Abstract
Pyrene degradation is known in bacteria. In this study, Mycobacterium sp. strain KMS was used to study the metabolites produced during, and enzymes involved in, pyrene degradation. Several key metabolites, including pyrene-4,5-dione, cis-4,5-pyrene-dihydrodiol, phenanthrene-4,5-dicarboxylic acid, and 4-phenanthroic acid, were identified during pyrene degradation. Pyrene-4,5-dione, which accumulates as an end product in some gram-negative bacterial cultures, was further utilized and degraded by Mycobacterium sp. strain KMS. Enzymes involved in pyrene degradation by Mycobacterium sp. strain KMS were studied, using 2-D gel electrophoresis. The first protein in the catabolic pathway, aromatic-ring-hydroxylating dioxygenase, which oxidizes pyrene to cis-4,5-pyrene-dihydrodiol, was induced with the addition of pyrene and pyrene-4,5-dione to the cultures. The subcomponents of dioxygenase, including the alpha and beta subunits, 4Fe-4S ferredoxin, and the Rieske (2Fe-2S) region, were all induced. Other proteins responsible for further pyrene degradation, such as dihydrodiol dehydrogenase, oxidoreductase, and epoxide hydrolase, were also found to be significantly induced by the presence of pyrene and pyrene-4,5-dione. Several nonpathway-related proteins, including sterol-binding protein and cytochrome P450, were induced. A pyrene degradation pathway for Mycobacterium sp. strain KMS was proposed and confirmed by proteomic study by identifying almost all the enzymes required during the initial steps of pyrene degradation.
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants. Bioremediation is broadly accepted as an effective tool for the degradation of PAHs to nontoxic compounds. Pyrene degradation pathways of mycobacteria, including Mycobacterium vanbaalenii PYR-1, M. flavescens PYR-GCK, Mycobacterium sp. strain RJGII-135, Mycobacterium sp. strain KR2, and Mycobacterium sp. strain AP1, have been studied and are proposed to be similar (3, 4, 16, 26, 28, 29). The proposed pathway is thought to be catalyzed by a number of enzymes. Pyrene is first oxidized in the K region by a dioxygenase to form cis-4,5-pyrene-dihydrodiol, which is rearomatized to form 4,5-dihydroxy-pyrene by dihydrodiol dehydrogenase. 4,5-Dihydroxy-pyrene is subsequently cleaved to yield phenanthrene-4,5-dicarboxylic acid by intradiol dioxygenase. Following loss of a carboxyl group by decarboxylase, 4-phenanthroic acid is formed. Oxidation of 4-phenanthoic acid by ring-hydroxylating dioxygenase produces 3,4-phenanthrene dihydrodiol-4-carboxylic acid, which is further transformed to 3,4-dihydroxyphenanthrene by dehydrogenase/decarboxylase. Once 3,4-dihydroxyphenanthrene is formed, it enters the phenanthrene degradation pathway (18). Two additional pathways have been proposed. One proposition is that pyrene hydroxylation takes place at the 1, 2 positions, leading to the formation of 4-hydroxy-perinaphthenone, which is a dead end product and so far has been found only in M. vanbaalenii PYR-1 cultures (3). Another pathway involves the accumulation of 6,6′-dihydroxy-2,2′-biphenyl-dicarboxylic acid in Mycobacterium sp. strain AP1 (29).
Pyrene-4,5-dione can be formed following the autooxidation of 4,5-dihydroxy-pyrene and is a pyrene degradation metabolite in several bacteria. It was observed as a pyrene metabolite accumulated in Sphingomonas yanoikuyae strain R1 (12). Significant amounts of this compound were formed when M. vanbaalenii PYR-1 was incubated with a high concentration of cis-4,5-pyrene-dihydrodiol, although it was not reported as an intermediate when M. vanbaalenii PYR-1 grew on pyrene (12). In addition, pyrene-4,5-dione accumulation was observed in aged whole-sediment microcosm incubations (6) and in slurry-phase reactors with soil suspension at 25% wt/vol (5). Moreover, it was identified to be a pyrene metabolite in the phagemid clone My6-pBK-CMV, which contained a dioxygenase gene when it was incubated with pyrene (13). Even though pyrene 4,5-dione is a growth substrate for M. vanbaalenii PYR-1 (12), its further degradation is not delineated.
Based on the proposed metabolic pathway for the degradation of pyrene and phenanthrene by Mycobacterium species (18), at least 15 enzymes are involved in the degradation of pyrene and the o-phthalate degradation of phenanthrene, with some enzymes being common to the degradation of both PAHs. While some of the enzymes, including dioxygenase, aldehyde dehydrogenase, putative monooxgenase, hydratase aldolase, and catalase-peroxidase (14, 18, 30), have been identified as PAH-induced proteins, some other enzymes, especially those involved in the initial steps of pyrene degradation, are not known.
Mycobacterium sp. strain KMS was isolated from vadose-zone soil of the Champion International Superfund site (Libby, MT) and has the ability to degrade pyrene and other PAHs (22). The genome was sequenced by the U.S. Department of Energy/Joint Genome Institute (JGI), and the draft sequence is available in the NCBI database. Due to the toxicity of pyrene-4,5-dione (25) and its possible presence during pyrene degradation in mycobacteria, it is important to identify this metabolite and determine its fate during pyrene degradation, as it was observed as an end product in some gram-negative cultures and may result in an increase in toxicity during in situ soil bioremediation (12). Furthermore, biostimulation and bioaugmentation strategies, where Mycobacterium sp. strain KMS is employed for PAH bioremediation, require an understanding of enzymatic mechanisms. Therefore, the objectives of this work reported here were (i) to determine the pyrene degradation pathway used by Mycobacterium sp. strain KMS by isolating and identifying the metabolites, (ii) to determine the capability of Mycobacterium sp. strain KMS to degrade pyrene-4,5-dione, (iii) to obtain the proteomic profile of Mycobacterium sp. strain KMS, and (iv) to identify PAH-induced proteins.
