Abstract
Although many secondary metabolites exhibiting important pharmaceutical and agrochemical activities have been isolated from myxobacteria, most of these microorganisms remain difficult to handle genetically. To utilize their metabolic potential, heterologous expression methodologies are currently being developed. Here, the Red/ET recombination technology was used to perform all required gene cluster engineering steps in Escherichia coli prior to the transfer into the chromosome of the heterologous host. We describe the integration of the complete 57-kbp myxothiazol biosynthetic gene cluster reconstituted from two cosmids from a cosmid library of the myxobacterium Stigmatella aurantiaca DW4-3/1 into the chromosome of the thus far best-characterized myxobacterium, Myxococcus xanthus, in one step. The successful integration and expression of the myxothiazol biosynthetic genes in M. xanthus results in the production of myxothiazol in yields comparable to the natural producer strain.
Myxobacteria belong to the δ-group of the proteobacteria and represent a rich source of secondary metabolites with important applications in the pharmaceutical and agrochemical industries (11). These microorganisms and their ability to synthesize natural products have been intensively investigated in recent years (3, 32). To date, approximately 100 different natural products have been characterized, and novel secondary metabolites continue to be discovered (19, 29, 31, 34). Epothilones, soraphen, myxochromid, chivosazol, tubulysin, and disorazol are some examples of myxobacterial natural products exhibiting antibacterial, antifungal, or cytotoxic activities (5, 9, 16, 17, 33, 46). The biosynthetic genes responsible for the formation of a number of myxobacterial secondary metabolites have been cloned, and the molecular mechanisms of the underlying biosyntheses are currently under study (3). Some myxobacterial strains are able to produce the compound myxothiazol, which is active against numerous fungi and some gram-positive bacteria (10). This natural product acts on the cytochrome bc1 complex and is widely in use as an effective inhibitor of the respiratory chain (2, 10, 40). Myxothiazol A (Fig. 1d) was isolated from different strains of the genera Angiococcus, Stigmatella, and Myxococcus and rarely from the genera Cystobacter and Corallococcus. In addition to myxothiazol A, a similar compound—myxothiazol Z—was found in a few strains of M. fulvus (38; K. Gerth, unpublished data). Due to the biotechnological importance and the highly unusual biosynthesis of myxothiazol, the biochemical principles underlying the formation of the compound are subject to intensive investigations (28, 36, 43). Myxothiazols contain the β-methoxyacrylate pharmacophore and differ in their terminal functional groups bound to the C1 atom (Fig. 1d). Remarkable properties of the myxothiazol molecule include a bis-thiazole moiety, an unusual starter unit isovaleryl-coenzyme A (CoA) derived from either leucine or hydroxymethylglutaryl-CoA (22, 23), and a terminal amide structure (in myxothiazol A).
FIG. 1.
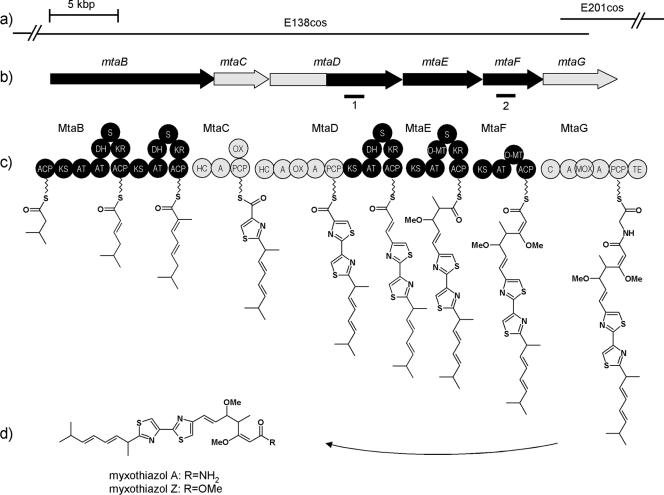
Gene arrangement and domain organization of the mta gene cluster from S. aurantiaca DW4/3-1 and a hypothetical biosynthesis of myxothiazol. (a) The cosmids used for the gene cluster reconstitution (E201cos and E138cos) are shown as lines. (b) Genes involved in biosynthesis are shown as arrows. Black shading indicates regions encoding PKS modules; gray shading indicates regions encoding NRPS modules. Bars 1 and 2 show repeats in the nucleotide sequence. (c) MtaB-MtaG are corresponding proteins with the domain organization. ACP, acylcarrier protein domain; KS, β-ketoacyl-ACP synthase; AT, acyltransferase; KR, β-ketoacyl-ACP reductase; DH, dehydratase; O-MT, methyltransferase; S, spacer region; TE, thioesterase; HC, heterocyclization; C, condensation; PCP, peptidyl carrier protein; A, adenylation; Ox, oxidation; MOX, monooxygenase. (d) Structure of myxothiazol.
