Abstract
Astaxanthin is a high-value carotenoid which is used as a pigmentation source in fish aquaculture. Additionally, a beneficial role of astaxanthin as a food supplement for humans has been suggested. The unicellular alga Haematococcus pluvialis is a suitable biological source for astaxanthin production. In the context of the strong biotechnological relevance of H. pluvialis, we developed a genetic transformation protocol for metabolic engineering of this green alga. First, the gene coding for the carotenoid biosynthesis enzyme phytoene desaturase was isolated from H. pluvialis and modified by site-directed mutagenesis, changing the leucine codon at position 504 to an arginine codon. In an in vitro assay, the modified phytoene desaturase was still active in conversion of phytoene to ζ-carotene and exhibited 43-fold-higher resistance to the bleaching herbicide norflurazon. Upon biolistic transformation using the modified phytoene desaturase gene as a reporter and selection with norflurazon, integration into the nuclear genome of H. pluvialis and phytoene desaturase gene and protein expression were demonstrated by Southern, Northern, and Western blotting, respectively, in 11 transformants. Some of the transformants had a higher carotenoid content in the green state, which correlated with increased nonphotochemical quenching. This measurement of chlorophyll fluorescence can be used as a screening procedure for stable transformants. Stress induction of astaxanthin biosynthesis by high light showed that there was accelerated accumulation of astaxanthin in one of the transformants compared to the accumulation in the wild type. Our results strongly indicate that the modified phytoene desaturase gene is a useful tool for genetic engineering of carotenoid biosynthesis in H. pluvialis.
Of all the carotenoids used as food supplements for humans, for food coloration, and as pigmentation sources for egg yolk, the ketocarotenoid astaxanthin (3,3′-dihydroxy-diketo-β,β-carotene-4,4′-dione) is the most important from the biotechnological point of view (4, 22). Besides the use of this compound as a colorant, it has been hypothesized that supplementation with astaxanthin might be a practical and beneficial strategy in management of human health due to its neuroprotective potential, its immunomodulating potential, and its antioxidant potential (13). Today most astaxanthin is produced by total chemical synthesis and is sold at a price of $2,500 kg−1 (16); the high price and increasing demand for this compound as a food supplement provide a good opportunity for naturally produced astaxanthin.
Biosynthesis of astaxanthin has been observed in only a limited number of organisms, including some marine bacteria, the yeast Xanthophyllomyces dendrorhous, and some green algae (16). The unicellular green alga Haematococcus pluvialis accumulates the highest level of astaxanthin (up to 4%/g [dry weight]) and seems to be a very promising source of natural astaxanthin (3). A promising strategy for further improving the astaxanthin yield of H. pluvialis is genetic engineering of the carotenoid biosynthesis pathway.
There have been two reports of successful transformation of H. pluvialis by particle bombardment using the β-galactosidase gene (lacZ) (26, 41). The reporter gene was transiently expressed under control of the simian virus 40 promoter or the β-carotene ketolase promoter. Unfortunately, stable transformation was not obtained. Moreover, to date there have been no reports of a dominant selective transformation system for H. pluvialis. For establishment of a stable transformation system several genetic tools are needed. These tools include a reporter gene, a strong promoter, an adapted termination signal, and a suitable method for stable DNA transfer into the cells. For H. pluvialis no strong promoters have been isolated yet. A further problem known from transformation of other green algae is that foreign genes that are introduced (e.g., into Chlamydomonas reinhardtii) exhibit low transcript levels despite being fused to strong endogenous promoters (24). It has been suggested that such failures of exogenous gene expression might be due to mismatches in codon usage patterns (34), epigenic silencing by DNA methylation or changes in chromatin domain structure (1, 15), and/or rapid mRNA degradation (7). These hurdles might be overcome by selecting a modified endogenous gene from the organism to be transformed (19, 29, 31).
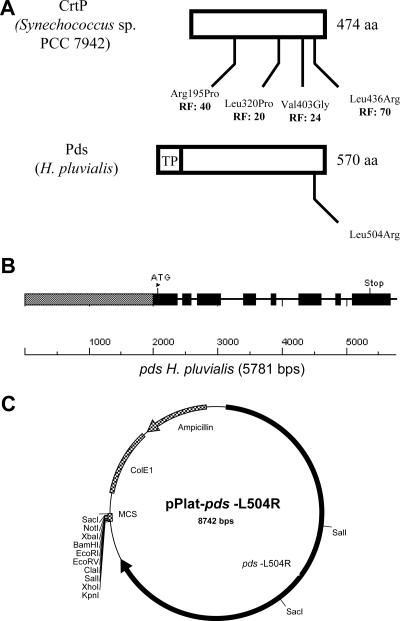
Sequence information for genes that could be adapted as dominant selective markers after appropriate genetic modification is not available for H. pluvialis. The limited sequence information that is available includes information for most of the carotenoid biosynthesis genes (12, 17, 21, 23, 37, 39, 40). One of these genes, the phytoene desaturase (Pds) gene, is the target for the bleaching herbicide norflurazon (5, 8, 25, 33). For the phytoene desaturase (CrtP) from Synechococcus sp. strain PCC 7942 it is known that four known amino acid changes lead to norflurazon resistance, as shown in Fig. 1A. Since H. pluvialis is very sensitive to this herbicide and there are high levels of sequence similarity between the Synechococcus sp. and H. pluvialis proteins, changing one of the amino acids in Fig. 1A should be a promising strategy for engineering norflurazon resistance to the Haematococcus Pds and use of the modified pds gene as a dominant selectable marker and a reporter gene for transformation of H. pluvialis.
FIG. 1.
(A) Comparison of the CrtP phytoene desaturase from Synechococcus sp. strain PCC 7942, including the four known amino acid changes and their resistance factors (RF) for the bleaching herbicide norflurazon, with the Pds from H. pluvialis. The ChloroP program identified a chloroplast transit peptide (TP) at the N terminus. The modified codon at position 504 of Pds from H. pluvialis, leading to an amino acid change from leucine to arginine, corresponds to codon 436 in CrtP from Synechococcus sp. (B) Intron and exon structure of the pds gene. The promoter sequence is indicated by a gray box, the exon sequences are indicated by black boxes, and the intron sequences are indicated by lines. (C) Map of H. pluvialis transformation vector pPlat-pds-L504R with restriction sites indicated. The engineered pds gene (pds-L504R) is inserted into the NaeI site of the vector pBluescript SK(−). The construct also contains the ampicillin resistance gene, the E. coli origin of replication (ColE1), and a multiple cloning site (MCS). The sequence of this vector has been deposited in the GenBank database under accession number DQ404589.