MATERIALS AND METHODS
Chemicals.
Pyrene (99%) was purchased from Fluka (Buchs, Switzerland). cis-4,5-Pyrenedihydrodiol was kindly provided by Michael Aitken of the University of North Carolina at Chapel Hill. Pyrenol (1-hydroxypyrene, 99%) and phthalic acid (99%) were purchased from Aldrich Chemical Company (Milwaukee, WI). All solvents used (methanol, acetone, acetonitrile, ethyl acetate, and methylene chloride) were high-performance liquid chromatography (HPLC) grade or the equivalent and were purchased from Sigma-Aldrich (St. Louis, MO). The basal salt medium (BSM) and Luria broth (LB) used were the same as described by Miller et al. (22). The deuterated solvent methanol-d4 (99.8%) was purchased from Sigma-Aldrich (St. Louis, MO), and dimethyl sulfoxide-D6 was purchased from Acros Organics (Morris Plains, NJ). Pyrene-4,5-dione and phenanthrene-4,5-dicarboxylic acid were synthesized according to Yong and Funk's procedure (31).
Urea, thiourea, dithiothreitol (DTT), iodoacetamide, CHAPs {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, bromophenol blue, nuclease mix, glycerol, sodium dodecyl sulfate (SDS), Tris, pharmalyte, low-molecular-weight calibration kits, agarose, Immobiline DryStrips, and DryStrip cover fluid were purchased from Amersham Biosciences (Piscataway, NJ). ASB-14 was purchased from Calbiochem (Nottingham, United Kingdom). A bovine serum albumin standard, bicinchoninic acid protein assay kit, Compat-Able protein assay preparation reagent set, and Imperial protein stain were purchased from Pierce (Rockford, IL). Protease inhibitor cocktail was purchased from Roche Diagnostics Corporation (Indianapolis, IN). Criterion precast 12.5% Tris-HCl gel, Tris-glycine-SDS running buffer, and a SDS-polyacrylamide gel electrophoresis (PAGE) power unit were purchased from Bio-Rad (Hercules, CA).
Bacteria and growth conditions.
Mycobacterium sp. strain KMS cells were grown in BSM+ (a 9:1 mixture of BSM and LB) for 5 days to stationary phase, pelleted, and washed twice with sterile distilled deionized water. The suspension was used as an inoculum.
In order to identify pyrene degradation intermediates, replicate batch cultures were grown in 2.8-liter flasks containing 1 liter BSM+ and either 4.95 mM pyrene or 64 μM pyrene-4,5-dione. After 20 ml Mycobacterium sp. strain KMS inoculum was added to the flasks, incubation was initiated by putting the flasks on a rotary shaker at 120 rpm at 25°C in the dark. Uninoculated flasks and flasks without pyrene or pyrene-4,5-dione served as controls.
For the proteomic study, three sets of 2-liter flasks were employed. Pyrene and pyrene-4,5-dione solution in methylene chloride was added to the first and second set of two flasks, respectively. The third set of two flasks served as a control without PAH added. After methylene chloride evaporated completely from the flasks, 800 ml BSM+ was added to each flask together with 30 ml Mycobacterium sp. strain KMS inoculum. The concentrations of pyrene and pyrene-4,5-dione were 120 and 60 μM, respectively. The cultures were incubated at 28°C on a rotary shaker at 150 rpm in the dark.
Isolation of pyrene and pyrene-4,5-dione metabolites.
After each culture was incubated for 1 week, the whole culture was passed through glass wool in a separatory funnel to remove residual pyrene or pyrene-4,5-dione. The pH was then adjusted to 1.5 with 4 M HCl. Metabolites were isolated from the culture using modified, cold, continuous liquid-liquid extraction (CLLE) (20). Briefly, the culture flask was not heated to prevent the degradation of metabolites. Fresh ethyl acetate was added to the culture flask continuously from the top.
After CLLE was extracted, furnace-dried anhydrous sodium sulfate was added to the receiving flask to remove the water. The ethyl acetate solution was then evaporated to dryness in a rotary evaporator at 30°C. The residue was then dissolved in methanol for analysis by HPLC.
Identification of pyrene and pyrene-4,5-dione metabolites.
A modular HPLC system (Shimadzu Corp., Kyoto, Japan), consisting of three liquid chromatograph (LC) 6A pumps, an auto sampler (SIL-10A), system controller (SCL-10A), and dual-wavelength UV detector (SPD-6A), was used. The program for HPLC was previously described in detail (19).
The concentrated extract was first analyzed using an analytical column to obtain the chromatographic profile. Individual peaks in selected regions were then isolated by manual fractionation, using a semi-preparative column. Repeated injections of the concentrated extract and subsequent collection resulted in the accumulation of small amounts of individual metabolites. Isolated metabolites were subjected to the following analysis.
1H NMR spectra were recorded on a Bruker ARX 400 MHz spectrometer with a 5-mm broadband probe. Chemical shifts were reported in relation to the solvent used under the following conditions: temperature, 298° K; sweep width, 7,246 Hz; data points, 32,768.
Mass spectra were recorded using a Finnigan MAT LCQ (Thermo, San Jose, CA) mass spectrometer (MS) in the atmospheric pressure chemical ionization mode, operating with a vaporizer temperature of 450°C, a discharge current of 5 μA, and a capillary temperature of 200°C.
