Abstract
1,1,1-Trichloroethane (1,1,1-TCA) is a common groundwater pollutant as a result of improper disposal and accidental spills. It is often found as a cocontaminant with trichloroethene (TCE) and inhibits some TCE-degrading microorganisms. 1,1,1-TCA removal is therefore required for effective bioremediation of sites contaminated with mixed chlorinated organics. This study characterized MS, a 1,1,1-TCA-degrading, anaerobic, mixed microbial culture derived from a 1,1,1-TCA-contaminated site in the northeastern United States. MS reductively dechlorinated 1,1,1-TCA to 1,1-dichloroethane (1,1-DCA) and then to monochloroethane (CA) but not further. Cloning of bacterial 16S rRNA genes revealed among other organisms the presence of a Dehalobacter sp. and a Desulfovibrio sp., which are both phylogenetically related to known dehalorespiring strains. Monitoring of these populations with species-specific quantitative PCR during degradation of 1,1,1-TCA and 1,1-DCA showed that Dehalobacter proliferated during dechlorination. Dehalobacter growth was dechlorination dependent, whereas Desulfovibrio growth was dechlorination independent. Experiments were also performed to test whether MS could enhance TCE degradation in the presence of inhibiting levels of 1,1,1-TCA. Dechlorination of cis-dichloroethene (cDCE) and vinyl chloride (VC) in KB-1, a chloroethene-degrading culture used for bioaugmentation, was inhibited with 1,1,1-TCA present. When KB-1 and MS were coinoculated, degradation of cDCE and VC to ethene proceeded as soon as the 1,1,1-TCA was dechlorinated to 1,1-DCA by MS. This demonstrated the potential application of the MS and KB-1 cultures for cobioaugmentation of sites cocontaminated with 1,1,1-TCA and TCE.
The heavy industrial use of trichloroethene (TCE) as a chlorinated solvent peaked in the early 1970s, when the compound was shown to have both adverse human and adverse environmental effects (12). Consequentially, a seemingly safer alternative, 1,1,1-trichloroethane (1,1,1-TCA), was widely adopted. While acute exposure to large doses of 1,1,1-TCA is known to depress the central nervous system, no link to cancer through chronic exposure has been demonstrated (4). However, 1,1,1-TCA is an ozone-depleting substance, so its manufacture and use have been phased out as of 1996 in accordance with the Montreal Protocol on Substances that Deplete the Ozone Layer (48). Nonetheless, due to a legacy of historical accidental spillage and improper disposal, 1,1,1-TCA is a common groundwater contaminant. In the United States, 1,1,1-TCA has been found as a contaminant in at least 459 sites (30% of total) designated to the National Priorities List (NPL) by the U.S. Environmental Protection Agency (data from a search of the NPL database in January 2006).
1,1,1-TCA undergoes both abiotic and biotic degradation under natural conditions. Abiotically, the compound is dehydrohalogenated to 1,1-dichloroethene (1,1-DCE) and acetic acid, with a half-life of >2.8 years (24, 50). Biological degradation by aerobic microorganisms can occur cometabolically, with the production of 2,2,2-trichloroethanol, trichloroacetic acid, and dichloroacetic acid (26). However, because impacted groundwater is often anoxic, anaerobic degradation of 1,1,1-TCA has greater relevance for site remediation. The high density and low solubility of 1,1,1-TCA allow it to form dense non-aqueous-phase layers deep in the subsurface, making anaerobic conditions particularly important. Fortunately, several anaerobic microorganisms, including methanogens (e.g., references 1 and 10) and sulfate reducers (10), can reductively dechlorinate 1,1,1-TCA cometabolically, usually resulting in the partially dechlorinated daughter products 1,1-dichloroethane (1,1-DCA) and monochloroethane (CA). Several years ago, an organism (strain TCA1) phylogenetically related to the tetrachloroethene (PCE)- and TCE-respiring Dehalobacter restrictus that coupled 1,1,1-TCA and 1,1-DCA degradation to growth was isolated (45). It is not known how widespread 1,1,1-TCA-dehalorespiring organisms are, but these studies clearly illustrate the potential for using biological processes to remediate sites contaminated with 1,1,1-TCA.
While 1,1,1-TCA can be biologically degraded, it has also been observed to inhibit some anaerobic biological processes, including methanogenesis (2, 11, 43) and reductive dechlorination (17). Inhibition of reductive dechlorination is significant because many sites are contaminated by multiple chlorinated constituents, and it is often the more toxic compounds, such as PCE and TCE, that drive remedial efforts; bioremediation of chlorinated, ethene-contaminated sites has proven to be a successful technology (19), but its success can be limited by cocontaminants (32). The most prevalent inhibitory cocontaminants are chloroform and 1,1,1-TCA, both of which interfere with the complete detoxification of chlorinated ethenes (17). Given that 1,1,1-TCA and TCE are found as cocontaminants in at least 310 (20%) USEPA NPL sites (data from a search of the NPL database in May 2006), the inhibitory effects of 1,1,1-TCA are a real obstacle to bioremediation efforts.
In this study, a mixed anaerobic microbial culture that reductively dechlorinates 1,1,1-TCA to 1,1-DCA and CA is described. This culture, known as MS, was enriched from sediment derived from a 1,1,1-TCA-contaminated site in the northeastern United States. 16S rRNA gene cloning and quantitative PCR (qPCR) were used to identify the dechlorinating organism and to demonstrate that 1,1,1-TCA degradation occurs metabolically. The potential to overcome 1,1,1-TCA-mediated inhibition of reductive dechlorination of chlorinated ethenes by simultaneous bioaugmentation with the MS and KB-1 cultures was also investigated.
MATERIALS AND METHODS
Establishment of enrichment cultures and subcultures.
Anaerobic microcosms were constructed in 2001 with groundwater and solids from a northeastern United States industrial area contaminated with high concentrations of 1,1,1-TCA (38 μM) and TCE (8 μM). Subsurface cores were collected from two locations in the saturated anoxic zone at a depth of approximately 30 feet. The cores consisted of fine and silty sand and were combined and homogenized prior to being dispensed into microcosm bottles. Microcosm preparation was conducted in a disposable glove bag under an N2 headspace. Homogenized subsurface material (30 g) and site groundwater (100 ml) were anaerobically transferred to sterile bottles (250 ml), purged with N2-CO2 (80%/20%) to remove volatile organic compounds, and then amended with a carbon source mixture of methanol, ethanol, acetate, and lactate (a mixture of equal electron equivalents of each compound, referred to as “MEAL”) as the electron donor and 1,1,1-TCA (0.04 mM) as the electron acceptor. The original microcosms and subsequent enrichment cultures were maintained statically in an anaerobic chamber (Coy Laboratory Products, Madison, WI) filled with a CO2-H2-N2 (10%/10%/80%) gas mixture. After dechlorination activity was observed (5 months), transfer cultures were prepared in defined anaerobic mineral medium containing vitamins (18). These transfer cultures were amended with 1,1,1-TCA at initial aqueous concentrations ranging from 0.08 to 0.20 mM. MEAL was provided to these cultures as an electron donor at concentrations ranging from 0.27 to 1.33 mM, each compound representing approximately 10 times the number of electron equivalents required for dechlorination. These amendments were calculated on the assumption that two electron equivalents of donor are required per dechlorination step and that methanol, ethanol, acetate, and lactate provide 6, 12, 8, and 12 electron equivalents per mol, respectively. When dechlorinating activity ceased, 20 to 50% of the culture was replaced with fresh mineral medium (12 times since 2001). This enrichment culture is referred to as MS.