In this paper we describe isolation of the gene coding for the carotenoid biosynthesis enzyme Pds from H. pluvialis. After mutation of the pds gene, resulting in a norflurazon-resistant phytoene desaturase, this gene was used for successful nuclear transformation and genetic engineering of the carotenoid biosynthesis pathway for accelerated astaxanthin biosynthesis.
MATERIALS AND METHODS
H. pluvialis strain, growth conditions, and media.
H. pluvialis Flotow NIES-144 was obtained from the National Institute for Environmental Studies, Tsukuba, Japan, and was grown in basal medium containing acetate (18). Cultures were incubated at 22°C in Erlenmeyer flasks without aeration using a dark-light cycle consisting of 12 h of low light (20 μmol photons m−2 s−1, provided by Osram L65W/25S universal white lamps) and 12 h of darkness for 3 days (final cell density, approximately 3.5 × 105 cells/ml). The cultures were shaken manually once a day. For high-light treatment, H. pluvialis was grown with 175 μmol photons m−2 s−1 continuously as indicated below. For strong-light treatment, H. pluvialis cells were grown with 700 μmol photons m−2 s−1. For transformation protocols, optimal H. pluvialis medium (11) supplemented with 2.42 g/liter Tris-acetate (pH 7.2) (OHA) was used. After transformation, cells were plated on solid OHA containing 5 μM norflurazon.
Construction of genomic DNA library, screening, and DNA sequencing.
For genomic DNA isolation, H. pluvialis cells were collected by centrifugation after growth for 3 days. The cells were frozen and subsequently converted to a powder under liquid nitrogen using a mortar and pestle. Genomic DNA was isolated by using a Plant DNA isolation kit (Roche) according to the manufacturer's instructions. A genomic DNA library was constructed using a SuperCos I vector kit (Stratagene) according to the manufacturer's instructions. Screening of the genomic DNA library for isolation of the pds gene was carried out by colony hybridization using a specific pds cDNA probe according to the instructions for the DIG nonradioactive nucleic acid labeling and detection system (Roche). Probe labeling and hybridization were carried out as described previously (39). Positive clones were purified, and cosmid DNA was isolated using a High Pure plasmid isolation kit (Roche) and analyzed by Southern blotting. After restriction of DNA by enzyme digestion, a 5.78-kb XbaI/XhoI fragment harboring a pds gene fragment was isolated and cloned in the pBluescript SK(−) vector (Stratagene). The resulting plasmid was designated pBluescript-pds. The nucleotide sequence of the H. pluvialis pds gene was determined for both strands using an ABI Prism dye terminator cycle sequencing Ready Reaction kit (Perkin-Elmer). Nucleotide and derived amino acid sequences were analyzed using the Lasergene program (DNASTAR, Inc.). The intron-exon organization of the pds gene is shown in Fig. 1B.
Construction of the transformation vector pPlat-pds-L504R.
The mutated form of the pds gene from H. pluvialis (with codon position 504 changed from a leucine codon to an arginine codon) was constructed by site-directed mutagenesis using the vector pBluescript-pds and primers 5′-CC AAG CAG AAG TAC CGC GCC TCC ATG GAG GG-3′ and 5′-CCC TCC ATG GAG GCG CGG TAC TTC TGC TTG G-3′ (the codons used for the change from leucine to arginine are underlined). For mutagenesis a Quickchange mutagenesis kit (Stratagene) was used. The plasmid with the modified pds gene was designated pBluescript-pds-L504R. To verify the nucleotide change, the H. pluvialis pds gene was partially sequenced using an ABI Prism dye terminator cycle sequencing Ready Reaction kit (Perkin-Elmer). To maintain the multiple cloning site for the final transformation vector, the mutated pds gene was amplified by PCR using primers 5′-CGC GAT ATC GGT GAG GGG TTC AAG TGC C-3′ and 5′-CCG ATA TCG TTT GAA TTT TGG CTT GTT TGC-3′. In this step, the terminal XhoI and XbaI restriction sites of the pds gene were eliminated, and two EcoRV sites were introduced into the PCR product. By restriction digestion with EcoRV the PCR product was cloned into the NaeI site of a pBluescript SK(−) vector. The resulting transformation vector was designated pPlat-pds-L504R (Fig. 1C).
cDNA synthesis.
To recover the mutation in the cDNA pool of the wild-type (WT) pds and mutated pds (Pds-L504R) transcripts, total RNA was isolated and cDNA synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. For specific PCR amplification of pds cDNAs, primers 5′-ACT CTA GAA ATG CAG ACA ACA ATG CGT GGC-3′ and 5′-CCC GGA TTC AAT AGT ATA CAC CAC AAG C-3′ were used. The resulting PCR products were sequenced to verify the nucleotide change at codon position 504 in the cDNA pool of the H. pluvialis pds transcripts.
Transformation and growth of Escherichia coli.
E. coli strain BL21(DE3) (Novagene) was transformed with individual plasmids for expression of Pds from H. puvialis in its wild-type form (WT Pds) or mutated form (Pds-L504R). For production of phytoene as a substrate in the enzyme reaction, E. coli JM101 cells harboring plasmid pACCRT-EB were used (28). Cell were grown in LB medium supplemented with 100 μg/ml ampicillin and 50 μg/ml chloramphenicol. Isopropylthiogalactose (1 mM) was added to cultures of the enzyme-expressing strains after the optical density at 600 nm (1-cm light path) reached 0.6. Cultures were incubated at 25°C overnight. Plasmid pBlue-pdsWT, which mediated overexpression of the WT Pds of H. pluvialis, was constructed by reverse transcription-PCR of the corresponding cDNA using primers 5′-ACT CTA GAA ATG CAG ACA ACA ATG CGT GGC-3′ and 5′-CCC GGA TTC AAT AGT ATA CAC CAC AAG C-3′ and cloning in frame into the XbaI/EcoRI sites of the vector pBluescript SK(−) (Stratagene).