Although sequence information is increasing rapidly, genetic manipulation systems required for the analysis of each natural product producing myxobacterium are poorly established. The genes involved in the formation of myxothiazol have been identified, and the nucleotide sequence of the whole biosynthetic gene cluster has been determined from Stigmatella aurantiaca DW4/3-1 (36). It has been shown that the biosynthesis of myxothiazol is catalyzed by a combined megasynthetase consisting of polyketide synthases (PKS) and nonribosomal peptide synthetases (NRPS), encoded by genes mtaB to mtaG (Fig. 1b). While mtaB, mtaE, and mtaF are PKS encoding genes, mtaC and mtaG encode NRPS, and the gene mtaD encodes a hybrid protein containing both PKS and NRPS modules. The coding region responsible for the biosynthesis of myxothiazol A spans about 60 kbp on the S. aurantiaca chromosome (36). The biosynthesis occurs in a multistep process (Fig. 1c) that includes the condensation of activated isovalerate, malonate, methylmalonate, and cysteine units (22, 41) In addition, the biosynthetic activity of PKS and NRPS is dependent on the 4′-phosphopantetheinyl (P-pant) transferase MtaA, which is also required for the formation of other secondary metabolites in S. aurantiaca DW4/3-1 (8, 35).
The role of the biosynthetic genes involved in myxothiazol formation was investigated by construction of various mutants of the producer strain S. aurantiaca DW4-3/1 (43). Although extensive efforts were applied to establish a markerless mutagenesis system for this strain enabling the construction of chromosomal point mutations or in frame deletions/insertions, almost all mutants with alterations in mtaB, mtaE, mtaF, and mtaH did not produce altered compounds with expected structures (43). This finding may be explained by the fact that this time-consuming mutagenesis system only allowed the construction of a very limited number of mutants. However, a recent report stresses that numerous combinations of splice sites have to be made in order to find active ones (7).
One of the strategies for the engineering of natural product biosynthesis, especially if the manipulation on the chromosome in the producer strain is difficult or impossible, is the heterologous expression of the corresponding gene cluster. Such studies, in combination with biochemical and genetic analyses of biosynthetic gene clusters from myxobacteria, will make the manipulation of the biosyntheses more feasible, as has already been shown for some other bacterial genera, e.g., the well-investigated erythromycin gene cluster, as well as the pikromycin or aureothin biosynthetic genes (1, 14, 30). Different strategies have been used for heterologous expression (for a review, see reference 44), which in most cases aimed at the definition of conditions to improve the yield of the natural product. Myxobacterial gene clusters responsible for the synthesis of epothilone, soraphen, and myxochromid have been successfully expressed in heterologous hosts with different impact on the product yields compared to the natural producer (18, 39, 45, 49).
In the present study we used a combination of Red/ET recombineering and classical cloning procedures for the reconstitution (“stitching”) of the whole myxothiazol biosynthetic gene cluster from two cosmids. Further modifications such as promoter exchange and conjugation cassette integration, were done by Red/ET recombination as well (see Fig. 1 to 3). The genetically manipulated gene cluster was then introduced into the chromosome of the related myxobacterial strain M. xanthus DZF1 to achieve production levels similar to those of the natural producer.
FIG. 3.
