus inactivation by nonionizing (253.7-nanometer-wavelength [UV]) and ionizing (gamma) radiation. Appl. Environ. Microbiol. 70:5089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duizer, E., P. Bijkerk, B. Rockx, A. De Groot, F. Twisk, and M. Koopmans. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 70:4538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbeej, B., and L. A. Eriksson. 2003. On the formation of cyclobutane pyrimidine dimers in UV-irradiated DNA: why are thymines more reactive? Photochem. Photobiol. 78:159-167. [DOI] [PubMed] [Google Scholar]
- 8.Enriquez, C. E., M. Abbaszadegan, I. L. Pepper, K. J. Richardson, and C. P. Gerba. 1993. Poliovirus detection in water by cell culture and nucleic acid hybridization. Water Res. 27:1113-1118. [Google Scholar]
- 9.Golmohammadi, R., K. Valegard, K. Fridborg, and L. Liljas. 1993. The refined structure of bacteriophage MS2 at 2.8 A resolution. J. Mol. Biol. 234:620-639. [DOI] [PubMed] [Google Scholar]
- 10.Harm, W. 1980. Biological effects of ultraviolet radiation, p. 217. In IUPAB Biophysics Series. Cambridge University Press, Cambridge, United Kingdom.
- 11.Hegedus, M., K. Modos, G. Ronto, and A. Fekete. 2003. Validation of phage T7 biological dosimeter by quantitative polymerase chain reaction using short and long segments of phage T7 DNA. Photochem. Photobiol. 78:213-219. [DOI] [PubMed] [Google Scholar]
- 12.Hijnen, W. A., E. F. Beerendonk, and G. J. Medema. 2006. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res. 40:3-22. [DOI] [PubMed] [Google Scholar]
- 13.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 14.Koopmans, M., C. H. von Bonsdorff, J. Vinje, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuluncsics, Z., D. Perdiz, E. Brulay, B. Muel, and E. Sage. 1999. Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts. J. Photochem. Photobiol. B 49:71-80. [DOI] [PubMed] [Google Scholar]
- 16.Kurosaki, Y., H. Abe, H. Morioka, J. Hirayama, K. Ikebuchi, N. Kamo, O. Nikaido, H. Azuma, and H. Ikeda. 2003. Pyrimidine dimer formation and oxidative damage in M13 bacteriophage inactivation by ultraviolet C irradiation. Photochem. Photobiol. 78:349-354. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, G. D., S. L. Molloy, G. E. Greening, and J. Dawson. 2000. Influence of environmental factors on virus detection by RT-PCR and cell culture. J. Appl. Microbiol. 88:633-640. [DOI] [PubMed] [Google Scholar]
- 18.Lytle, C. D., and J. L. Sagripanti. 2005. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J. Virol. 79:14244-14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, J. F., T. M. Straub, I. L. Pepper, and C. P. Gerba. 1994. Cell culture and PCR determination of poliovirus inactivation by disinfectants. Appl. Environ. Microbiol. 60:4203-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier, A., D. Tougianidou, A. Wiedenmann, and K. Botzenhart. 1995. Detection of poliovirus by cell culture and by PCR after UV disinfection. Water Sci. Technol. 31:141-145. [Google Scholar]
- 21.Meng, Q. S., and C. P. Gerba. 1996. Comparative inactivation of enteric Adenoviruses, poliovirus, and coliphages by ultraviolet irradiation. Water Res. 30:2665-2668. [Google Scholar]
- 22.Moore, N. J., and A. B. Margolin. 1994. Efficacy of nucleic acid probes for detection of poliovirus in water disinfected by chlorine, chlorine dioxide, ozone, and UV radiation. Appl. Environ. Microbiol. 60:4189-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newland, S., G.-A. Shin, and M. D. Sobsey. 2001. Comparative inactivation of Norwalk virus, poliovirus 1, and coliphage MS2 in water by low pressure UV radiation. First International Congress on Ultraviolet Technology, Washington, D.C.
- 24.Nuanualsuwan, S., and D. O. Cliver. 2002. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 104:217-225. [DOI] [PubMed] [Google Scholar]
- 25.Nuanualsuwan, S., T. Mariam, S. Himathongkham, and D. O. Cliver. 2002. Ultraviolet inactivation of feline calicivirus, human enteric viruses and coliphages. Photochem. Photobiol. 76:406-410. [DOI] [PubMed] [Google Scholar]
- 26.Nuanualsuwan, S., and D. O. Cliver. 2003. Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl. Environ. Microbiol. 69:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin, G.-A., and M. D. Sobsey. 2003. Reduction of Norwalk virus, poliovirus 1, and bacteriophage MS2 by ozone disinfection of water. Appl. Environ. Microbiol. 69:3975-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonet, J., and C. Gantzer. 2006. Degradation of poliovirus 1 genome by chlorine dioxide. J. Appl. Microbiol. 100:862-870. [DOI] [PubMed] [Google Scholar]
- 29.Sobsey, M. D., D. A. Battigelli, G.-A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Sci. Technol. 38:91-94. [Google Scholar]
- 30.Sommer, R., A. Cabaj, W. Pribil, and T. Haider. 1997. Influence of lamp intensity and water transmittance on the UV disinfection of water. Water Sci. Technol. 35:113-118. [Google Scholar]
- 31.Sommer, R., T. Haider, A. Cabaj, W. Pribil, and M. Lhotsky. 1998. Time dose reciprocity in UV disinfection of water. Water Sci. Technol. 38: 145-150. [Google Scholar]
- 32.Sommer, R., G. Weber, A. Cabaj, J. Wekerle, G. Keck, and G. Schauberger. 1989. UV-inactivation of microorganisms in water. Zentralbl. Hyg. Umweltmed. 189:214-224. [PubMed] [Google Scholar]
- 33.Tars, K., M. Bundule, K. Fridborg, and L. Liljas. 1997. The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses. J. Mol. Biol. 271:759-773. [DOI] [PubMed] [Google Scholar]
- 34.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Water Care Services Ltd. 2002. Pilot plant investigations, surrogate study, results and recommendations. Disinfection review group report. Water Care Services Ltd., Auckland, New Zealand.
- 36.Yamada, Y., A. Shigeta, and K. Nozu. 1973. Ultraviolet effects on biological function of RNA phage MS2. Biochim. Biophys. Acta 299:121-135. [DOI] [PubMed] [Google Scholar]