Pds-L504R from H. pluvialis was constructed by site-directed mutagenesis using the vector pBlue-pdsWT and primers 5′-CC AAG CAG AAG TAC CGC GCC TCC ATG GAG GG-3′ and 5′-CCC TCC ATG GAG GCG CGG TAC TTC TGC TTG G-3′ (codon 504 for the change from leucine to arginine is underlined). The resulting plasmid was designated pBlue-pds-L504R. For overexpression of WT Pds and Pds-L504R the pET system (Novagene) was used. Therefore, plasmids pBlue-pdsWT and pBlue-pds-L504R were restriction digested, and the corresponding cDNAs were cloned in frame into the SacI/XhoI sites of the expression vector pET24b. The resulting plasmids were pET24b-pdsWT and pET24b-pds-L504R.
Pds assay, including substrate and enzyme preparation.
Extraction of phytoene, preparation of the enzyme, and the Pds enzyme assay were performed as described by Breitenbach and coworkers (6). To the Pds assay mixture, norflurazon was added at concentrations ranging from 0.001 μM to 0.5 μM for the WT Pds enzyme and from 0.5 μM to 20 μM for the engineered Pds-L504R enzyme.
Transformation protocol.
Haematococcus cells were grown in basal medium for 3 days under the standard conditions (see above). Cells were concentrated by centrifugation and resuspended to a density of approximately 1 × 108 cells/ml in OHA. One milliliter of concentrated cells was used for each bombardment and plated on nylon filters on OHA plates. For transformation, the Biolistic PDS-1000/He system was used (Bio-Rad).
Fifty microliters of an M 17 tungsten particle solution (60 mg/ml in H2O) was mixed with 2 μl of a DNA solution (1 μg/μl), 50 μl of 2.5 M CaCl2, and 20 μl of 0.1 M spermidine base. The mixture was incubated for 10 min at room temperature and centrifuged for 10 s, and the pellet was resuspended in 250 μl ethanol. After additional centrifugation for 10 s the pellet was resuspended in 50 μl ethanol. Twenty microliters of DNA-coated particles was layered on a macrocarrier.
Plates were bombarded from a distance of 7.5 cm under a vacuum of 25 mm Hg using 1,350-lb/in2 rupture disks. After transformation, the cell-containing nylon disks were placed in 50 ml OHA for regeneration for 24 h using a dark-light cycle that included 12 h of low light (20 μmol photons m−2 s−1). After this the cells were concentrated by centrifugation and plated on 10 8-cm-diameter petri dishes containing selective OHA. Colonies appearing after 1 month were picked and restreaked at least three times on selective OHA.
Northern and Southern blot analysis.
Probe labeling and hybridization were carried out according to the instructions for the DIG nonradioactive nucleic acid labeling and detection system (Roche). The H. pluvialis pds (EMBL GenBank accession no. X86783) and bhy (EMBL GenBank accession no. AF162276) cDNAs were used as probes. For analysis of vector integration into the nuclear genome of H. pluvialis, an additional hybridization probe for the ampicillin resistance cassette was used.
For Northern blot analysis the H. pluvialis cells were collected by centrifugation after 3 days of growth. The cells were frozen and subsequently converted to a powder under liquid nitrogen using a mortar and pestle. RNA was then isolated using the miniprep RNA extraction procedure described previously (36). The Northern blot analysis was performed as described previously (39).
For Southern blot analysis, genomic DNA was isolated by using a Plant DNA isolation kit (Roche) according to the manufacturer's instructions. The genomic DNA (5 μg) was XbaI/XhoI digested, and the DNA fragments were separated by electrophoresis on a 0.8% agarose gel, transferred to a positively charged nylon membrane (Roche), and hybridized with the pds cDNA probe or the ampicilin resistance gene probe in the presence of 50% formamide at 42°C.
Isolation of proteins and Western blot analysis.
The polyclonal antibody against Pds was kindly provided by C. Hagen (12). For protein isolation, H. pluvialis transformants were collected by centrifugation after growth for 3 days. The cells were frozen and subsequently converted to a powder under liquid nitrogen using a mortar and pestle. The proteins were precipitated, and 75 μg of each transformant protein extract was separated on a 12% sodium dodecyl sulfate-polyacrylamide gel, electroblotted onto a polyvinylidene difluoride membrane (pore size, 0.45 μm; Amersham Biosciences Europe GmbH, Freiburg, Germany), and probed with anti-Pds antibody at a 1:5,000 dilution and a horseradish peroxidase-linked anti-rabbit immunoglobulin antibody at a 1:20,000 dilution. Signals were visualized by using the enhanced chemiluminescence Western blotting detection system and Hyperfilm enhanced chemiluminescence (Amersham Biosciences). A second, identically loaded gel was run as a control and stained with Coomassie brilliant blue R250 (not shown).
Carotenoid and chlorophyll analysis.
For carotenoid analysis in the green state, cells grown under strong light were instantly frozen in liquid nitrogen and freeze-dried. Extracts were obtained by using methanol containing 6% (wt/vol) KOH at 60°C for 20 min. The extracts were partitioned against 10% (vol/vol) diethyl ether in petrol, and the carotenoid content was determined by high-performance liquid chromatography (HPLC) on a 25-cm C18 Vydac 218TP54 column with methanol as the mobile phase at 20°C (10). Reference carotenoids were isolated from cyanobacteria (38) or pepper leaves (35). Carotenoid extraction, HPLC analysis, and quantification of astaxanthin and astaxanthin esters from H. pluvialis were carried as described previously (39). Chlorophyll was extracted in a similar way using methanol without KOH and was quantified as described by Lichtenthaler and Wellburn (20). Carotenoid and chlorophyll values are expressed below as means ± standard deviations for four independent determinations.
Chlorophyll fluorescence measurement.
Screening of transformants for changes in fluorescence parameters was performed using the Walz pulse-amplitude modulation fluorescent imaging system (model IMAG; Walz, Effeltrich, Germany). Transformants and WT cells were grown on solid OHA without norflurazon for 3 weeks. Before measurement the cells were adapted for 4 h in the dark. A saturating pulse (800 ms) of blue light (2,400 μmol photons m−2 s−1) was provided by light-emitting diode lamps for determination of the maximum fluorescence in the dark-adapted state (Fm). Actinic light (180 μmol photons m−2 s−1) was provided by blue light-emitting diode lamps. The measuring beam was switched to 2 Hz during saturating pulses and during actinic illumination. Nonphotochemical quenching (NPQ) was calculated using (Fm − Fm′)/Fm′, where Fm′ is the maximum fluorescence in the light-adapted state, and the imaging software provided.