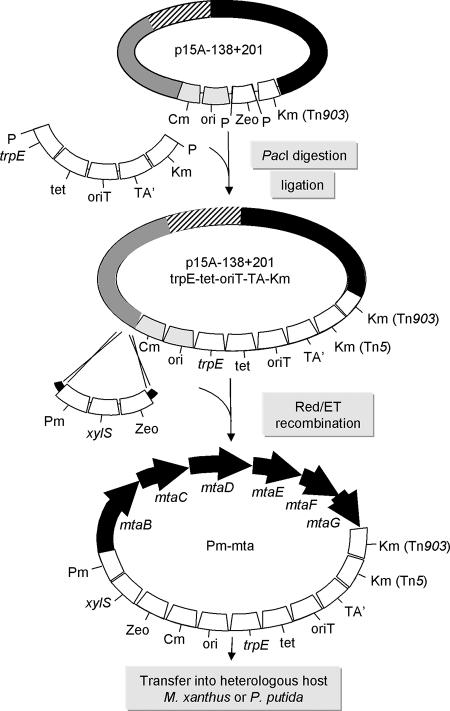
Modification of the construct containing the whole gene cluster for the transfer into the heterologous host. ░⃞, Start of the biosynthetic cluster with upstream region; ▨, overlapping region; ▪, end of the cluster and downstream region; P, PacI restriction site; Cm, chloramphenicol resistance selection marker; Km, kanamycin resistance selection marker derived from Tn903 and Tn5; Zeo, zeocin resistance selection marker; tet, tetracycline resistance selection marker, ori, p15A origin of replication; oriT, origin of transfer; TA′, fragment of the TA encoding gene from M. xanthus; trpE, homologous target from P. putida; Pm, P. putida toluate catabolic pathway promoter; xylS, regulator of the promoter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strains harboring plasmids or cosmids used in the present study were grown either at 37°C or at 30°C (for Red/ET protein expression) in LB (26) medium supplemented with the required antibiotics (kanamycin [Km], 50 μg/ml; zeocin [Zeo], 25 μg/ml; chloramphenicol [Cm], 25 μg/ml). The cells of M. xanthus DZF1 (27) were grown at 30°C in CTT medium (15), which was supplemented with Km (50 μg/ml) for the myxothiazol-producing strain.
Cloning by Red/ET recombination.
The Red/ET recombination technique was described previously (47). In the present study this method was used for the “stitching” of the biosynthetic gene cluster onto one vector. Electrocompetent E. coli cells containing a Red/ET protein expression plasmid (ET+) and the parent molecule to be modified were electroporated with 0.3 μg of a linear DNA fragment (modification cassette), which was obtained either by PCR or by restriction. The selection of transformants was carried out depending on the selection marker located on the cassette. DNA isolated from clones that arose after electroporation was tested by restriction analysis.
PCRs were performed with HotStarTaq polymerase (QIAGEN, Hilden, Germany) according to the manufacturer's protocol, and an optimal annealing temperature was chosen for each primer pair. The PCR product was used directly without purification for further applications.
First, the myxothiazol gene cluster was stitched onto one construct. The mta genes from cosmid E138 were cloned into the minimal linear cloning vector p15A-Cm obtained by PCR using pACYC184 as a template as described previously (48). The oligonucleotides used for subcloning the mta genes from cosmid E138 (Fig. 1 and 2b) were 5′-TCTATCCAACTGAGCTACGGGGCCAGACAGGGGGCGGTTTATCGCAGGTCTAGAGCATTAATCATATGTTACGCCCCGCCCTGCCACTC-3′ and 5′-TCCAGAAGCTGGCCTCCGTGCTGCGCCAGGAGGGCCTCGCTGCGGCCCGCACGCCACTAGTATTAATTGGCTTAAGTTAATAAGATGATCTTCTTGAG-3′.
FIG. 2.