Gas chromatography (GC)-MS spectra were obtained by using a Finnigan MAT GCQ system (Thermo, San Jose, CA) equipped with an ion trap mass filter and a DB-5 capillary column (0.25 μm by 30 m; J & W Scientific, Rancho Cordova, CA). The program and parameters for GC-MS analysis were the same as those described by Heitkamp et al. (9).
Cell lysis and proteome extraction.
After 1 week, when the cultures reached an optical density of 0.5 at 600 nm, they were passed through glass wool to remove residue PAHs and centrifuged. The cell pellets were resuspended in a mixture of 900 μl of lysis buffer (7 M urea, 2 M thiourea, 4% CHAPs, 1% ASB-14, 40 mM DTT, and 2% pharmalyte [pH 5 to 8]), 100 μl of protease inhibitor cocktail (1 tablet in 1 ml distilled deionized water), and 10 μl of nuclease mix. Then the mixtures were transferred to 2-ml screw-cap and precooled microfuge tubes containing 200 μm acid-washed zirconium beads from OPS Diagnostics (Lebanon, NJ). Cell disruption was performed on a mini-beadbeater (Glen Mills, Clifton, NJ), which was set at 4,800 rpm with a 1-min cycle. After five cycles of bead beating and cooling on ice, the lysed solutions were transferred to 2-ml centrifuge tubes and centrifuged twice at 17,000 × g for 15 min (IEC Microlite Microcentrifuge, Waltham, MA). Then the supernatant was ready for protein quantification and subsequent two-dimensional (2-D) PAGE analysis.
Pretreatment with the Pierce Compat-Able protein assay preparation reagent set was employed to remove interferences from DTT and thiourea in lysis buffer. Protein concentrations were determined by using a UV-visible spectrophotometer (DU 640B; Beckman, Fullerton, CA) at a wavelength of 562 nm, based on the bicinchoninic acid protein assay.
Isoelectric focusing.
Protein solutions containing 200 μg proteins were first treated with acetone to remove interferences, following the protocol recommended by Pierce (Pierce Biotechnology, Inc. Rockford, IL). After acetone treatment, 200 μl of rehydration buffer solutions from Amersham Biosciences were added to the protein pellets, which were then vortexed until they dissolved.
Immobiline DryStrips (11 cm, pH 4 to 7) were used in this study. Active rehydration was employed on an Ettan IPGphor from Amersham Biosciences, followed by the isoelectric focusing program recommended by the manufacturer.
2-D PAGE.
Precast polyacrylamide criterion gels (12.5%) were used to separate proteins in the 10- to 100-kDa range. After the strips were equilibrated with DTT and iodoacetamide in SDS equilibration buffer, they were transferred to criterion gels. A low-molecular-mass (Mr) calibration marker in 0.5% agarose solution was applied to the acidic end of the strips. Separation was performed under 200-V constant voltage at 4°C. After 1 h of electrophoresis, the gels were treated with Imperial protein stain according to the protocol recommended by the manufacturer.
Protein identification by mass spectrometry.
Gel image comparison and analysis were conducted using Progenesis software (Progenesis PG 220, v. 2006). Protein spots were considered to be induced if their normalized spot densities in a PAH-treated sample were found to be consistently increased by more than twofold compared to those of the control sample. A protein spot was regarded as unmatched or newly detected if it was apparent only in PAH-treated samples and was not detectable in the control sample. After the 2-D gels were analyzed, selected features were robotically excised, using the Etten Spot Picker (GE Healthcare Bio-Science Corp., Piscataway, NJ). Subsequent gel plugs were digested with trypsin, using a published protocol (10). The resultant peptide pools were analyzed using nano-LC-tandem mass spectrometry (MS-MS) on a Q-Tof Primer tandem mass spectrometer (Waters, Manchester, United Kingdom) following the protocol recommended by the manufacturer.
RESULTS
Identification of pyrene degradation metabolites.
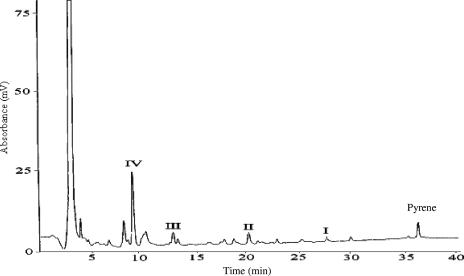
The HPLC elution profile of the CLLE-extractable pyrene residue in the spent medium of Mycobacterium sp. strain KMS is shown in Fig. 1. Profiles of controls without pyrene or without Mycobacterium sp. strain KMS showed no metabolite peaks. The most polar fraction of the HPLC, which eluted in 5 min, was further fractioned and analyzed by GC-MS for identification of possible intermediates. Two polar metabolites, phthalic acid and 3,4-dihydroxybenzoic acid, were detected after derivatization with diazomethane by comparing the GC retention times and mass spectra with those of standards. Later eluting peaks (I to IV) were first analyzed by GC-MS and LC-MS, and then sufficient mass of each peak was obtained for nuclear magnetic resonance (NMR) analysis. These peaks were identified as follows. Peak I was identified as 1-hydroxypyrene by comparing the HPLC and GC-MS retention times and mass spectrum patterns with those of a standard. Peak II was identified as pyrene-4,5-dione, based on the same HPLC retention time, MS, and 1H NMR spectrum patterns as those of the standard. Peak III was identified as cis-4,5-pyrene-dihydrodiol in the same manner as peak II. Peak IV was identical to the synthesized standard of phenanthrene-4,5-dicarboxylic acid, based on evidences from MS and 1H NMR analyses.
FIG. 1.
HPLC chromatogram showing UV absorbance at 254 nm of CLLE-extractable pyrene residues in Mycobacterium sp. strain KMS culture after a 1-week exposure to pyrene. (I) 1-hydroxypyrene; (II) pyrene-4,5-dione; (III) cis-4,5-pyrene-dihydrodiol; (IV) phenanthrene-4,5-dicarboxlylic acid.