Nucleotide sequence accession number.
The nucleotide sequence of the transformation vector pPlat-pds-L504R has been deposited in the GenBank database under accession number DQ404589.
RESULTS
Isolation of the H. pluvialis nuclear gene coding for Pds.
The norflrazon resistance encoded by the mutated pds gene was chosen as a suitable selection marker. Furthermore, transformation with this gene should modify carotenogenesis in H. pluvialis. For isolation of the pds gene, a genomic DNA library of H. pluvialis was screened using a specific hybridization probe for the pds cDNA. Eight different clones with genomic fragments between 30 and 45 kbp long harboring the putative pds gene were collected. Using XbaI/XhoI digestion, a 5.78-kb fragment was isolated and cloned in the corresponding restriction sites of the pBluescript SK(−) vector (Stratagene). The DNA fragment was sequenced, and alignment with the known pds cDNA of H. pluvialis revealed a 2-kb promoter region, seven introns, and a 423-bp 3′-untranslated region. The intron and exon organization is shown in Fig. 1B.
Pds-L504R exhibited higher resistance to the bleaching herbicide norflurazon in the in vitro assay than WT Pds exhibited.
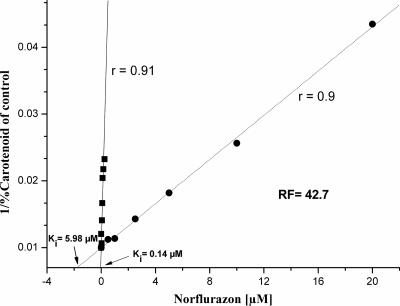
The desaturase activities of an E. coli recombinant WT Pds and Pds-L504R of H. pluvialis (with codon 504 changed from a leucine codon to an arginine codon) were measured by determining the conversion of phytoene to ζ-carotene after addition of different concentrations of norflurazon using an in vitro assay. A Dixon plot of the reciprocal of product formation versus the concentration of the inhibitor norflurazon resulted in a straight line with an r value of 90% for WT Pds and an r value of 91% for Pds-L504R (Fig. 2) (9). From the intercepts with the x axis the ki values for norflurazon as an inhibitor were calculated to be 0.14 μM and 5.98 μM for the heterologously expressed WT Pds and Pds-L504R, respectively. Compared to the wild-type enzyme the mutated form exhibited 43-fold-higher resistance.
FIG. 2.
Dixon plot of the reciprocal of in vitro phytoene-to-ζ-carotene conversion versus concentration of the inhibitor norflurazon for recombinant WT Pds (▪) and Pds-L504R (•) of H. pluvialis. The ki values for norflurazon inhibition were calculated from the intercepts with the x axis to be 0.14 μM and 5.98 μM for the heterologously expressed WT Pds and Pds-L504R, respectively. RF, resistance factor.
Analysis of Haematococcus transformants.
After transformation of H. pluvialis with the vectors pPlat-pds-L504R and pBluescript-pds by particle bombardment and a regeneration period, the cells were plated on norflurazon-containing OHA plates. After 1 month of growth, colonies appeared in samples that were transformed with the pPlat-pds-L504R vector. No growth was visible when the pBluescript-pds vector, harboring the WT pds gene, was used for transformation. It was difficult to estimate the efficiency of transformation by particle bombardment on a cell basis because many of the cells were outside the target zone. In these experiments using ∼108 cells and 1 μg of DNA, generally 100 transformants were recovered. Therefore, the transformation efficiency was estimated to be approximately 10−6 cells/μg DNA. Although the transformation rate is rather low compared to that of C. reinhardtii, transformations with similar rates were reproducible and the transformants obtained grew well under selective and nonselective conditions.
Transformants have additional copies of pds-L504R in the nuclear genome.
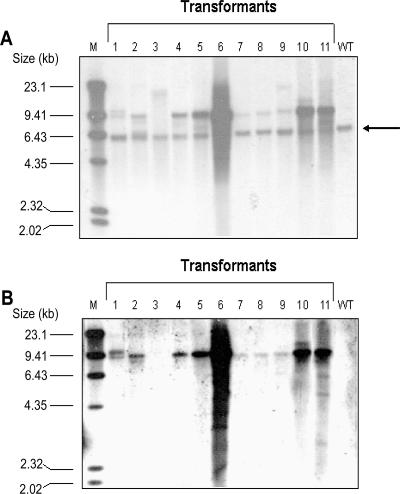
After growth for 1 month, the colonies were restreaked at least three times on norflurazon-containing plates and then incubated in liquid media. To determine if the resistance to norflurazon was due to stable nuclear integration of the engineered pds gene, Southern blot analysis of the transformants was performed with hybridization probes for the pds gene and the ampicillin resistance cassette of the transformation vector. DNA was isolated from cultures of 11 individual transformants, digested with restriction enzymes, and hybridized with a probe for the pds gene (Fig. 3A). The endogenous pds gene was detected in all transformants and the WT H. pluvialis strain; the size, 5.78 kb, corresponded to the size of the XhoI/XbaI fragment that was isolated and subcloned from the genomic DNA library of H. pluvialis.
FIG. 3.
Southern blot analysis of H. pluvialis transformants P1 to P11 and H. pluvialis WT. The DNA from transformants and WT were digested with XbaI and XhoI and electrophoresed on a 0.8% agarose gel. For detection, a pds-specific cDNA probe (A) or a specific probe for the ampicillin resistance cassette (B) was used. The arrow indicates the position of the 5.78-kb XbaI/XhoI fragment containing the endogenous pds gene. The numbers on the left indicate the sizes of the labeled λ HindIII fragments used as markers (lane M).