To test whether hydrogen could act as a sole electron donor and to initiate the isolation of the 1,1,1-TCA-degrading microorganism, a subculture of MS, with hydrogen as the electron donor and 1,1,1-TCA (0.07 mM) as the electron acceptor, was established in 160-ml bottles containing 100 ml of mineral medium amended with 5 mM acetate and 6 ml of H2-CO2 (80%/20%). Upon the degradation of two amendments of 1,1,1-TCA, a 5% transfer into a new bottle was made. Five transfers have been performed to date in this manner. This subculture is referred to MS/H2.
In some experiments, KB-1, a TCE-dechlorinating enrichment culture used commercially for bioaugmentation (SiREM, Guelph, ON, Canada), was also used. This culture has been maintained on TCE and methanol as an electron donor for 8 years (17) and is dominated by Dehalococcoides (16).
Substrate range study.
The ability of the MS culture to degrade 1,2-dichloroethane (1,2-DCA), 1,1,2-trichloroethane (1,1,2-TCA), PCE, TCE, cis-dichloroethene (cDCE), and vinyl chloride (VC) over a 6-week period was tested. Duplicate screw-top vials (45 ml) with Mininert septa (VICI Precision Sampling, Baton Rouge, LA) were filled with 10 ml of mineral medium and 10 ml of MS culture. These were amended with each potential chlorinated electron acceptor at an aqueous concentration of 0.06 to 0.10 mM (10 mg/liter) and MEAL as an electron donor, with each MEAL component at a concentration representing approximately 10 times the number of electron equivalents required for dechlorination. Uninoculated controls were prepared for each electron acceptor.
The maximum concentration of 1,1,1-TCA that could be degraded by MS was determined by inoculating 5 ml mineral medium with 5 ml of MS culture in 17-ml screw-top vials with Mininert septa. Duplicate vials were amended with increasing volumes of a saturated aqueous 1,1,1-TCA solution (1,400 mg/liter), and MEAL was added as an electron donor, with each component at a concentration representing approximately 10 times the number of electron equivalents required for dechlorination.
16S rRNA gene cloning.
To identify the bacterial populations present, bacterial 16S rRNA genes were cloned from MS following extraction of total genomic DNA, as previously described (25). Thirty-seven 16S rRNA gene clones were sequenced by the University Health Network Research DNA Sequencing Facility (Toronto, ON, Canada) with the primer 8f (38), and the closest sequence match was identified with a BLASTN search of GenBank (www.ncbi.nlm.nih.gov/blast).
Time course experiment.
The growth of putative dechlorinating microorganisms during degradation of 1,1,1-TCA and 1,1-DCA was monitored. Screw-top bottles (250 ml) with Mininert septa were filled with either 160 ml (for 1,1,1-TCA treatments and no-electron-acceptor controls) or 120 ml (for 1,1-DCA treatments) mineral medium. Three bottles were amended with either neat 1,1,1-TCA (initial aqueous concentration of 0.19 mM) or neat 1,1-DCA (0.26 mM) and MEAL (each component at a concentration representing approximately 5 times the number of electron equivalents required for dechlorination, for a total of 20 times the number of electron equivalents). Other bottles were amended with MEAL only, as a no-electron-acceptor control. A 1.5% (vol/vol) inoculum of MS was added. Immediately after inoculation, 40 ml of culture was removed for DNA extraction (time point 0). For the 1,1,1-TCA treatments, DNA was subsequently extracted from 40-ml samples from all bottles when 1,1,1-TCA was completely degraded to 1,1-DCA (time point 1), when 1,1-DCA was 60% degraded to CA (time point 2), and when 1,1-DCA was fully degraded to CA (time point 3). Samples from the no-electron-acceptor bottles were taken at the same times as those from the 1,1,1-TCA treatment bottles. For the 1,1-DCA treatments, DNA was extracted when the 1,1-DCA was 60% degraded to CA (time point 1) and when 1,1-DCA was fully degraded to CA (time point 2). All bottles were reamended with MEAL (with a total of 5 times the number of electron equivalents) after the DNA extractions at time point 1 to ensure that the electron donor was not limiting.
For DNA extractions, culture was transferred to anaerobic conical centrifuge tubes (50 ml) (Fisher Scientific, Toronto, ON, Canada) and centrifuged at 2,300 × g for 50 min at 4°C. The pellet was collected and DNA extracted with a MoBio UltraClean soil DNA kit according to the manufacturer's alternative protocol, except that the DNA was finally eluted with 5 mM Tris-HCl, pH 8.0. The copies of Dehalobacter and Desulfovibrio 16S rRNA genes in the extracted DNA were analyzed by quantitative PCR (see below).
The growth yield of each organism was determined by assuming 100% DNA extraction efficiency (16). Yield was calculated by first determining how many moles of the chlorinated compound (1,1,1-TCA or 1,1-DCA) were degraded between two time points, considering both liquid and headspace in the bottle and taking into account mass removed with DNA extraction. Changes in 16S rRNA gene copy number were calculated for the same time periods. A yield in units of 16S rRNA gene copies per mole of chlorinated compound degraded during a single reductive dechlorination step was then determined.
Coaugmentation studies.
Coaugmentation studies were conducted to evaluate interactions between 1,1,1-TCA and TCE dechlorination because 1,1,1-TCA can inhibit methanogenesis (2, 11, 43) and TCE dechlorination (17). A comprehensive treatment matrix was prepared to compare the dechlorination of 1,1,1-TCA alone, TCE alone, or 1,1,1-TCA and TCE together and was inoculated with either MS, KB-1, or both MS and KB-1. Duplicate screw-top vials (45 ml) with Mininert septa were filled with 15 ml (for single-culture inoculation treatments) or 10 ml (for coinoculation treatments) of mineral medium and 5 ml each of MS and/or KB-1. Vials were amended with either 1,1,1-TCA (initial aqueous concentration of 0.30 mM), TCE (0.30 mM), or both (each at 0.30 mM). The MEAL electron donor mixture was added to provide approximately 20 times the number of electron equivalents required for dechlorination. Uninoculated controls were prepared for each electron acceptor. The vials were reamended with MEAL when dechlorination stalled.