Identification of pyrene-4,5-dione culture metabolites.
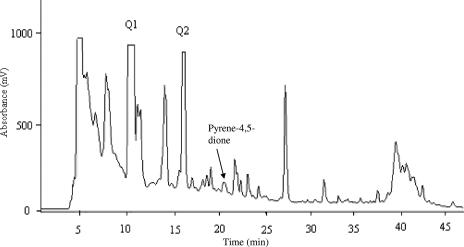
Pyrene-4,5-dione was degraded by Mycobacterium sp. strain KMS within 1 week (Fig. 2). By comparing chromatograms with and without pyrene-4,5-dione, it was established that almost all the peaks in the pyrene-4,5-dione culture were from pyrene-4,5-dione degradation. Two peaks, Q1 and Q2, were chosen for further identification.
FIG. 2.
HPLC chromatogram showing UV absorbance at 254 nm of an acidic extract from Mycobacterium sp. strain KMS culture with pyrene-4,5-dione added. The Q1 and Q2 fractions were obtained by using a semi-preparative HPLC column and were identified as phenanthrene-4,5-dicarboxylic acid and 4-phenanthroic acid, respectively.
Compound Q1 was found to be the major metabolite present, based on mass analysis, which may indicate that the degradation of Q1 is rate limiting or slow. Identification of Q1 as phenanthrene-4,5-dicarboxylic acid was based on MS and 1H NMR analyses, which proved to be identical to those of pyrene culture peak IV, and those of the standard phenanthrene-4,5-dicarboxylic acid.
Metabolite Q2 has the same UV-visible absorption spectrum as that of 4-phenanthroic acid identified in the pyrene degradation pathway of M. vanbaalenii PYR-1 (9). The atmospheric pressure chemical ionization MS spectrum shows (M − H)− at m/z 221 under negative ion mode, corresponding to a molecular weight of 222. An aliquot of metabolite Q2 was derivatized with diazomethane and analyzed by capillary column GC-MS. The GC-MS and the 1H NMR spectra in methanol-D4 of Q2 were the same as those reported in Heitkamp et al. (9).
2-D PAGE.
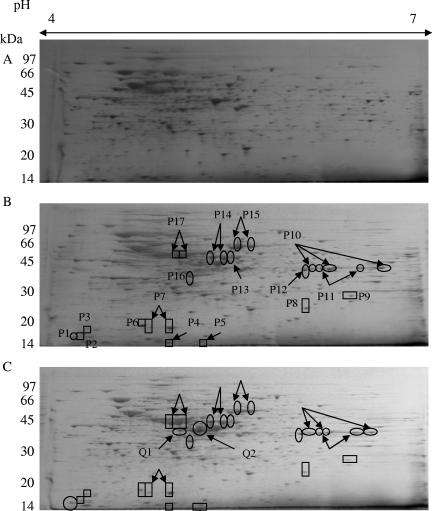
Protein spots were well distributed and resolved on the 12.5% gel (Fig. 3). A total of more than 700 spots was detected in the protein extracts of the control without PAH added (Fig. 3A) and the two PAH treatments (Fig. 3B and C) with similar spot patterns. Protein identification was accomplished by LC-MS-MS analysis. Proteins sharing a high degree of similarity with the existing database were identified. These results were confirmed by a Mascot search using the peak list files generated from the LC-MS-MS analysis. The proteins that were identified are summarized in Table 1.
FIG. 3.
2-D PAGE images of samples treated with pyrene or pyrene-4,5-dione at a pI range of 4 to 7. The molecular mass marker is 14 to 97 kDa. (A) Mycobacterium sp. strain KMS with no PAH added; (B) with pyrene added; (C) with pyrene-4,5-dione added. ○, proteins induced at a more than twofold-higher level than for the control; □, newly detected proteins. P1 to P14, Q1, and Q2 were selected for further identification.
TABLE 1.
Identification of proteins induced or newly detected by PAH treatments
| Protein no. | Protein identification | NCBI accession no. | Spot density in indicated culture
|
Mascot score | No. of peptides matched | % Coverage | Observed migrationa
|
Theoretical migrationb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Pyrene added | Quinone added | Mr (kDa) | pI | Mr (kDa) | pI | ||||||
| P1 | 4Fe-4S ferredoxin, iron-sulfur binding | ZP_01282568 | 0.06 | 0.25 | 0.26 | 153 | 2 | 46 | 16.3 | 4.3 | 10.5 | 4.5 |
| P2 | Aromatic-ring-hydroxylating dioxygenase, β subunit | ZP_01282558 | NDc | 0.15 | 0.17 | 569 | 8 | 59 | 16.7 | 4.4 | 18.9 | 4.6 |
| P3 | Aromatic-ring-hydroxylating dioxygenase, β subunit | ZP_01286731 | ND | 0.17 | 0.13 | 49 | 2 | 14 | 18.3 | 4.4 | 18.8 | 4.5 |
| P4 | Sterol binding protein | ZP_01282557 | ND | 0.47 | 0.42 | 602 | 20 | 86 | 15.6 | 5.0 | 14.3 | 5.0 |
| P5 | Hypothetical protein MkmsDRAFT_0077 | ZP_01286732 | ND | 0.66 | 0.50 | 637 | 20 | 87 | 16.7 | 5.3 | 14.3 | 5.1 |
| P6 | Aromatic-ring-hydroxylating dioxygenase, β subunit | ZP_01286725 | ND | 0.26 | 0.22 | 471 | 12 | 59 | 19.5 | 4.8 | 19.5 | 4.9 |
| P7 | Aromatic-ring-hydroxylating dioxygenase, β subunit (nidB2) | AAT51747 | ND | 1.06 | 0.68 | 525 | 10 | 60 | 18.7 | 5.1 | 19.4 | 5.1 |
| P8 | Aromatic-ring-hydroxylating dioxygenase, β subunit | ZP_01282769 | ND | 0.20 | 0.19 | 420 | 7 | 39 | 24.9 | 6.1 | 22.4 | 5.7 |