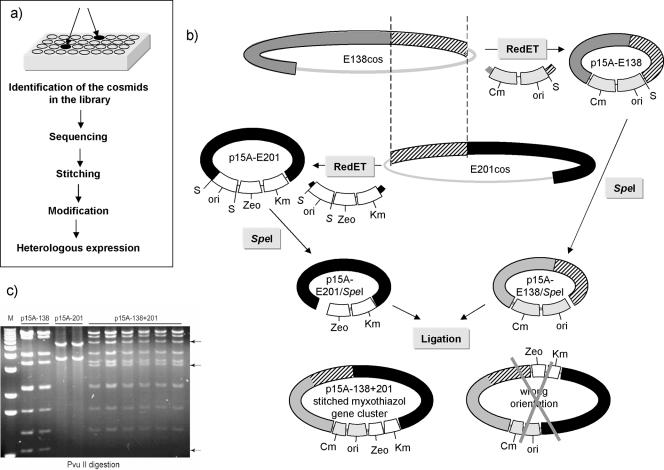
(a) General workflow for the expression of the complete biosynthetic gene cluster in a heterologous host. (b) Stitching of the myxothiazol biosynthetic gene cluster. Cm, chloramphenicol resistance selection marker; Km, kanamycin resistance selection marker derived from Tn903; Zeo, zeocin resistance selection marker; ori, p15A origin of replication; ░⃞, start of the biosynthetic cluster with upstream region; ▨, overlapping region; ▪, end of the cluster and downstream region; S, SpeI restriction site. (c) Analysis of the correct stitched construct by digestion with PvuII.
The mta genes from cosmid E201 were subcloned into p15A-Zeo-Km minimal vector, which was obtained after the cloning of the zeocin resistance gene flanked with PacI restriction sites in pACYC177 and using this construct as a template for PCR to generate p15A-Zeo-Km minimal linear vector as described previously (48). The oligonucleotides used for subcloning the mta genes from cosmid E201 were 5′-TCGGATGCATGAAGAAGAGGGGCGGTTGATCGCCCGTCTCTTGGATGCGCACTAGTTAATAAGATGATCTTCTTGAG-3′ and 5′-TGCGCATCTTCAATACCTACGGGCCGAGGATGCGGTTGAAGGATTAGAAAAACTCATCGAGCATC-3′.
For the integration into the chromosome of M. xanthus a fragment of the myxovirescin biosynthetic gene cluster (37) (TA′) was amplified by PCR using the primers OP19 (CTATCTGCTCAAGCTTCACG) and OP20 (AACAGTGAGAGGTTCGGGTG). This fragment has been cloned as a 1.7-kb BamHI/HindIII fragment into a pGEM-7Zf(−) (Promega) derivative containing oriT from the RP4 mob region and the Tn5 kanamycin resistance gene (S. Beyer, unpublished data). The resulting plasmid was named pOPB18. A functional cassette containing trpE-tetR-oriT was removed from p15A-oriT-trpE-tetR (45) by BamHI digestion and inserted into the BamHI site in pOPB18. The resulting cassette, trpE-tetR-oriT-TA′-Km in pOPB18, was subcloned into the p15A-Cm minimum vector using Red/ET recombination to form p15A-Cm-trpE-tetR-oriT-TA′-Km. The oligonucleotides used for the construction of p15A-Cm-trpE-tetR-oriT-TA′-Km were 5′--GCCAAGTAATATCACGA GGCCCTTTCGTCTTCAAGAATTCGCGGCCGCTTAATTAAGAGAATTACAACTTATATCGT-3′ and 5′-CAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGACTTAATTAATTACGCCCCGCCCTGCCACTC-3′.
PacI sites were engineered into both ends of the trpE-tetR-oriT-TA′-neo cassette (indicated in boldface), and the complete fragment was next introduced into the PacI site present in the stitched myxothiazol gene cluster backbone (Fig. 3). A toluic acid-inducible Pm promoter with its regulator gene xylS839 and a zeocin selection marker gene were inserted in front of the mtaB gene by Red/ET recombination (Fig. 3). The oligonucleotides used for the insertion of the Pm-xylS-zeocin cassette were 5′-CGAGATGCACACGGACCGTGTGCTGACCAAGGACGGATGCGGCGGGTCAGGGCAT TTAAATCTGGGATC-3′ and 5′-AGTTGGCCGGTGCTCCGGCCAACTCCCCCAACACCATGTTCTGTCGACATGTTC ATGACTCCATTATTATTG-3′.