This experiment was repeated using lower concentrations of 1,1,1-TCA (0.03 mM) and TCE (0.03 mM) and less inoculum (10% of each culture). Electron donor mixture concentrations were decreased correspondingly. Each treatment was prepared in triplicate.
Quantitative PCR.
qPCR for enumerating Dehalobacter 16S rRNA gene copies was conducted as described previously (25), except that 20-μl reaction mixtures were used, with each reaction mixture containing 10 μl of SYBR green JumpStart Taq ReadyMix, 7.2 μl of sterile water, 2 μl of a DNA template, and 0.5 μM each of the forward and reverse primers. Desulfovibrio-specific qPCR was performed in the same manner by using primers DSB1180F (5′-CCTAGGGCTACACACGTACTAA-3′) (22) and DSB1405R (5′-CCGGCTTCGGGTAAAACCAG-3′) with an annealing temperature of 61°C. Calibration was performed with serial dilutions of a known quantity of M13r-/T7f-amplified fragments of Dehalobacter or Desulfovibrio 16S rRNA gene-containing plasmids generated in the cloning study described above. The dynamic range for qPCR for both targeted 16S rRNA genes was 2 × 103 to 4 × 108 16S rRNA gene copies/reaction. DNA concentrations were determined by UV absorbance measurement (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE).
Analytical procedures.
For culture maintenance and the time course experiment, chlorinated ethanes, ethenes, methane, and ethene were measured by injecting a 300-μl-headspace sample onto a Hewlett-Packard 5890 Series II gas chromatograph (GC) fitted with a GSQ column (J&W Scientific), as previously described by Grostern et al. (25) for chlorinated ethanes and Duhamel et al. (17) for chlorinated ethenes. In coaugmentation experiments, the former GC method was used to analyze vials amended with 1,1,1-TCA only, while the latter GC method was used to analyze vials amended with TCE only. To analyze headspace samples for vials amended simultaneously with 1,1,1-TCA and TCE, the following method was used: the carrier gas pressure was initially 70 kPa, and the oven temperature was programmed to hold at 50°C for 90 s and then increase to 155°C at 30°C/min and then increase to 190°C at 4°C/min and hold for 2 min.
For substrate range experiments, chlorinated ethanes, ethenes, methane, and ethene were analyzed either in a headspace sample as described above or in a 1-ml liquid sample that was mixed with 5 ml of acidified water in a headspace vial (10 ml), using an HP 7694 headspace sampler (Hewlett Packard, Mississauga, ON, Canada) connected to an HP 5890A gas chromatograph (Hewlett Packard) fitted with a GSQ column and a flame ionization detector, as described previously (25).
Nucleotide sequence accession numbers.
The cloned Dehalobacter and Desulfovibrio sequences identified in these cultures were deposited in GenBank with the following accession numbers: for the Dehalobacter sp. in MS, DQ663785; for the Desulfovibrio sp. in MS, DQ663786.
RESULTS
Culture development and characterization.
Microcosms established with aquifer material and groundwater and amended with MEAL demonstrated dechlorination of 1,1,1-TCA to 1,1-DCA after 70 days and further dechlorination to CA after 130 days incubation. No degradation of CA was observed. Methanogenesis was absent in the presence of 1,1,1-TCA but commenced once 1,1,1-TCA was depleted and only 1,1-DCA remained. Active microcosms were successively transferred (12 transfers to date) into defined mineral medium, and the rate of dechlorination progressively increased from about 0.6 to over 13 μmol/liter/day.
To identify the most abundant species as well as possible dechlorinating organisms in the enriched cultures, bacterial 16S rRNA gene fragments were cloned with general bacterial primers 4 years after the initial microcosms were established. Only bacterial rRNA genes were targeted because our goal was to investigate organisms that degrade the chlorinated compounds through dehalorespiration, which is energy yielding and growth supporting. To date, no archaeal species has been shown to respire with chlorinated compounds, although cometabolic dechlorination has been observed. The closest GenBank matches for the partially sequenced clones included Dehalobacter (10 clones), Clostridium (10 clones), Desulfovibrio (6 clones), Sedimentibacter (1 clone), Spirochaete (1 clone), and clones distantly related to Aminomonas (1 clone), Oscillosporia (2 clones), and Streptomyces (1 clone). One operational taxonomic unit (five clones) could not be related to any cultured microorganisms. The Dehalobacter and Desulfovibrio sequences, in addition to being phylogenetically related to known dechlorinators, were among the most abundant in the clone library. The Dehalobacter sequence had 1,374/1,384 bp identity (99.3%) to Dehalobacter restrictus (27) and 1,353/1,384 bp identity (97.8%) to Dehalobacter strain TCA1 (45), while the Desulfovibrio sequence had 1,163/1,303 bp identity (89.3%) to Desulfovibrio strain 2BP-48 (21). Subsequent analysis focused on Dehalobacter as the putative dechlorinating organism and Desulfovibrio as a control for nondechlorinating activity.
Substrate range studies were performed on the culture to test its ability to degrade select commercial chlorinated ethanes and ethenes. Under the conditions tested, dechlorination of PCE, TCE, cDCE, VC, and 1,2-DCA was not observed in 40 days. However, 1,1,2-TCA was stoichiometrically transformed to VC via dihaloelimination in 8 days (data not shown). Concentrations of up to 1.5 mM 1,1,1-TCA (200 mg/liter) were completely dechlorinated to CA. Between 1.5 mM and 2.2 mM (300 mg/liter) 1,1,1-TCA, dechlorination proceed only as far as 1,1-DCA during the 2-month observation period. Dechlorination was completely inhibited above 2.2 mM 1,1,1-TCA.
A common characteristic of many dehalorespiring bacteria is their ability to use dihydrogen as an electron donor. Therefore, the ability of H2 to support reductive dechlorination in an MS subculture was tested. Transfer cultures sustained dechlorination of 1,1,1-TCA upon successive transfers when amended with only acetate as a carbon source and H2 as an electron donor. Methanogenic activity was no longer observed after two transfers. However, after three transfers, the ability to reductively dechlorinate 1,1-DCA to CA was lost, despite the addition of more electron donors and prolonged incubation (data not shown).
1,1,1-TCA and 1,1-DCA degradation time course experiments.
The potential for growth-linked dechlorinating activity by the Dehalobacter and Desulfovibrio populations identified in the clone library was investigated with a degradation time course experiment. Vials amended with MEAL as an electron donor and either 1,1,1-TCA or 1,1-DCA as an electron acceptor were inoculated with MS culture; no-electron-acceptor controls were also prepared. The Dehalobacter and Desulfovibrio populations were monitored during dechlorination by analyzing them for the presence of each organism with species-specific qPCR following DNA extraction. No degradation of the tested chlorinated compounds was observed in uninoculated controls.