| P9 | Phthalate dihydrodiol dehydrogenase | AAQ91917 | ND | 0.16 | 0.11 | 525 | 10 | 42 | 28.6 | 6.4 | 28.3 | 5.5 |
| P10 | Ring-hydroxylating dioxygenase, alpha subunit (Rieske [2Fe-2S] region) | ZP_01282559 | 0.04 | 0.73 | 0.50 | 231 | 10 | 17 | 40.8 | 6.6 | 45.0 | 6.0 |
| P11 | Glycosyl hydrolase, BNR | ZP_01282560 | 0.05 | 0.45 | 0.57 | 812 | 111 | 50 | 40.8 | 6.4 | 39.1 | 5.9 |
| P12 | Epoxide hydrolase-like alpha/beta hydrolase fold | ZP_01282545 | 0.16 | 0.60 | 0.50 | 542 | 14 | 46 | 40.2 | 6.0 | 42.4 | 5.6 |
| P13 | Ring-hydroxylating dioxygenase, alpha subunit (Rieske [2Fe-2S] region) | ZP_01282563 | 0.08 | 0.59 | 0.72 | 1087 | 25 | 56 | 46.4 | 5.6 | 50.2 | 5.3 |
| P14 | Aromatic-ring-hydroxylating dioxygenase, alpha subunit (nidA) | AAQ95208 | 0.03 | 0.90 | 0.99 | 906 | 16 | 37 | 47.0 | 5.4 | 50.1 | 5.3 |
| P15 | Flavoprotein-like fumarate reductase/succinate dehydrogenase (FAD- dependent oxidoreductase) | ZP_01282569 | 0.06 | 1.70 | 1.41 | 1227 | 23 | 37 | 57.8 | 5.6 | 73.8 | 5.8 |
| P16 | PhdG (hydratase-aldolase) | AAT51745 | ND | 0.38 | 0.50 | 1160 | 19 | 65 | 36.3 | 5.2 | 36.8 | 5.2 |
| P17 | Aldehyde dehydrogenase (NAD+) | ZP_01286724 | ND | 1.11 | 1.44 | 555 | 8 | 27 | 47.0 | 5.1 | 52.0 | 5.0 |
| Q1 | Zinc-containing alcohol dehydrogenase superfamily (alcohol dehydrogenase) | ZP_01283066 | 0.09 | 0.03 | 0.36 | 1468 | 27 | 64 | 41.2 | 5.1 | 50.2 | 5.3 |
| Q2 | Cytochrome P450 | ZP_01283067 | 0.21 | 0.25 | 3.04 | 1323 | 34 | 65 | 41.6 | 5.3 | 47.6 | 5.2 |
The observed migrations Mr and pI were based on 2-D calibration during gel image analysis.
The theoretical migrations Mr and pI were based on Mascot searches.
ND, nondetectable.
Several proteins of interest gave rise to a doublet or triplet of spots with similar Mrs and slightly different pIs due to either posttranslational modification or minor amino acid changes (23). Thus, 19 proteins were identified out of 26 spots (Fig. 3 and Table 1). Of the 19 proteins, there were 7 induced and 10 newly detected proteins in the pyrene-treated sample and 9 induced and 10 newly detected proteins in the pyrene-4,5-dione-treated sample. Comparison of the two treatments showed that all induced and newly detected proteins in the pyrene-treated sample were also observed in the pyrene-4,5-dione-treated sample. However, there were two additional proteins that were expressed in elevated amounts upon exposure to pyrene-4,5-dione only.
P1 was identified as an iron-sulfur binding protein in Mycobacterium sp. strain KMS. P2 matched to a beta subunit of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS. P3 matched to a beta subunit of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS, Mycobacterium sp. strain JLS, M. flavescens PYR-GCK, and M. vanbaalenii PYR-1, with the same Mascot score, matched peptides, and sequence coverage but slightly different Mrs and pIs. A Mascot search of P4 gave only one significant hit, which was a sterol-binding protein in Mycobacterium sp. strain KMS. A search of P5 also gave only one significant hit to a hypothetical protein, MkmsDRAFT_0077, in Mycobacterium sp. strain KMS. P6 matched to a beta subunit of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS.
P7 was identified as similar to a beta subunit of aromatic-ring-hydroxylating dioxygenase (nidB and nidB2 in M. vanbaalenii PYR-1 and nidB in Mycobacterium sp. strain MCS). P8 matched to another beta subunit of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS, with different Mrs and pIs than those of P2, P3, P6, and P7. P9 matched to a phthalate dihydrodiol dehydrogenase in M. vanbaalenii PYR-1. P10 was a triplet and matched to an alpha subunit, the Rieske (2Fe-2S) region of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS, Mycobacterium sp. strain JLS, M. flavescens PYR-GCK, and M. vanbaalenii PYR-1, with slightly different Mrs and pIs. P11 was a doublet and was identified as a glycosyl hydrolase Bacterial neuraminidase repeat (BNR) in Mycobacterium sp. strain KMS and Mycobacterium sp. strain JLS. P12 matched to an epoxide hydrolase-like alpha/beta hydrolase fold in Mycobacterium sp. strain KMS.