All transformants tested had additional copies of the engineered pds-L504R gene that were larger than the endogenous WT pds gene. To verify integration of the transgenes into the nuclear genome of H. pluvialis, additional Southern blot analysis using a hybridization probe for the ampicillin resistance cassette of the transformation vector was performed (Fig. 3B). The data support the results shown in Fig. 3A, where the pds cDNA was used as a hybridization probe. Almost all transformants (10 of the 11 transformants tested) yielded one or more bands that were visible evidence for transgene integration into the genome. No ampicillin resistance cassette band was detected in transformant P3 and WT H. pluvialis, although transformant P3 was about 20-fold more resistant to norflurazon than the WT in an assay with different norflurazon concentrations on OHA plates (data not shown). Transformant P3 produced the additional bands between 9.41 and 23.1 kb in the Southern blot using the pds cDNA hybridization probe (Fig. 3A). This finding indicates that there was a loss of the ampicillin resistance cassette and perhaps parts of the transformation vector during the integration of the transgene into the nuclear genome of H. pluvialis.
The transformation stability was analyzed using plasmid pBluescript-pds-L504R harboring the engineered pds-L504R gene in the multiple cloning site. After growth for 20 weeks (approximately 80 generations; growth rate, 0.58 day−1) under nonselective conditions, 70% of the transformants analyzed retained norflurazon resistance and exhibited stable transgene integration in the genome (data not shown).
Transformants show increased pds expression levels compared to the WT.
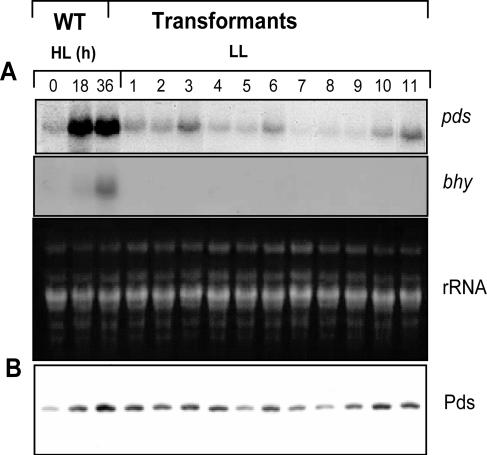
To monitor whether the additional copies of the transgene in the genomes of the 11 transformants analyzed changed the level of expression of the pds mRNA, a Northern blot analysis was carried out. For further comparison of the pds expression levels in the different transformants and the WT, total RNA was extracted from WT cells before and after 18 h and 36 h of high-light treatment. Recently, it was demonstrated that the expression of genes encoding enzymes involved in carotenoid biosynthesis is up-regulated in response to increased light intensities (37, 39). Exposure of low-light-grown WT cultures to higher light intensities resulted in accumulation of pds transcripts (Fig. 4A). The pds transcript levels in the transformants grown under low-light conditions were analyzed (Fig. 4A). Seven of 11 transformants exhibited unchanged basal expression of the pds gene under the conditions tested. Transformants P3, P6, P10, and P11 contained increased levels of pds transcripts compared to the WT (Fig. 4A). To obtain further information about whether the up-regulation of the pds transcripts was due to stress effects or to additional copies of the pds gene in the nuclear genome, the expression of the bhy gene was determined under the same conditions (Fig. 4B). In previous experiments, it was shown that bhy gene expression was detected only under stress conditions (37, 39). In all transformants, expression of the bhy gene was below the level of detection. However, light induction of the bhy gene was observed in the WT cells included as a stress control. Since the transcript level does not necessarily match the protein level, Western blot analyses using Pds antibodies were performed. The results revealed that the amount of Pds correlated with the level of its transcripts.
FIG. 4.
(A) Transcript analysis of the phytoene desaturase gene (pds) and carotenoid hydroxylase gene (bhy) in wild-type H. pluvialis cells after 0, 18, and 36 h of high-light (HL) treatment and in transformants P1 to P11 under low-light (LL) conditions. For comparison, total RNA (rRNA) was stained with ethidium bromide. (B) Western analysis of phytoene desaturase protein (Pds) levels.
Transformants have a mixture of WT pds and pds-L504R transcripts.
To obtain further information about the expression of transgenic pds-L504R, as well as the functionality of the promoter and terminator region of the 5.7-kb pds gene, cDNA synthesis and subsequent sequencing of the pds transcript pool were performed. The cDNAs of wild-type H. pluvialis and of transformants P3 and P11 were investigated by sequencing (data not shown). No additional mutations were found in the pds cDNA except for a mixture of CTG and CGC at codon position 504 for transformants P3 and P11. The WT pds cDNA obtained was identical to that present in the EMBL GenBank. These findings indicate that the 2-kb promoter region of the engineered 5.7-kb pds gene fragment isolated from an H. pluvialis genomic DNA library is sufficient to drive transgene expression.
Carotenoids, chlorophylls, and nonphotochemical quenching of selected transformants.
Analysis of norflurazon-resistant Synechococcus sp. mutants revealed that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis in cyanobacteria (8). Furthermore, previous results indicated that heterologous expression of the phytoene desaturase gene (crtI) from Erwinia uredovora in tobacco changed the composition of leaf carotenoids (27). To obtain more information about these processes in H. pluvialis, transformants were grown under low light intensities or for 1 h in the presence of strong light, and carotenoid concentrations and compositions in green cells were determined (Table 1). Transformants P3 and P11 both had a higher carotenoid content than the WT. Additionally, the chlorophyll concentration in transformant P3 was lower, resulting in a carotenoid/chlorophyll ratio which was significantly higher than that in the WT. Subsequent HPLC analysis of the carotenoid compositions of transformants P3 and P11 and the WT illuminated for 1 h with strong light revealed that for both transformants there were significant increases in the lutein, zeaxanthin, and β-carotene levels.
TABLE 1.