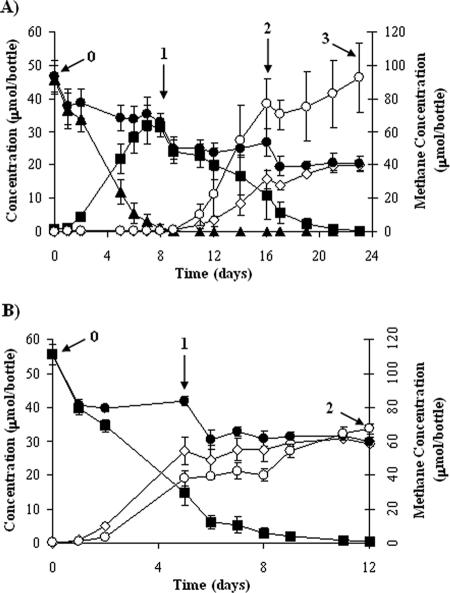
In the 1,1,1-TCA-amended bottles, degradation of 1,1,1-TCA occurred sequentially. First, 1,1,1-TCA was reductively dechlorinated with no lag to 1,1-DCA in 10 days (Fig. 1A). At this point, DNA was extracted from an aliquot of the 1,1,1-TCA and no-electron-acceptor treatment bottles. 1,1-DCA was reductively dechlorinated to CA in a further 14 days; DNA was extracted during mid-1,1-DCA degradation and when 1,1-DCA degradation was completed. Mass decreases immediately following sampling time points (Fig. 1) were due to sample removal for DNA extraction. Methanogenesis did not occur while 1,1,1-TCA was present, although it commenced during 1,1-DCA degradation. Dechlorination in the 1,1-DCA treatment bottles started with no lag and was complete in 12 days (Fig. 1B). Methanogenesis occurred throughout 1,1-DCA degradation.
FIG. 1.
Degradation profiles for 1,1,1-TCA and 1,1-DCA time course experiments. (A) Degradation in 1,1,1-TCA-amended cultures. (B) Degradation in 1,1-DCA-amended cultures. Closed triangles, 1,1,1-TCA; squares, 1,1-DCA; open diamonds, CA; closed circles, total chlorinated ethanes; open circles, methane. Numbers indicate the approximate time points for DNA extraction in the respective experiments (see the text). Each curve shows the mean value for three bottles. Error bars show the standard deviations for three bottles.
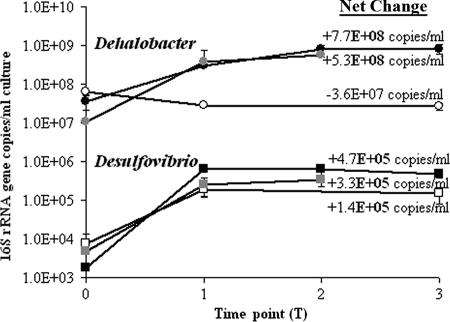
qPCR monitoring of the Dehalobacter population during dechlorination revealed that growth occurred, as shown by a 10-fold increase in 16S rRNA gene copies, during both 1,1,1-TCA and 1,1-DCA degradation in the 1,1,1-TCA amendment treatment bottles (Fig. 2). Growth during 1,1-DCA degradation was confirmed for the 1,1-DCA amendment treatments (Fig. 2). No proliferation of Dehalobacter was seen in the no-electron-acceptor treatment bottles, indicating that chlorinated compounds can be used as electron acceptors for growth by this population. These results agree with previous findings for Dehalobacter strains (27, 45, 49, 53).
FIG. 2.
Dehalobacter and Desulfovibrio growth during 1,1,1-TCA and 1,1-DCA degradation. Circles represent Dehalobacter; squares represent Desulfovibrio. Open symbols represent controls (amended with a donor only); black symbols represent bottles amended with 1,1,1-TCA; gray symbols represent bottles amended with 1,1-DCA. 16S rRNA gene copy values are mean values for three bottles and are expressed as numbers of copies per ml of culture. Numbers on the right represent net increases (+) or decreases (−) in 16S rRNA gene copies/ml from the start to the end of the experiment for each target organism in each treatment bottle. Error bars show the standard deviations for three bottles.
qPCR monitoring of the Desulfovibrio population revealed growth in all treatment bottles (Fig. 2), including the no-electron-acceptor control. Hence, this population is not strictly dependent on a chlorinated compound as an electron acceptor for growth. However, in the 1,1,1-TCA and 1,1-DCA treatment bottles, overall increases in 16S rRNA gene copies for Desulfovibrio were considerably less (∼1,600-fold) than those for Dehalobacter. Additionally, the absolute concentration of Desulfovibrio was 3 orders of magnitude lower than that for Dehalobacter in all treatment bottles. Therefore, the Desulfovibrio population's direct involvement in dechlorination, if any, was likely minor compared to the energy-yielding dehalorespiration performed by Dehalobacter in this experiment.
Assuming that most of the dechlorination was performed by Dehalobacter, growth yields for 1,1,1-TCA and 1,1-DCA could be determined by calculating the moles of the electron acceptor dechlorinated between the extraction time points, taking into account mass lost due to DNA sampling. The yields for Dehalobacter during dechlorination were (9.4 ± 1.7) × 108 16S rRNA gene copies/μmol 1,1,1-TCA degraded to 1,1-DCA (n = 3) and (2.4 ± 1.2) × 109 16S rRNA gene copies/μmol 1,1-DCA degraded to CA (n = 6). Sun et al. (45) determined a yield of 5.60 g of cells (dry weight) per mol of Cl− evolved for Dehalobacter strain TCA1. Given the cell dimensions of strain TCA1 (a short rod with a diameter of 0.4 to 0.6 μm and a length of 1.0 to 2.0 μm) reported by Sun et al. (45), approximating the cell shape to that of a cylinder, and assuming that cell density is equal to that of water and that a cell is 80% water, a yield of 8.10 × 107 cells/μmol of Cl− evolved was calculated. For the MS culture, an average yield of (1.4 ± 0.3) × 109 cells/μmol of Cl− evolved was calculated for the 1,1,1-TCA-to-CA experiment, assuming that the number of 16S rRNA gene copies in the Dehalobacter genome is one. These yields can also be compared to results from our previous study (25) on a Dehalobacter-containing culture that degraded 1,1,2-TCA (Table 1). The yield for the Dehalobacter described herein is highest on a comparable per-electron-equivalent-reduced basis and is similar to the yields obtained for a mixed culture dechlorinating TCE at a high rate.
TABLE 1.