P13 matched to an alpha subunit, the Rieske (2Fe-2S) region of aromatic-ring-hydroxylating dioxygenase in Mycobacterium sp. strain KMS with different Mrs and pIs than those of P10. P14 was a doublet and was identified as an alpha subunit of aromatic-ring-hydroxylating dioxygenase (nidA) in Mycobacterium sp. strain KMS. It also matched to the alpha subunit of aromatic-ring-hydroxylating dioxygenase (nidA) in Mycobacterium sp. strain JLS, Mycobacterium sp. strain S65, M. frederiksbergense FAn9T, Mycobacterium sp. strain CH-2, Mycobacterium sp. strain MHP-1, M. gilvum, and M. vanbaalenii PYR-1. P15 was a doublet that matched to fumarate reductase-succinate dehydrogenase flavoprotein-like flavin adenine dinucleotide (FAD)-dependent oxidoreductase in Mycobacterium sp. strain KMS, Mycobacterium sp. strain JLS, and M. flavescens PYR-GCK and to a dehydrogenase in M. vanbaalenii PYR-1. P16 matched to PhdG-hydratase-aldolase in M. vanbaalenii PYR-1. P17 was a doublet and matched to aldehyde dehydrogenases (nidD) in Mycobacterium sp. strain KMS and M. vanbaalenii PYR-1.
Q1 matched to a zinc-containing alcohol dehydrogenase belonging to an alcohol dehydrogenase superfamily in Mycobacterium sp. strain KMS. Q2 was identical to cytochrome P450 in Mycobacterium sp. strain KMS.
DISCUSSION
Isolation and identification of pyrene degradation intermediates revealed similar compounds that were reported for other mycobacteria. cis-4,5-Pyrene-dihydrodiol has been found in several other bacterial species. The formation of this intermediate is through the addition of two oxygen atoms by dioxygenase, which was confirmed by the identification of the dioxygenase genes in Mycobacterium sp. strain KMS (7), M. vanbaalenii PYR-1 (13), and Mycobacterium sp. strain 6PY1 (18) and the enzyme in M. vanbaalenii PYR-1 (13) and Mycobacterium sp. strain KMS (this study).
Dioxygenase from several Mycobacterium species, including M. vanbaalenii PYR-1, M. flavescens PYR-GCK, M. frederiksbergense FAn9T, Mycobacterium sp. strain RJGII-135 (1), Mycobacterium sp. strain KMS, Mycobacterium sp. strain JLS, and Mycobacterium sp. strain MCS (7), has been proposed as a naphthalene-induced dioxygenase system. Dioxygenase enzyme is a multicomponent protein that includes an electron transport chain and a terminal dioxygenase (21). The terminal dioxygenase is composed of large alpha and small beta subunits (11, 21). The alpha subunit is the catalytic component and contains two conserved regions, the Rieske (2Fe-2S) center and the mononuclear iron binding domain, which are involved in the consecutive electron transfer to the dioxygen molecule (24).
The large alpha and small beta subunits, the Rieske center, and the ferredoxin of dioxygenase have been identified in this study. This is the most complete study that has been undertaken to indicate the presence of different components of dioxygenase in one gel and demonstrated that Mycobacterium sp. strain KMS has the capability to highly express these components to degrade PAHs.
This study also showed that there were five different small subunits (P2, P3, P6, P7, and P8) and two different Rieske (2Fe-2S) regions of the alpha subunit (P10 and P13) of aromatic-ring-hydroxylating dioxygenase, which may indicate that multiple copies of dioxygenase exist and are expressed during pyrene degradation. This observation is supported by the genomic sequences of Mycobacterium sp. strain KMS, Mycobacterium sp. strain JLS, Mycobacterium sp. strain MCS, M. vanbaalenii PYR-1, and M. flavescens PYR-GCK, each of which contains more than 70 dioxygenase genes (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). Multiple copies of the dioxygenase gene were also reported for Mycobacterium sp. strain 6PY1 (18).
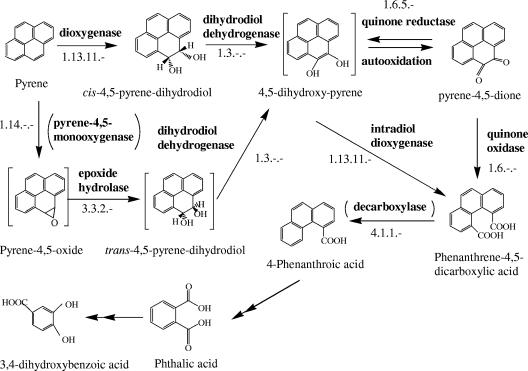
trans-4,5-Pyrene-dihydrodiol, reported in previous studies (9, 29), was not identified in this study, but the enzyme responsible for its formation, epoxide hydrolase, was identified. Therefore, this metabolite was included in the pyrene degradation pathway (Fig. 4). On contig 39 of the genome sequence of Mycobacterium sp. strain KMS, the epoxide hydrolase gene is clustered with other PAH-degrading genes.
FIG. 4.
Proposed pyrene degradation pathway of Mycobacterium sp. strain KMS. Single arrows represent one step; the double arrows indicate multiple steps. Enzymes and their EC numbers are listed for each single step. The intermediates shown in brackets and the enzymes in parentheses are not identified in this study.
1-Hydroxypyrene has been identified as a pyrene degradation intermediate in Mycobacterium sp. strain KMS (this study), M. vanbaalenii PYR-1 (9), and Pseudomonas sp. XPW2 cultures (32). It has been isolated from and identified in PAH-contaminated soil after active bioremediation (27). The cytochrome P450 pipA gene of M. vanbaalenii PYR-1 was proposed to be responsible for the formation of 1-hydroxypyrene (2). Cytochrome P450 was detected in the pyrene-treated sample but not at an elevated level. The function of this protein requires further investigation.