Electroporation of M. xanthus and colony PCR.
The myxothiazol gene cluster was introduced into the chromosome of the M. xanthus DZF1 by electroporation. Electrocompetent cells were prepared from 10 ml of culture after three washes with ice-cold water, and the cell pellet was resuspended in 100 μl of H2O. A total of 100 μl of the cell suspension was mixed with up to 1 μg of DNA and electroporated (GenePulser Xcell; Bio-Rad) at 25 μF, 650 V, and 400 Ω using 0.1-cm electroporation cuvettes. After electroporation, 900 μl of CTT medium was added, the cells were resuspended in 5 ml of CTT medium, and the Erlenmeyer flasks were incubated at 30°C on a rotary shaker at 160 rpm overnight. After centrifugation the cells were resuspended in 0.5 ml of CTT medium, and 100 or 400 μl was added to 4 ml of soft agar (CTT) and plated for selection on agar plates supplemented with 50 μg of kanamycin/ml. Typically, kanamycin-resistant colonies appeared after 7 days and were tested for growth in liquid medium.
Obtained clones were tested by colony PCR as follows. A single colony was resuspended in 5 μl of H2O, followed by incubation at 95°C for 10 min. Then, 2 μl of the resulting suspension was used as a PCR template using Taq polymerase (Gibco-BRL) according to the manufacturer's protocol. Myxothiazol-specific primers were designed to detect different parts of the gene cluster to verify the integration of the whole biosynthetic gene cluster into the chromosome. For amplification of a mtaB fragment, we used OP59 (5′-GAACGTGGTCGTCTCGGGAG-3′) and OP60 (5′-CGAATCACCAGCCCGGAGAC-3′); for amplification of a mtaE fragment, we used OP126 (5′-TCAAGCCGGATGAGGTCTAC-3′) and OP127 (5′-CTTGGACACGGTATCGAGGT-3′); and for amplification of a mtaG fragment, we used OP128 (5′-CTCTTCTTCATGCATCCGAC-3′) and OP129 (5′-CCGGTACATCTGAACCTGCT-3′).
Analysis of the heterologous myxothiazol production in M. xanthus.
M. xanthus containing the myxothiazol biosynthetic gene cluster was inoculated from an overnight culture (5%) and incubated in 300-ml flasks containing 50 ml of CTT medium supplemented with kanamycin (50 μg/ml) and with 2% XAD 16 adsorber resin (Rohm und Haas, Frankfurt, Germany) for 4 days at 30°C (200 rpm).
The cells and the resin were harvested by centrifugation and extracted with acetone and methanol. Solvents were removed in vacuo, and the residue was dissolved in 1 ml of methanol. An aliquot of 5 μl was analyzed by high-pressure liquid chromatography-mass spectrometry (HPLC-MS); an Agilent 1100 series solvent delivery system coupled to Bruker HCTplus ion trap mass spectrometer was used. Chromatographic separation was carried out on an RP column Nucleodur C18 (125 by 2 mm, 3-μm particle size; Macherey and Nagel) equipped with a precolumn C18 (8 × 3 mm, 5 μm). The mobile-phase gradient (solvent A [water containing 0.1% formic acid] and solvent B [acetonitrile containing 0.1% formic acid]) was linear from 5% B at 2 min to 95% B within 30 min at a flow rate of 0.4 ml/min.
Detection was carried out in positive ionization mode. Myxothiazol A was identified by comparison to the retention time and the MS2 pattern of the authentic reference standard (m/z [M+H]+ = 488). For kinetic studies, 10-ml cultures were inoculated (two replicates) and harvested after different incubation times, and extracts were prepared as described above.
For quantitative analysis, samples were separated on a gradient starting at 80% solvent B and running to 95% solvent B at 5 min, with elution of myxothiazol A occurring at 3 min. Quantitation was carried out in manual MS2 mode. Ions of m/z [M+H]+ = 488 were collected and subjected to fragmentation. The intensities of the characteristic fragment ions m/z = 456 and m/z = 439 were summed up, and peak integration was carried out utilizing the Bruker QuantAnalysis v1.6 software package. A calibration curve was established from serial dilutions of myxothiazol A down to 0.1 μg/ml. Samples under investigation were diluted as required to fit the dynamic range of the method.