Comparison of yields for selected Dehalobacter and Dehalococcoides spp. during dehalorespiration
| Organism group (culture)a | Processb | ΔG°′c | Yieldd | Source or reference |
|---|---|---|---|---|
| Dehalobacter sp. (MS) | 1,1,1-TCA→DCA→CA | −115.8 | (7.0 ± 1.5) × 108 | This study |
| Dehalococcoides sp. (KB-1/VC) | VC→ethene | −75.0 | (2.8 ± 0.7) × 108 | 16 |
| Dehalobacter sp. (WL) | 1,1,2-TCA→VC | −103.1 | (1.5 ± 1.4) × 108 | 25 |
| Dehalococcoides sp. (WL) | VC→ethene | −75.0 | (4.4 ± 1.3) × 107 | 25 |
| Dehalobacter strain TCA1 | 1,1,1-TCA→DCA→CA | −115.8 | 4.0 × 107e | 45 |
Except for Dehalobacter strain TCA1, all organisms were in mixed cultures.
Each dechlorination step (indicated by →) requires two electron equivalents. Except for 1,1,2-TCA→VC, all indicated processes result in release of one Cl− ion per dechlorination step.
Gibbs free energies of formation based on the work of Dolfing and Janssen (13). Values are kJ/electron equivalents for overall reactions, with H2 used as an electron donor.
Number of 16S rRNA gene copies/μmol electron equivalents reduced.
Assuming that the genome of Dehalobacter strain TCA1 has one copy of the 16S rRNA gene.
Coaugmentation studies.
The potential for the MS culture to relieve 1,1,1-TCA inhibition of TCE degradation by the KB-1 culture was investigated. Replicate vials were amended with 1,1,1-TCA, TCE, or 1,1,1-TCA and TCE and were inoculated with MS, TCE-grown KB-1, or MS and KB-1, with MEAL as the electron donor mixture. This experiment was performed twice, with different starting concentrations for the chlorinated compounds and different strengths of inoculum. In the first experiment, a 25% inoculum was used, and the targeted concentrations were 0.30 mM (40 mg/liter) for each of the chlorinated compounds. No degradation was observed in the uninoculated controls. While complete degradation of 1,1,1-TCA to 1,1-DCA and then to CA was observed in 8 days in the MS-inoculated vials, no degradation by-products were detected in 1,1,1-TCA-amended vials that were inoculated with KB-1, confirming this culture's inability to degrade this compound (data not shown). In TCE-amended vials inoculated with KB-1, complete dechlorination of TCE to ethene occurred in 20 days, whereas in the MS-inoculated vials, TCE was not degraded.
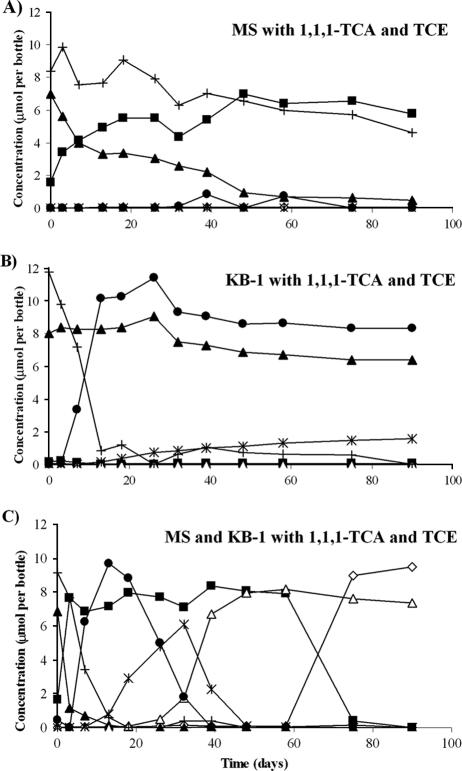
Vials amended simultaneously with 1,1,1-TCA and TCE showed more-complex degradation patterns. In vials inoculated with MS only, 1,1,1-TCA was slowly dechlorinated to 1,1-DCA (93% in 90 days) in the presence of TCE, and no further dechlorination to CA was observed (Fig. 3A). This indicates that 1,1,1-TCA degradation and, in particular, 1,1-DCA degradation by the MS culture were inhibited by TCE. Conversely, in vials inoculated with KB-1 only, no degradation of 1,1,1-TCA was observed, while TCE dechlorination stalled at cDCE and VC; no ethene was produced, even when an electron donor was provided (Fig. 3B). These results were consistent with the previous finding for KB-1 (17) that 1,1,1-TCA inhibits the degradation of TCE daughter products rather than the reductive dechlorination of TCE itself.
FIG. 3.
Representative degradation profiles of 1,1,1-TCA- and TCE-amended vials in the coaugmentation study. (A) Vials inoculated with MS only. (B) Vials inoculated with KB-1 only. (C) Vials inoculated with both MS and KB-1. Closed triangles, 1,1,1-TCA; squares, 1,1-DCA; open diamonds, CA; crosses, TCE; circles, cDCE; asterisks, VC; open triangles, ethene.
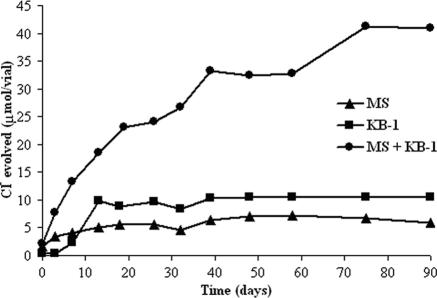
In vials amended with both MS and KB-1, dechlorination of TCE past VC did not occur until 1,1,1-TCA was fully converted to 1,1-DCA (Fig. 3C). With the 1,1,1-TCA converted, degradation of the chlorinated ethenes to ethene proceeded quickly, but degradation of 1,1-DCA stalled. After the chlorinated ethenes were fully converted to ethene, dechlorination of 1,1-DCA to CA eventually proceeded. The benefit of coaugmentation is most vividly illustrated in Fig. 4, which presents the data for the cumulative chloride evolved (as calculated from the data for the disappearance of all chlorinated substrates) with the different inocula.
FIG. 4.
Representative cumulative chloride production in 1,1,1-TCA- and TCE-amended vials from the coaugmentation study. Triangles represent vials inoculated with MS only. Squares represent vials inoculated with KB-1 only. Circles represent vials inoculated with both MS and KB-1.
To investigate whether these results were concentration dependent, this experiment was repeated with a lower inoculum (10%) of each culture and significantly lower concentrations (0.03 mM) of 1,1,1-TCA and TCE (4 mg/liter and 5 mg/liter, respectively). Degradation and inhibition patterns were very similar to what was observed with the higher starting concentrations: in the 1,1,1-TCA- and TCE-amended vials inoculated with both MS and KB-1, chlorinated ethene degradation again stalled at cDCE and VC until the 1,1,1-TCA was fully converted to 1,1-DCA, and 1,1-DCA degradation did not proceed until after the chlorinated ethenes were fully converted to ethene. These results confirmed that the observed degradation and inhibition patterns were not concentration dependent.