Pigment contents of green H. pluvialis cells treated for 1 h with strong light
| Cells | Total carotenoid concn (mg/g [dry wt])a | Chlorophyll concn (mg/g [dry wt])a | Carotenoid concn/ chlorophyll concn | Carotenoid composition (μg/g [dry wt])
|
Zeaxanthin/ (violaxanthin + antheraxanthin + zeaxanthin) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neoxanthin | Violaxanthin | Antheraxanthin | Lutein | Zeaxanthin | ß-Carotene | Violaxanthin + antheraxanthin + zeaxanthin | |||||
| WT | 2.35 ± 0.24 | 31.72 ± 2.41 | 7.4 | 37 ± 6 | 79 ± 1 | 91 ± 4 | 1,464 ± 97 | 142 ± 18 | 532 ± 46 | 312 | 0.46 |
| P3 | 2.65 ± 0.30 | 26.28 ± 3.47 | 10 | 36 ± 5 | 76 ± 2 | 90 ± 6 | 1,596 ± 36 | 202 ± 20 | 648 ± 30 | 368 | 0.55 |
| P11 | 3.00 ± 0.28 | 31.32 ± 2.40 | 9.6 | 18 ± 4 | 65 ± 7 | 55 ± 3 | 1,920 ± 99 | 214 ± 11 | 728 ± 84 | 334 | 0.64 |
The total carotenoid and chlorophyll concentrations were similar prior to high-light illumination.
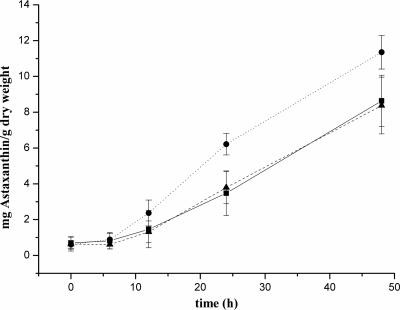
In addition, we investigated whether the phytoene desaturation step is a rate-limiting step for astaxanthin synthesis. Therefore, the transformants were grown for 3 days under low-light conditions and then exposed to continuous high-light illumination, and astaxanthin accumulation was determined. During the time course, transformant P3 had a visible red phenotype after 8 h of exposure to high light, whereas the other transformant and the WT remained green (Fig. 5). Quantification of astaxanthin in transformants P3 and P11 and the WT cells after 48 h of high-light illumination confirmed that the high-light treatment led to enhanced astaxanthin levels in transformant P3 (11.4 ± 0.9 mg/g [dry weight]), whereas the levels in the WT (8.6 ± 1.4 mg/g [dry weight]) and in transformant P11 (8.4 ± 1.6 mg/g dry weight) were 26% lower.
FIG. 5.
Astaxanthin formation after exposure of H. pluvialis WT (▪) and transformants P3 (•) and P11 (▴) to continuous high light.
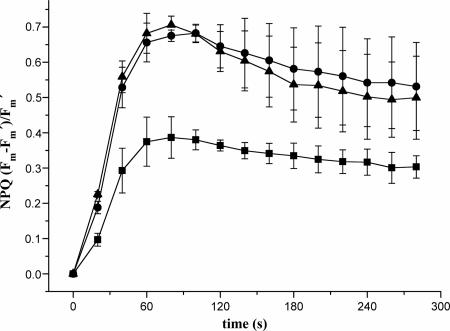
Chlorophyll fluorescence was determined with dark-adapted H. pluvialis WT and transformants. The time course of NPQ is shown in Fig. 6. For WT cells the maximum value (0.38) was reached after 70 s. For transformants P3 and P11 the maxima were reached after the same time; however, the NPQ value was almost twofold higher for both of these transformants.
FIG. 6.
Kinetics of NPQ of dark-adapted H. pluvialis WT (▪) and transformants P3 (•) and P11 (▴).
DISCUSSION
The development of a transformation system depends on the availability of suitable strong promoters, termination signals, and promising reporter or resistance genes. At present, no appropriate genetic tools are available for H. pluvialis. The pds/crtP gene from plants, algae, and cyanobacteria is susceptible to the herbicide norflurazon, and four mutations in the enzyme from the cyanobacterium Synechoccocus sp. strain PCC 7942 are known to result in resistance (8). Since alignment of the amino acid sequences of CrtP/Pds from Synechococcus sp. and H. pluvialis revealed a high level of sequence similarity in the areas where the mutations were found, we used a direct genetic approach, changing Leu504 to Arg in the phytoene desaturase of H. pluvialis (Fig. 1A). By in vitro enzyme analysis, it was shown that the engineered Pds-L504R was active and exhibited 43-fold-higher resistance to norflurazon than the WT enzyme exhibited (Fig. 2). This decrease in norflurazone susceptibility was sufficient for selection of H. pluvialis transformed with the mutated pds gene.
After biolistic transformation of H. pluvialis and following selection on norflurazon-containing medium, the integration of the transformation vector used, pPlat-pds-L504R, was examined by Southern blot analysis (Fig. 3). Using a pds cDNA probe revealed that all transformants except the WT had additional copies of the pds gene in the nuclear genome. For the 11 transformants that were analyzed we do not fully understand whether the transforming DNA was linked to the pds locus by homologous recombination or the integration was due to heterologous recombination events in the genome. It was estimated for C. reinhardtii that homologous recombination events take place at a ratio of 1:100 for a mutated protophorphyrinogen oxidase gene (ppx1) when the biolistic method is used (31). A homologous recombination event due to a single crossover should have resulted in a 6.4-kb band and an 8.11-kb band in the Southern blot using the pds cDNA probe. A homologous recombination event due to a double crossover should have resulted in a loss of the vector backbone and therefore the ampicillin resistance cassette. It was difficult to estimate the correct sizes of the different bands that hybridized with the pds cDNA and the ampicillin gene probe. Transformant P2 produced signals that could match this assumption, but in general strong signals were found at a vector size of 8.7 kb. It is more likely that the transforming DNA underwent complex rearrangement and concatemer formation before integration, resulting in a complex tandem array of pPlat-pds-L504R DNA that was stable after integration into the genome. This has previously been reported for the integration of transgenes into the genome of the green alga Volvox carteri that takes place as tandem repeats due to homologous recombination of the plasmids forming circular concatemeric structures prior integration into the genome (1, 14). Interestingly, transformants P5, P6, P10, and P11 showed very strong signals at 8.7 kb, matching the vector size, 8.74 kb. To exclude the possibility that the intact plasmid was maintained, the genomic DNA of these four transformants and the WT was restriction digested with BstBI. This restriction enzyme did not cut the vector sequence. Using the pds hybridization probe, the endogenous pds gene was detected in all transformants and the WT H. pluvialis strain at 9.4 kb (data not shown). The four transformants had additional copies of the engineered pds-L504R gene whose sizes were about 23 kb and larger (data not shown).