4,5-Dihydroxypyrene was not identified in this study due to the same reasons as reported by others (9, 12, 15). However, dihydrodiol dehydrogenase was identified as a phthalate dihydrodiol dehydrogenase, which also indicates that the later stage of pyrene degradation follows the phthalate pathway as an alternative to the Evans pathway (3).
Pyrene-4,5-dione was identified to be a pyrene degradation metabolite of Mycobacterium sp. strain KMS. This is the first study to show that a Mycobacterium species transforms pyrene to quinone. Formation of pyrene-4,5-dione is a result of nonenzymatic autooxidation of 4,5-dihydroxypyrene. In this study, pyrene-4,5-dione was degraded when it was added directly to a Mycobacterium sp. strain KMS culture. Identification of degradation products from pyrene-4,5-dione revealed the presence of two major intermediates, including phenanthrene-4,5-dicarboxylic acid and 4-phenanthroic acid. Flavoprotein-like, FAD-dependent oxidoreductase was identified and proposed to be involved in pyrene-4,5-dione-related reactions.
Pyrene-4,5-dione may be reduced back to 4,5-dihydroxypyrene by quinone reductase (PQR) as reported for Mycobacterium sp. strain PYR100 and M. vanbaalenii PYR-1 (15, 17). The activity of PQR in Mycobacterium sp. strain KMS remains unknown and is under further investigation. Even with the existence of PQR in Mycobacterium sp. strain KMS, the possibility of further oxidation of pyrene-4,5-dione cannot be excluded, considering its redox-reactive property. Hammel et al. (8) reported that 9,10-phenanthrene quinone was oxidized to form 2, 2′-diphenic acid by hydrogen peroxide in ligninolytic fungus. When 4,5-dihydroxypyrene is autooxidized to pyrene-4,5-dione, reactive oxygen species are released, which may further oxidize pyrene-4,5-dione and break the central ring.
In the transformation from pyrene to pryene-4,5-dione, at least two enzymes are involved, dioxygenase and dihydrodiol dehydrogenase. However, these two proteins were also found in the pyrene-4,5-dione-treated sample. This observation was explained by the genomic sequence of contig 66 of Mycobacterium sp. strain KMS, where oxidoreductase clusters with the beta subunit of dioxygenase and the iron-sulfur region of the large alpha subunit of dioxygenase. In other words, since genes encoding these proteins are on the same operon, when Mycobacterium sp. strain KMS was exposed to pyrene-4,5-dione and the oxidoreductase gene was expressed, all the other genes on this operon should also be expressed.
Decarboxylase, proposed to transform phenanthrene-4,5-dicarboxylic acid to 4-phenanthroic acid, was not identified in this study and other published works. However, searching the genomic sequence of Mycobacterium sp. strain KMS revealed 24 copies of the decarboxylase gene. The reason for not identifying this protein may be that its gene has a low level of expression.
The hydratase aldolase identified in this study may be involved in the transformation of trans-4-(1′-hydroxynaphth-2′-yl)-2-oxobut-3-enoic acid to 1-hydroxy-2-naphthoic acid and trans-2′-carboxybenzalpyruvic acid to 2-carboxybenaldehyde, which takes place at the later stage of pyrene degradation. This protein was also identified as a pyrene-induced protein in Mycobacterium sp. strain 6PY1 (18).
The genes encoding several proteins, including aldehyde dehydrogenase, sterol binding protein, the glycosyl hydrolase BNR, and the hypothetical MkmsDRAFT_0077, cluster with other PAH-degrading genes on either contig 66 or contig 39. Even though their exact functions were not known at the time this paper was submitted for publication, they must be related to PAH degradation somehow.
Two proteins, alcohol dehydrogenase and cytochrome P450, were highly expressed with the pyrene-4,5-dione treatment but not with the pyrene treatment. Investigation of their functions is under way.
Overall, this paper provided a detailed study of pyrene degradation intermediates and the proteins involved in pyrene degradation by Mycobacterium sp. strain KMS. The identification of pyrene-4,5-dione and its further degradation products not only helps to understand the pyrene degradation pathway, but also makes Mycobacterium sp. strain KMS a suitable candidate for in situ bioremediation of PAH-contaminated sites. This study has shown that Mycobacterium sp. strain KMS must have unique mechanisms to tolerate the toxicity of pyrene-4,5-dione and further degrade it to less-toxic chemicals.
All the enzymes required in the initial steps of pyrene degradation were identified except decarboxylase and pyrene-4,5-monooxygenase. Identification of these proteins confirms the proposed pyrene degradation pathway and defines the biochemical mechanisms of pyrene degradation.
Acknowledgments
We acknowledge financial support for this study from the NSF Phytoremediation Project (IBN-0346539), the Inland Northwest Research Alliance (INRA), and the Huntsman Environmental Research Center (HERC) at Utah State University.