DISCUSSION
Recent literature focusing on the biodegradation of 1,1,1-TCA is sparse relative to that devoted to the degradation of the chlorinated ethenes. Whereas at least 25 chloroethene-respiring anaerobic isolates (in the genera Desulfomonile, Desulfuromonas, Dehalococcoides, Dehalobacter, Desulfitobacterium, Sulfospirillum, and Geobacter) have been reported, only a single 1,1,1-TCA-respiring anaerobic bacterium has been isolated. Although this research bias may be due to the higher human toxicity of chlorinated ethenes, the widespread contamination of groundwater by 1,1,1-TCA warrants increased examination of the microorganisms and conditions required for its removal. This is especially important given that 1,1,1-TCA is a common cocontaminant with TCE and that 1,1,1-TCA can inhibit chlorinated ethene-degrading organisms, resulting in the undesirable accumulation of vinyl chloride.
The lack of reported 1,1,1-TCA-enriched anaerobic cultures makes it difficult to compare the degradation characteristics of the MS culture to those of other cultures. Adamson and Parkin (2) showed that a lactate- and PCE-enriched culture could transform 20 μM 1,1,1-TCA to 1,1-DCA and partially to CA in less than a day, despite no prior exposure; higher concentrations of 1,1,1-TCA were not tested. Similarly, a lactate-enriched methanogenic culture derived from digested sludge could degrade 2 μM 1,1,1-TCA cometabolically, but again, higher concentrations were not tested (23). Dehalobacter strain TCA1 could completely dechlorinate 450 μM (60 mg/liter) 1,1,1-TCA to CA in 5 weeks (45); no degradation characteristics have been reported for the mixed culture from which this isolate was obtained. Other studies (8, 42, 43, 54) have reported the degradation of significant amounts of 1,1,1-TCA, but these concentrations were less than those tested in the present study. Therefore, while the current study represents the highest reported 1,1,1-TCA concentration degraded, little data exist for comparison.
A culture-independent approach was used to identify the putative active dechlorinating organism(s) in MS. Organisms related to Dehalobacter and Desulfovibrio were identified by bacterial 16S rRNA gene cloning, and because of their phylogenetic relationship to known dehalorespiring organisms, their growth was tracked during dechlorination of 1,1,1-TCA and 1,1-DCA. Growth of the Dehalobacter sp. in MS was found to be dependent on a chlorinated substrate, consistent with the growth requirements of other Dehalobacter strains (5, 25, 27, 45, 53). Several reported Desulfovibrio strains have the ability to reductively dehalogenate brominated or chlorinated compounds, and this occurs through either a cometabolic (5) or a dehalorespiratory (6, 21, 44) process. Given that the Desulfovibrio strain in MS grew in the absence of 1,1,1-TCA and 1,1-DCA, this strain is not dependent on a halogenated substrate for growth; rather, it was likely growing fermentatively on the ample electron donors provided. Growth in the presence of the chlorinated compounds was slightly higher than that in the absence of these compounds, presumably as a result of a syntrophic relationship between Desulfovibrio and Dehalobacter, mediated by interspecies H2 transfer, similar to that seen between Desulfitobacterium frappieri TCE1 and Desulfovibrio fructosivorans (14).
The narrowness of the substrate range observed for the MS culture is analogous to that observed for Dehalobacter isolates (27, 45, 53) and other reported Dehalobacter-containing enrichment cultures (25, 30, 40, 49, 51). For example, the only tested chlorinated compounds that Dehalobacter strain TCA1 can dehalorespire are 1,1,1-TCA and 1,1-DCA (45), while Dehalobacter restrictus strains PER-K23 and TEA can dehalorespire only PCE and TCE to cDCE (27, 53). Therefore, while it is known that Dehalobacter has a narrow electron donor range (hence the species name “restrictus”), it seems that a restricted chlorinated electron acceptor range is also a general property of this genus. A determining factor may be the presence of small numbers of narrow-substrate-range reductive dehalogenase (RDase) genes in the Dehalobacter genomes. A PCR-based study by Regeard et al. (37) identified four putative or verified RDase genes in Dehalobacter restrictus PER-K23 genomic DNA. While the genome of this organism has not been sequenced, and thus the total number of putative RDase genes is unknown, this number of RDase genes can be compared to that found in recently sequenced genomes of Dehalococcoides and Desulfitobacterium species. Dehalococcoides ethenogenes strain 195 (41) and Dehalococcoides strain CBDB1 (31) have an astounding 17 and 32 putative RDase genes, respectively, and the organisms have been shown to degrade at least 25 (20, 33-35) and 16 (3, 7, 28, 29) chlorinated compounds, respectively. Desulfitobacterium hafniense strains Y51 (36) and DCB-2 (data from the D. hafniense DCB-2 whole-genome shotgun project; GenBank accession number AAAW00000000) have two and nine putative RDase genes, respectively, and are known to degrade seven (47) and two (9) chlorinated compounds, respectively. Evidently, the D. restrictus PER-K23 putative RDase gene number is lower than those of many dechlorinating organisms studied, which correlates with its narrow chlorinated-substrate range.
The loss of 1,1-DCA-degrading ability in the MS/H2 transfers indicates that the dechlorination processes of 1,1,1-TCA and 1,1-DCA differ; this may be at either the organism or the enzyme level. While this study demonstrated growth of Dehalobacter during dechlorination of both 1,1-DCA and 1,1,1-TCA, it is possible that actually two or more distinct strains of Dehalobacter possessing near-identical 16S rRNA genes were present in the enrichment culture. Their growths would not be distinguished with the Dehalobacter-specific qPCR primers used here. Although the presence of multiple Dehalobacter strains in mixed culture has not been reported, similar occurrences have been observed in chlorinated ethene-degrading cultures containing multiple strains of Dehalococcoides with identical or near-identical 16S rRNA genes (16, 39, 46, 52). In these cases, differentiation of the strains present came either by isolation or through identification of reductive dehalogenase genes whose population copy numbers were significantly different from the total Dehalococcoides 16S rRNA gene copy number. Alternatively, the loss of 1,1-DCA-dechlorinating ability may be due to different RDases acting on 1,1,1-TCA and 1,1-DCA, where a cofactor required for active 1,1-DCA RDase is not produced under the H2-acetate conditions, either by the Dehalobacter sp. or by another organism in the mixed culture. A dependence on another organism for unknown factors influencing dechlorination has been previously reported by van Doesburg et al. (49), who observed that a Dehalobacter sp. could not degrade β-hexachlorocyclehexane in the absence of a Sedimentibacter sp.