The changed expression level of the pds gene under standard growth conditions was not linked to stress induction, as shown by the bhy gene expression (Fig. 4). It is more likely that the altered expression of the pds gene was due to multiple copies of the gene in the genomes of the transformants. Interestingly, the strength of expression was somehow linked to the copy number of the transgene, but there seemed to be additional positional effects. The integration locus of the introduced DNA in the genome seemed to be an important factor in determining the transgenic pds gene expression. Western blot analysis revealed that the pds gene transcript and protein level matched the degree of expression. This result indicated that under standard growth conditions there was no posttranscriptional regulation of the pds gene expression.
Furthermore, analysis of total pds gene transcripts was performed for the WT strain and transformants P3 and P11. The two transformants had a mixture of WT and transgene transcripts, indicating that the promoter was sufficient to drive transgenic pds-L504R gene expression and that the endogenous pds gene was intact.
The transformants were analyzed not only to determine transcription and protein expression but also to determine possible effects of increased phytoene desaturase levels on carotenoid metabolism. Carotenoid analysis of transformants P3 and P11 showed that the total carotenoid concentration was higher in P11 on a dry weight basis, whereas the value was lower for P3 (Table 1). Since the chlorophyll content was also lower, a pleiotropic effect on the chloroplast can be assumed. Related to the chlorophyll content, the total carotenoid content was also increased compared with the wild-type level. Both transformants synthesized higher levels of zeaxanthin and lutein than the WT synthesized (Table 1). This is in accordance with the findings that heterologous expression of a bacterial phytoene desaturase in tobacco changed the composition of leaf carotenoids (27) and changed the carotenoid formation in tomato toward β-carotene at the expense of lycopene (32). Under light stress conditions, transformant P3 showed enhanced accumulation of astaxanthin, whose level after 48 h of exposure to high light was 26% higher than the levels in the WT and transformant P11 (Fig. 5). All our results indicate that the phytoene-desaturating step is rate limiting for carotenoid synthesis in the chloroplast of H. pluvialis under high-light conditions. The degree of nonphotochemical quenching is related to the zeaxanthin content and the amount of zeaxanthin relative to the amount of all xanthophyll cycle carotenoids calculated by using the zeaxanthin/(zeaxanthin + antheraxanthin + violaxanthin) ratio (2). These zeaxanthin-related parameters were both increased in transformants P3 and P11 (Table 1). In accordance with the higher zeaxanthin concentration, NPQ values were almost twofold higher for the transformants (Fig. 6). Consequently, NPQ measurement may be used as a convenient screening procedure for transformants obtained with the pPlat-pds-L504R vector. Video imaging of chlorophyll fluorescence has been shown to be successful for detecting Chlamydomonas xanthophyll cycle mutants (30). The results reported here clearly demonstrate that the mutated pds gene can be used as a dominant selective marker for stable nuclear transformation of H. pluvialis. Thus, our pPlat-pds-L504R transformation vector (Fig. 1C) resembles a basic tool for genetic modification of this alga. In addition, we describe for the first time specific genetic engineering of the carotenoid biosynthesis pathway leading to increased carotenoid biosynthesis and accelerated accumulation of astaxanthin in H. pluvialis. The work described here resembles a proof of concept for genetic engineering of the carotenoid biosynthetic pathway in H. pluvialis toward increasing astaxanthin production.
Acknowledgments
This work was possible because of generous support from I. Adamska, University of Konstanz, Germany, and the Peter and Traudl Engelhorn Stiftung.
We are grateful to R. Miller-Sulger for excellent technical assistance and to H. Linden for assistance with preparation of the manuscript. We thank C. Hagen, University of Jena, Germany, for providing the Pds antibody and P. Kroth, University of Konstanz, Germany, for use of the Biolistic PDS-1000/He system.
Footnotes
Published ahead of print on 29 September 2006.
REFERENCES
- 1.Babinger, P., I. Kobl, W. Mages, and R. Schmitt. 2001. A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res. 29:1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baroli, I., and K. K. Niyogi. 2000. Molecular genetics of xanthophyll-dependent photoprotection in green algae and plants. Phil. Trans. R. Soc. Lond. B 355:1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boussiba, S. 2000. Carotenogenesis in the green alga Haematococcus pluvialis: cellular physiology and stress response. Physiol. Plant. 108:111-117. [Google Scholar]
- 4.Boussiba, S., L. Fan, and A. Vonshak. 1992. Enhancement and determination of astaxanthin accumulation in green alga Haematococcus pluvialis. Methods Enzymol. 213:386-391. [Google Scholar]
- 5.Böger, P., and G. Sandmann. 1990. Modern herbicides affecting typical plant processes, p. 174-216. In W. S. Bowers, W. Ebing, D. Martin, and R. Wegler (ed.), Chemistry of plant protection, vol. 6. Springer Publ., Berlin, Germany. [Google Scholar]
- 6.Breitenbach, J., C. F. Zhu, and G. Sandmann. 2001. Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. J. Agric. Food Chem. 49:5270-5272. [DOI] [PubMed] [Google Scholar]
- 7.Cerutti, H., A. M. Johnson, N. W. Gillham, and J. E. Boynton. 1997. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics 145:97-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamovitz, D., G. Sandmann, and J. Hirschberg. 1993. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 268:17348-17353. [PubMed] [Google Scholar]
- 9.Dixon, M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55:170-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eppler, K. S., L. C. Sander, R. G. Ziegler, S. A. Wise, and N. E. Craft. 1992. Evaluation of reversed-phase liquid-chromatographic columns for recovery and selectivity of selected carotenoids. J. Chromatogr. 20:89-101. [DOI] [PubMed] [Google Scholar]
- 11.Fábregas, J., A. Dominguez, M. Regueiro, A. Maseda, and A. Otero. 2000. Optimization of culture medium for the continuous cultivation of the microalga Haematococcus pluvialis. Appl. Microbiol. Biotechnol. 53:530-535. [DOI] [PubMed] [Google Scholar]