We thank G.-Q. Chen and P. Dobrowolski for helping with the identification of pyrene metabolites. Also appreciated is the collaboration with the research group of Carl E. Cerniglia of the Division of Microbiology, National Center for Toxicological Research, U.S. Food and Drug Administration, Jefferson, Arkansas.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Brezna, B., A. A. Khan, and C. E. Cerniglia. 2003. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol. Lett. 223:177-183. [DOI] [PubMed] [Google Scholar]
- 2.Brezna, B., O. Kweon, R. L. Stingley, J. P. Freeman, A. A. Khan, B. Polek, R. C. Jones, and C. E. Cerniglia. 2005. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 11:1-11. [DOI] [PubMed] [Google Scholar]
- 3.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 4.Dean-Ross, D., and C. E. Cerniglia. 1996. Degradation of pyrene by Mycobacterium flavescens. Appl. Microbiol. Biotechnol. 46:307-312. [DOI] [PubMed] [Google Scholar]
- 5.Fara, F., S. Berselli, P. Conte, A. Piccolo, and L. Marchetti. 2004. Effect of humic substances and soya lecithin on the aerobic bioremediation of a soil historically contaminated by polycyclic aromatic hydrocarbons (PAHs). Biotechnol. Bioeng. 88:214-223. [DOI] [PubMed] [Google Scholar]
- 6.Guthrie-Nichols, E., A. Grasham, C. Kazunga, R. Sangaiah, A. Gold, J. Bortiatynski, M. Salloum, and P. Hatcher. 2003. The effect of aging on pyrene transformation in sediments. Environ. Toxicol. Chem. 22:40-49. [PubMed] [Google Scholar]
- 7.Hall, K. M. C., D. L. Sorensen, A. J. Anderson, and R. C. Sims. 2005. Development of a catabolically significant genetic probe for polycyclic aromatic hydrocarbon-degrading mycobacteria in soil. Biodegradation 16:475-484. [DOI] [PubMed] [Google Scholar]
- 8.Hammel, K. E., W. Z. Gai, B. Green, and M. A. Moen. 1992. Oxidative degradation of phenanthrene by the liginolytic fungus Phanerochaete chrysoporium. Appl. Environ. Microbiol. 58:1832-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitkamp, M. A., J. P. Freeman, D. W. Miller, and C. E. Cerniglia. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimenez, C. R., L. Huang, Y. Qiu, and A. L. Burlingame. 2003. In-gel digestion of proteins for MALDI-MS fingerprint mapping, p. 16.4.1-16.4.5. In J. E. Coligan, B. M. Dunn, H. L. Ploegh, D. W. Speicher, and P. T. Wingfield (ed.), Current protocols in protein science. Wiley, Hoboken, N.J. [DOI] [PubMed]
- 11.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 12.Kazunga, C., and M. D. Aitken. 2000. Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl. Environ. Microbiol. 66:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan, A. A., R.-F. Wang, W.-W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, S. J., R. C. Jones, C. J. Cha, O. Kweon, R. D. Edmondson, and C. E. Cerniglia. 2004. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics 4:3899-3908. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. H., K. H. Engesser, and C. E. Cerniglia. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch. Biochem. Biophys. 416:209-217. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y.-H., J. P. Freeman, J. D. Moody, K.-H. Engesser, and C. E. Cerniglia. 2005. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 67:275-285. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. H., J. D. Moody, J. P. Freeman, B. Brezna, K. H. Engesser, and C. E. Cerniglia. 2004. Evidence for the existence of PAH-quinone reductase and catechol-O-methyltransferase in Mycobacterium vanbaalenii PYR-1. J. Ind. Microbiol. Biotechnol. 31:507-516. [DOI] [PubMed] [Google Scholar]
- 18.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, Y.-N. 2003. Degradation and intermediates of pyrene by Mycobacterium sp. JLS, KMS, and MCS, isolated from soil at a former wood-preserving facility. M.S. thesis, Utah State University, Logan.
- 20.Liang, Y. N. 2006. Pyrene degradation by Mycobacterium sp. KMS: pathway, enzymatic mechanisms, and humic acid effect. Ph.D. dissertation, Utah State University, Logan.
- 21.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 22.Miller, C. D., K. Hall, Y.-N. Liang, K. Nieman, D. Sorensen, B. Issa, A. J. Anderson, and R. C. Sims. 2004. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading Mycobacterium isolates from soil. Microb. Ecol. 48:230-238. [DOI] [PubMed] [Google Scholar]
- 23.Packer, N. H., A. Pawlak, W. C. Kett, A. A. Gooley, J. W. Redmond, and K. L. Williams. 1997. Proteome analysis of glycoforms: strategies for the microcharacterisation of glycoproteins separated by 2D PAGE. Electrophoresis 18:452-460. [DOI] [PubMed] [Google Scholar]
- 24.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penning, T. M., M. E. Burczynski, C. F. Hung, K. D. McCoull, N. T. Palackal, and L. S. Tsuruda. 1999. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem. Res. Toxicol. 12:1-18. [DOI] [PubMed] [Google Scholar]
- 26.Rehmann, K., H. P. Noll, C. E. W. Steinberg, and A. A. Kettrup. 1998. Pyrene degradation by Mycobacterium strain KR2. Chemosphere 36:2977-2992. [DOI] [PubMed] [Google Scholar]
- 27.Roper, J. C., and F. K. Pfaender. 2001. Pyrene and chrysene fate in surface soil and sand microcosms. Environ. Toxicol. Chem. 20:223-230. [PubMed] [Google Scholar]
- 28.Schneider, J., R. Grosser, K. Jayasimhulu, W. Xue, and D. Warshawsky. 1996. Degradation of pyrene, benzo[a]anthracene and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 62:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vila, J., Z. Lopez, J. Sabate, C. Minguillon, A. M. Solanas, and M. Grifoll. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, R. F., D. Wennerstorm, W. W. Cao, A. A. Khan, and C. E. Cerniglia. 2000. Cloning, expression, and characterization of the katG gene, encoding catalase-peroxidase, from the polycyclic aromatic hydrocarbon-degrading bacterium Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 66:4300-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young, E. R. R., and R. L. Funk. 1998. A practical synthesis of pyrene-4,5-dione. J. Org. Chem. 63:9995-9996. [Google Scholar]
- 32.Zylstra, G. J., X. P. Wang, E. Kim, and V. A. Didolkar. 1994. Cloning and analysis of the genes for polycyclic aromatic hydrocarbon degradation. Ann. N. Y. Acad. Sci.-Recombinant DNA Technol. 721:386-398. [DOI] [PubMed] [Google Scholar]