The coaugmentation experiments described herein successfully demonstrated that if a microbial culture's ability to degrade a chlorinated compound is inhibited by the presence of a cocontaminant, that ability can be restored by removing the inhibiting cocontaminant via bioaugmentation with a second microbial culture. While the ideal situation for bioremediation involves the employment of a culture with a broad substrate range in order to degrade several cocontaminants simultaneously or sequentially, the reality is that a broad substrate range is a characteristic of highly complex microbial communities fed complex substrates (e.g., activated sludge), where many degrading populations are present. However, to obtain the large, consistent volumes of highly active culture required for the broad application of bioaugmentation for remediation of chloroorganic compounds, high-throughput culturing techniques that inevitably result in enrichment and consequently narrower substrate ranges must be employed. Therefore, the problem of degradation-inhibiting cocontaminants may necessitate bioaugmentation with two or more specialized microbial cultures. This study has shown that this strategy can, in principle, be successful.
These experiments helped to refine our understanding of the effects of complex mixtures of chlorinated compounds on the MS and KB-1 cultures. KB-1 is an ideal culture for these experiments not only because of its ability to fully degrade TCE but also because it can degrade 1,1-DCE (17), the abiotic breakdown product of 1,1,1-TCA. It was shown that the ability of KB-1 to reductively dechlorinate TCE to cDCE is not affected by 1,1,1-TCA but that subsequent degradation to VC and ethene is strongly, but reversibly, inhibited. KB-1 contains multiple dechlorinating organisms, including a Geobacter strain and at least two distinct Dehalococcoides strains (15, 17a). Geobacter grows during the reductive dechlorination of TCE to cDCE, but Dehalococcoides is likely responsible for subsequent degradation to ethene (15). Therefore, the stall at cDCE seen in the present study when KB-1 was exposed to both TCE and 1,1,1-TCA was due to direct or indirect inhibition of the Dehalococcoides populations in KB-1.
The inhibitory effect of TCE and its chlorinated degradation products on the degradation of 1,1,1-TCA and especially 1,1-DCA by the MS culture was an unexpected finding. These results revealed that a strategy consisting of first bioaugmenting a TCE-/1,1,1-TCA-cocontaminated site with MS to remove 1,1,1-TCA and then following with the application of KB-1 to remove the chlorinated ethenes could fail because these chlorinated ethenes may interfere with 1,1,1-TCA degradation. However, the inverse strategy, bioaugmenting the site with KB-1 to degrade TCE to cDCE, followed by bioaugmentation with MS to degrade 1,1,1-TCA, could result in remediation of the site. Additionally, the high chloroorganic concentrations treated in these experiments indicate that these cultures could be active in and near source zones, where high concentrations of the chlorinated compounds exist. Therefore, this study has contributed to a more informed application of these cultures for the bioremediation of chloroethane- and chloroethene-contaminated sites.
Acknowledgments
Initial microcosms were prepared by Sandra Dworatzek (SiREM, Guelph, ON, Canada), with the advice and assistance of Evan Cox (GeoSyntec Consultants, Guelph, ON, Canada). We thank Cristina Chornewich and Ekanki Saxena for conducting substrate range experiments and Joyce Dinglasan-Panlilio, YuSan Ong, and Susanna Lam for help with maintaining cultures. We are especially thankful to Melanie Duhamel (GeoSyntec Consultants) and Monisha Nandi (Golder Associates) for constructive comments on the manuscript.
The research was cofunded by the Natural Sciences and Engineering Research Council of Canada (NSERC Collaborative Research and Development Program) and GeoSyntec Consultants. A. Grostern was supported by an NSERC graduate fellowship.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Adamson, D. T., and G. F. Parkin. 1999. Biotransformation of mixtures of chlorinated aliphatic hydrocarbons by an acetate-grown methanogenic enrichment culture. Water Res. 33:1482-1494. [Google Scholar]
- 2.Adamson, D. T., and G. F. Parkin. 2000. Impact of mixtures of chlorinated aliphatic hydrocarbons on a high-rate, tetraehloroethene-dechlorinating enrichment culture. Environ. Sci. Technol. 34:1959-1965. [Google Scholar]
- 3.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 4.ATSDR. 2004. ToxFAQs for 1,1,1-trichloroethane agency for toxic substances and disease registry. [Online.] http://www.atsdr.cdc.gov/tfacts70.html. Accessed 31 January 2006.
- 5.Boyle, A. W., M. M. Häggblom, and L. Y. Young. 1999. Dehalogenation of lindane ([gamma]-hexachlorocyclohexane) by anaerobic bacteria from marine sediments and by sulfate-reducing bacteria. FEMS Microbiol. Ecol. 29:379-387. [Google Scholar]
- 6.Boyle, A. W., C. D. Phelps, and L. Y. Young. 1999. Isolation from estuarine sediments of a Desulfovibrio strain which can grow on lactate coupled to the reductive dehalogenation of 2,4,6-tribromophenol. Appl. Environ. Microbiol. 65:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. Lorenz, J. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., B. S. Ballapragada, J. A. Puhakka, S. E. Strand, and J. F. Ferguson. 1999. Anaerobic transformation of 1,1,1-trichloroethane by municipal digester sludge. Biodegradation 10:297-305. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen, N., and B. K. Ahring. 1996. Desulfitobacterium hafniense sp nov, an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46:442-448. [Google Scholar]
- 10.de Best, J. H., A. Hage, H. J. Doddema, D. B. Janssen, and W. Harder. 1999. Complete transformation of 1,1,1-trichloroethane to chloroethane by a methanogenic mixed population. Appl. Microbiol. Biotechnol. 51:277-283. [Google Scholar]
- 11.de Best, J. H., H. Jongema, A. Weijling, H. J. Doddema, D. B. Janssen, and W. Harder. 1997. Transformation of 1,1,1-trichloroethane in an anaerobic packed-bed reactor at various concentrations of 1,1,1-trichloroethane, acetate and sulfate. Appl. Microbiol. Biotechnol. 48:417-423. [Google Scholar]
- 12.Doherty, R. E. 2000. A history of the production and use of carbon tetrachloride, tetrachloroethylene, trichloroethylene and 1,1,1-trichloroethane in the United States: part 2—trichloroethylene and 1,1,1-trichloroethane, Environ. Forensics 1:83-93. [Google Scholar]
- 13.Dolfing, J., and D. B. Janssen. 1994. Estimates of Gibbs free energies of formation of chlorinated aliphatic compounds. Biodegradation 5:21-28. [Google Scholar]
- 14.Drzyzga, O., and J. C. Gottschal. 2002. Tetrachloroethene dehalorespiration and growth of Desulfitobacterium frappieri TCE1 in strict dependence on the activity of Desulfovibrio fructosivorans. Appl. Environ. Microbiol. 68:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duhamel, M. 2005. Community structure and dynamics of anaerobic chlorinated ethene-degrading enrichment cultures. Ph.D. thesis. Department of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, Ontario, Canada.