RESULTS
Cloning of the myxothiazol biosynthetic gene cluster.
The genes involved in myxothiazol biosynthesis have been located on two cosmids from a gene library of S. aurantiaca DW4/3-1 (36) (E138 and E201), and the end sequences were determined (Fig. 1a). Although cosmid E138 contains the start of the core biosynthetic gene cluster (mtaB gene) with the upstream region, the terminal part of the cluster including the mtaG gene (encoding a TE domain located at the C-terminal part of the MtaG protein) and the downstream region are located on cosmid E201. Both cosmids harbor overlapping regions enabling the use of Red/ET recombination to reconnect both parts of the gene cluster as shown in Fig. 2a and b).
Red/ET cloning is based on homologous recombination and, consequently, the repeats within the DNA regions may lead to difficulties and undesired recombinations. To avoid these problems, standard cloning techniques and Red/ET cloning were combined in our study. Moreover, the cosmid backbone was replaced by the p15A origin of replication so that the final construct containing the complete biosynthetic gene cluster was propagated at a lower copy number.
First, the cosmid backbones were replaced with cassettes containing replication ori—p15Aori—and the selection markers chloramphenicol acetyltransferase (Cm) for cosmid E138 (p15A-Cm) and zeocin plus kanamycin from Tn903 for cosmid E201 (p15A-Zeo-Km) (Fig. 2b). The zeocin resistance gene in p15A-Zeo-Km was flanked with PacI sites at both ends. During these steps SpeI restriction sites were included at strategic positions, and silent mutations were generated: the nucleotide sequence in mtaG, CGC ACG CCG CTC GTG CGC ATC (translation: Arg-Thr-Pro-Leu-Val-Arg-Ile), was changed to CGC ACG CCA CTA GTG CGC ATC (translation: Arg-Thr-Pro-Leu-Val-Arg-Ile). The changes are shown in boldface, and the SpeI site is underlined. Both resulting constructs—p15A-E138 and p15A-E201—were digested with this enzyme and ligated. The resulting construct containing all genes in the correct orientation (Fig. 2c) and, accordingly, the whole biosynthetic gene cluster was used for further modifications (Fig. 3).
In parallel, the functional cassette containing the origin of transfer (oriT) for conjugational DNA transfer, the tetracycline resistance gene, and a fragment of the trpE gene from the chromosome of Pseudomonas putida (trpE-tetR-oriT) was removed from p15A-oriT-trpE-tetR (45) by BamHI digestion and inserted into the BamHI site of pOPB18 to create pOPB18-oriT-trpE-tetR. The plasmid pOPB18 (see Materials and Methods) contains the modification cassette required for the integration of the genes into the heterologous host M. xanthus via conjugation (oriT, Km from Tn5 as a selection marker, and the TA′ fragment from the myxovirescin gene cluster). Therefore, the resulting region trpE-tetR-oriT-TA′-Km in pOPB18-oriT-trpE-tetR allows the integration into the chromosome of the heterologous hosts P. putida at trpE or M. xanthus at the TA′ region.
This trpE-tetR-oriT-TA′-Km cassette was subcloned into the p15A-Cm minimum vector by Red/ET recombination to form p15A-Cm-trpE-tetR-oriT-TA′-Km. In this way PacI sites were engineered into both ends of the cassette. The trpE-tetR-oriT-TA′-Km cassette needed for homologous integration into M. xanthus was then removed from the p15-Cm vector by restriction using the newly generated PacI sites and inserted into the PacI site of the stitched myxothiazol gene cluster backbone (replacing the zeocin resistance gene; Fig. 3). Finally, a Pm promoter, which is known to be toluic acid inducible in pseudomonads (24, 25), plus its regulator gene xylS839 and a zeocin selection marker gene, was inserted in front of the mtaB gene (encoding the loading module of the mta gene cluster; Fig. 3).