- 12.Grünewald, K., M. Eckert, J. Hirschberg, and C. Hagen. 2000. Phytoene desaturase is localized exclusively in the chloroplast and up-regulated at the mRNA level during accumulation of secondary carotenoids in Haematococcus pluvialis (Volvocales, Chlorophyceae). Plant Physiol. 122:1261-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerin, M., M. E. Huntley, and M. Olaizola. 2003. Haematococcus astaxanthin: applications for human health and nutrition. Trends Biotechnol. 21:210-216. [DOI] [PubMed] [Google Scholar]
- 14.Hallmann, A., A. Rappel, and M. Sumper. 1997. Gene replacement by homologous recombination in the multicellular green alga Volvox carteri. Proc. Natl. Acad. Sci. USA 94:7469-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobiak, T., W. Mages, B. Scharf, P. Babinger, K. Stark, and R. Schmitt. 2004. The bacterial paromomycin resistance gene, aphH, as a dominant selectable marker in Volvox carteri. Protist 155:381-393. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. A., and W. A. Schroeder. 1996. Microbial carotenoids. Adv. Biochem. Eng. Biotechnol. 53:119-178. [DOI] [PubMed] [Google Scholar]
- 17.Kajiwara., S., T. Kakizono, T. Saito, K. Kondo, T. Ohtani, N. Nishio, S. Nagai, and N. Misawa. 1995. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol. Biol. 29:343-352. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., T. Kakizono, and S. Nagai. 1993. Enhanced carotenoid biosynthesis by oxidative stress in acetate-induced cyst cells of a green unicellular alga, Haematococcus pluvialis. Appl. Environ. Microbiol. 59:867-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovar, J. L., J. Zhang, R. P. Funke, and D. P. Weeks. 2002. Molecular analysis of the acetolactate synthase gene of Chlamydomonas reinhardtii and development of a genetically engineered gene as a dominant selectable marker for genetic transformation. Plant J. 29:109-117. [DOI] [PubMed] [Google Scholar]
- 20.Lichtenthaler, H. K., and A. R. Wellburn. 1983. Determination of chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1111:591-592. [Google Scholar]
- 21.Linden, H. 1999. Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim. Biophys. Acta 1446:203-212. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz, R. T., and G. R. Cysewski. 2000. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 18:160-167. [DOI] [PubMed] [Google Scholar]
- 23.Lotan, T., and J. Hirschberg. 1995. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis FEBS Lett. 364:125-128. [DOI] [PubMed] [Google Scholar]
- 24.Lumbreras, V., D. R. Stevens, and S. Purton. 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441-447. [Google Scholar]
- 25.Martinez-Ferez, I., A. Vioque, and G. Sandmann. 1994. Mutagenesis of an amino acid responsible in phytoene desaturase from Synechocystis for binding of the bleaching herbicide norflurazon. Pestic. Biochem. Physiol. 48:185-190. [Google Scholar]
- 26.Meng, C. X., C. Y. Teng, P. Jiang, S. Qin, and C. K. Tseng. 2005. Cloning and characterization of beta-carotene ketolase gene promoter in Haematococcus pluvialis. Acta Biochim. Biophys. Sin. 37:270-275. [DOI] [PubMed] [Google Scholar]
- 27.Misawa, N., K. Masamoto, T. Hori, T. Ohtani, P. Böger, and G. Sandmann. 1994. Expression of Erwinia phytoene desaturase gene not only confers multiple resistance to herbicides interfering with carotenoid biosynthesis but also alters xanthophyll metabolism in transgenic plants. Plant J. 6:481-489. [Google Scholar]
- 28.Misawa, N., Y. Satomi, K. Kondo, A. Yokoyama, S. Kajiwara, T. Saito, T. Ohtani, and W. Miki. 1995. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 177:6575-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, J. A., P. B. Savereide, and P. A. Lefebvre. 1994. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14:4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niyogi, K. K., O. Bjorkman, and A. R. Grossman. 1997. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randolph-Anderson, B. L., R. Sato, A. M. Johnson, E. H. Harris, C. R. Hauser, K. Oeda, F. Ishige, S. Nishio, N. W. Gillham, and J. E. Boynton. 1998. Isolation and characterization of a mutant protoporphyrinogen oxidase gene from Chlamydomonas reinhardtii conferring resistance to porphyric herbicides. Plant Mol. Biol. 38:839-859. [DOI] [PubMed] [Google Scholar]
- 32.Römer, S., P. D. Fraser, J. W. Kiano, C. A. Shipton, N. Misawa, W. Schuch, and P. M. Bramley. 2000. Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18:666-669. [DOI] [PubMed] [Google Scholar]
- 33.Sandmann, G., and P. Böger. 1989. Inhibition of carotenoid biosynthesis by herbicides, p. 25-44. In P. Böger and G. Sandmann (ed.), Target sites of herbicide action. CRC Press, Boca Raton, FL.
- 34.Schiedlmeier, B., R. Schmitt, W. Muller, M. M. Kirk, H. Gruber, W. Mages, and D. L. Kirk. 1994. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 91:5080-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simkin, A. J., J. Breitenbach, M. Kuntz, and G. Sandmann. 2000. In vitro and in situ inhibition of carotenoid biosynthesis in Capsicum annuum by bleaching herbicides. J. Agric. Food Chem. 48:4676-4680. [DOI] [PubMed] [Google Scholar]
- 36.Sokolowsky, V., R. Kaldenhoff, M. Ricci, and V. E. A. Russo. 1990. Fast and reliable mini-prep RNA extraction from Neurospora crassa. Fungal Genet. Newslett. 37:41-43. [Google Scholar]
- 37.Steinbrenner, J., and H. Linden. 2001. Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol. 125:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiger, S., L. Schaefer, and G. Sandmann. 1999. High-light upregulation of carotenoids and their antioxidative properties in the cyanobacterium Synechocystis PCC 6803. J. Photochem. Photobiol. B 521:14-18. [Google Scholar]
- 39.Steinbrenner, J., and H. Linden. 2003. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol. Biol. 52:343-356. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Z., F. X. Cunningham, and E. Gantt. 1998. Differential expression of two isopentenyl pyrophosphate isomerases and enhanced carotenoid accumulation in a unicellular chlorophyte. Proc. Natl. Acad. Sci. USA 95:11482-11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng, C., S. Qin, J. Liu, D. Yu, C. Liang, and C. Tseng. 2002. Transient expression of lacZ in bombarded unicellular green alga Haematococcus pluvialis. J. Appl. Phycol. 14:497-500. [Google Scholar]