- 16.Duhamel, M., K. Mo, and E. A. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duhamel, M., S. Wehr, L. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-1,2-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 17a.Duhamel, M., and E. A. Edwards. 18 August 2006, posting date. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. [Online.] doi: 10.1111/j.1574-6941.2006.00191.x. [DOI] [PubMed]
- 18.Edwards, E. A., and D. Grbić-Galić. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ESTCP. 2006. Bioaugmentation for remediation of chlorinated ethenes: technology development, status, and research needs. U.S. Department of Defense. [Online.] http://www.estcp.org/Technology/upload/BioaugChlorinatedSol.pdf. Accessed 15 September 2006.
- 20.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 21.Fennell, D. E., S. Rhee, Y. Ahn, M. M. Häggblom, and L. J. Kerkhof. 2004. Detection and characterization of a dehalogenating microorganism by terminal restriction fragment length polymorphism fingerprinting of 16S rRNA in a sulfidogenic, 2-bromophenol-utilizing enrichment. Appl. Environ. Microbiol. 70:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fournier, M., C. Aubert, Z. Dermoun, M. C. Durand, D. Moinier, and A. Dolla. 2006. Response of the anaerobe Desulfovibrio vulgaris Hildenborough to oxidative conditions: proteome and transcript analysis. Biochimie 88:85-94. [DOI] [PubMed] [Google Scholar]
- 23.Gander, J. W., G. F. Parkin, and M. M. Scherer. 2002. Kinetics of 1,1,1-trichloroethane transformation by iron sulfide and a methanogenic consortium. Environ. Sci. Technol. 36:4540-4546. [DOI] [PubMed] [Google Scholar]
- 24.Gerkens, R. R., and J. A. Franklin. 1989. The rate of degradation of 1,1,1-trichloroethane in water by hydrolysis and dehydrochlorination. Chemosphere 19:1929-1937. [Google Scholar]
- 25.Grostern, A., and E. A. Edwards. 2006. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl. Environ. Microbiol. 72:428-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto, A., K. Iwasaki, N. Nakasugi, M. Nakajima, and O. Yagi. 2002. Degradation pathways of trichloroethylene and 1,1,1-trichloroethane by Mycobacterium sp. TA27. Biosci. Biotechnol. Biochem. 66:385-390. [DOI] [PubMed] [Google Scholar]
- 27.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and A. J. B. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 28.Hölscher, T., H. Görisch, and L. Adrian. 2003. Reductive dehalogenation of chlorobenzene congeners in cell extracts of Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 69:2999-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayachandran, G., H. Görisch, and L. Adrian. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411-416. [DOI] [PubMed] [Google Scholar]
- 30.Kengen, S. W. M., C. G. Breidenbach, A. Felske, A. J. M. Stams, G. Schraa, and W. M. de Vos. 1999. Reductive dechlorination of tetrachloroethene to cis-1,2-dichloroethene by a thermophilic anaerobic enrichment culture. Appl. Environ. Microbiol. 65:2312-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 32.Löffler, F. E., and E. A. Edwards. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274-284. [DOI] [PubMed] [Google Scholar]
- 33.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maymó-Gatell, X., Y. Chien, J. Gossett, and S. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetracholoethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 35.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes”. Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 36.Nonaka, H., G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui, and H. Yukawa. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188:2262-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regeard, C., J. Maillard, and C. Holliger. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107-118. [DOI] [PubMed] [Google Scholar]
- 38.Reysenbach, A. L., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Z. Wu, S. S. Koenigsberg, and F. E. Löffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlotelburg, C., C. von Wintzingerode, R. Hauck, F. von Wintzingerode, W. Hegemann, and U. B. Gobel. 2002. Microbial structure of an anaerobic bioreactor population that continuously dechlorinates 1,2-dichloropropane. FEMS Microbiol. Ecol. 39:229-237. [DOI] [PubMed] [Google Scholar]
- 41.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 42.Strand, S. E., J. V. Wodrich, and H. D. Stensel. 1991. Biodegradation of chlorinated solvents in a sparged, methanotrophic biofilm reactor. Res. J. Water Pollut. Control Fed. 63:859-867. [Google Scholar]
- 43.Suidan, M. T., A. M. Wuellner, and T. K. Boyer. 1991. Anaerobic treatment of a high-strength industrial-waste bearing inhibitory concentrations of 1,1,1-trichloroethane. Water Sci. Technol. 23:1385-1393. [Google Scholar]
- 44.Sun, B., J. R. Cole, R. A. Sanford, and J. M. Tiedje. 2000. Isolation and characterization of Desulfovibrio dechloracetivorans sp. nov., a marine dechlorinating bacterium growing by coupling the oxidation of acetate to the reductive dechlorination of 2-chlorophenol. Appl. Environ. Microbiol. 66:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, B., B. N. Griffin, H. L. Ayala-del-Rio, S. A. Hashsham, and J. M. Tiedje. 2002. Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298:1023-1025. [DOI] [PubMed] [Google Scholar]
- 46.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. E. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suyama, A., R. Iwakiri, K. Kai, T. Tokunaga, N. Sera, and K. Furukawa. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474-1481. [DOI] [PubMed] [Google Scholar]
- 48.UNEP. 2000. The Montreal protocol on substances that deplete the ozone layer. Secretariat for the Vienna Convention for the Protection of the Ozone Layer & The Montreal Protocol on Substances that Deplete the Ozone Layer. [Online.] http://hq.unep.org/ozone/Montreal-Protocol/Montreal-Protocol2000.shtml. Accessed 30 January 2006.
- 49.van Doesburg, W., M. H. A. van Eekert, P. J. M. Middeldorp, M. Balk, G. Schraa, and A. J. M. Stams. 2005. Reductive dechlorination of [beta]-hexachlorocyclohexane ([beta]-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 54:87-95. [DOI] [PubMed] [Google Scholar]
- 50.Vogel, T. M., and P. L. McCarty. 1987. Abiotic and biotic transformations of 1,1,1-trichloroethane under methanogenic conditions. Environ. Sci. Technol. 21:1208-1213. [Google Scholar]
- 51.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Gobel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waller, A. S., R. Krajmalnik-Brown, F. E. Löffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wild, A., R. Hermann, and T. Leisinger. 1996. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511. [DOI] [PubMed] [Google Scholar]
- 54.Wrenn, B. A., and B. E. Rittmann. 1996. Evaluation of a model for the effects of substrate interactions on the kinetics of reductive dehalogenation. Biodegradation 7:49-64. [Google Scholar]