Integration into M. xanthus DZF1 for heterologous expression.
The final construct (Pm-mta) containing the complete myxothiazol biosynthetic gene cluster was transferred into M. xanthus DZF1 by electroporation. After 7 days the colonies obtained on agar plates containing the kanamycin resistance gene were transferred into liquid medium, and the extracts of the cultures were analyzed by HPLC and HPLC-MS.
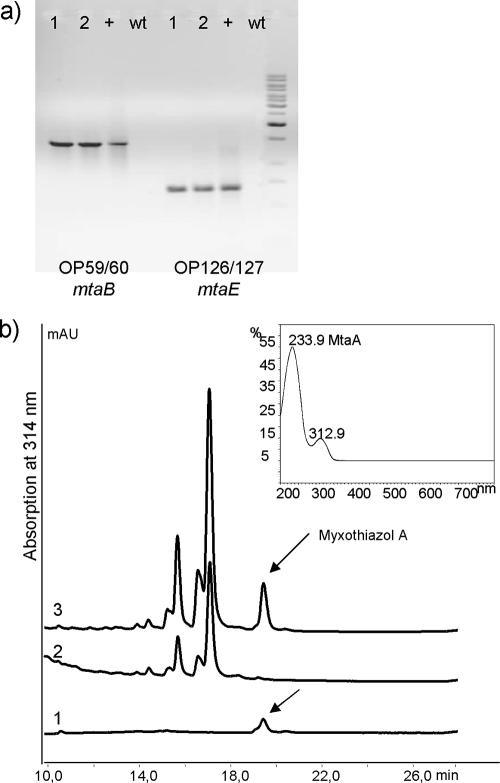
The successful integration of the cluster was verified by colony PCR using internal fragments of the genes mtaB, mtaE, and mtaG (Fig. 4a), as well as primers binding to the Pm promoter region. As expected, the amplification products could be detected only in recombinant clones containing the integrated myxothiazol biosynthetic genes.
FIG. 4.
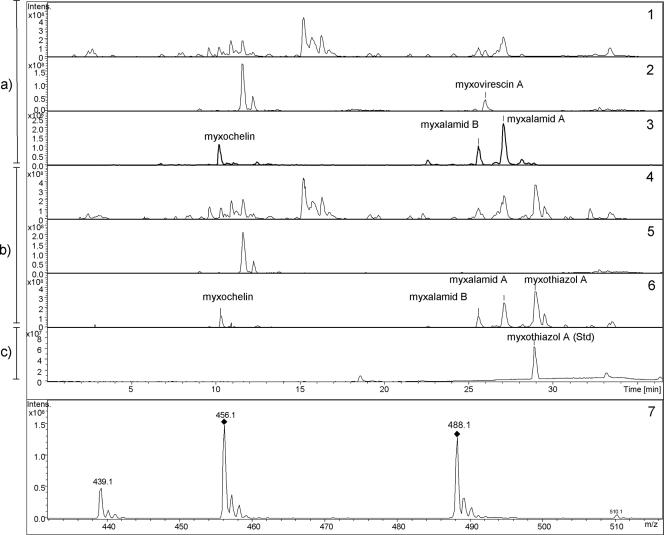
Insertion into the chromosome. (a) Colony PCR. Lanes: 1 and 2, mutant strain; +, positive control DNA; wt, wild-type M. xanthus DZF1 strain. (b) HPLC-diode array detector chromatogram. Curves: 1, authentic standard reference myxothiazol A; 2, extract of M. xanthus DZF1; 3, extract of M. xanthus DZF1::Pm-mta. Inset panel: UV spectrum of myxothiazol at 314 nm.
Analysis of myxothiazol production.
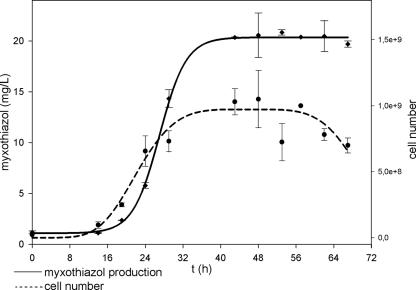
Positive clones were cultured in liquid medium, and myxothiazol production was verified by analysis of the extracts in comparison to an authentic reference standard (Fig. 4b). In the HPLC chromatogram an additional peak with the same retention time and the same UV spectra as the myxothiazol A reference was detected in the recombinant strain compared to the wild-type M. xanthus DZF1. Analyses of the mass spectra verified the biosynthesis of a metabolite with a mass spectrum identical to myxothiazol A (Fig. 5). As expected, in the mutant strain harboring the myxothiazol gene cluster, no production of myxovirescin could be detected because the mta gene cluster is integrated into the chromosome through homologous recombination in the TA region of M. xanthus responsible for myxovirescin biosynthesis (37). Within a range from 20 to 37°C, the optimal temperature for production was 30°C (data not shown). Therefore, this temperature was used in kinetic production studies. Myxothiazol production reached a maximum of 20 mg/liter after 28 to 32 h of incubation (Fig. 5 and 6).
FIG. 5.
(a) HPLC-MS base peak chromatograms indicating the presence of known secondary metabolites in extracts of wild-type M. xanthus DZF1. (b) In mutant strain M. xanthus DZF1::Pm-mta, the new peak corresponding to m/z = 488 (myxothiazol A) is shown, whereas no myxovirescin is detected. (c) Base peak chromatogram of authentic standard. The mass ranges for the base peak chromatograms were as follows: lines 1 and 4, m/z = 100 to 1,000; lines 2 and 5, m/z = 600 to 650; lines 3 and 6, m/z = 400 to 500. For line 7, the mass spectrum of myxothiazol A shows the fragments of m/z = 439 and m/z = 456.
FIG. 6.
Production kinetics of myxothiazol A in M. xanthus DZF1::Pm-mta at 30°C.
DISCUSSION
Increasing amounts of sequence information related to the biosynthesis of natural products are becoming available due to many microbial genome sequencing projects (4). To exploit these raw data, genetic and biochemical studies are needed to understand how the DNA information is transformed into a specific small molecule. These studies are a prerequisite for targeted modifications of the natural product or combinatorial biosynthesis, e.g., by mixing genes from different pathways. In many cases a suitable heterologous expression system is required, especially if the natural products are isolated from strains that grow slowly or are difficult to manipulate genetically (e.g., most myxobacterial strains). The biosynthetic genes for the important inhibitor of the respiratory chain myxothiazol A were isolated from S. aurantiaca DW4-3/1, and the nucleotide sequence of the complete gene cluster was determined (36). Myxothiazol biosynthesis is catalyzed by a PKS/NRPS megasynthetase with some interesting properties, including the incorporation of the unusual starter unit isovalerate, an exceptional domain organization in module 1 of MtaB (two AT domains), a superfluous oxidation domain in MtaC, the hybrid NRPS/PKS protein MtaD, and a lack of heptadiene isomerase activity that is presumably required for the formation of the irregularly located double bonds in myxothiazol (36, 43) (Fig. 1c). Although the genetic information about the mta genes is available from S. aurantiaca, biosynthetic modification in this strain is difficult and time-consuming, as demonstrated by Weinig et al. (43), who established a system for markerless mutagenesis. These experiments showed that alternative genetic methods are required to analyze the myxothiazol megasynthetase.
Many attempts have been made to improve secondary metabolite formation by combinatorial biosynthesis, e.g., by combining genes from different pathways, as well as by the expression of only single genes or relatively small parts of biosynthetic gene clusters. Obviously, expression of the complete biosynthetic gene cluster in suitable heterologous hosts would be highly advantageous, and the selection of the host organism is of great importance to achieve significant production levels (44). Factors such as GC content or codon usage of the genes of interest are key determinants for the successful production of secondary metabolites. The large size of the biosynthetic gene clusters and the possibility that the heterologous host may not synthesize required precursors for the biosynthetic machinery are additional difficulties associated with the heterologous expression of secondary metabolites. Since the PKS and NRPS proteins must be posttranslationally modified for their activity (20), the heterologous strain must possess a P-pant transferase required for the modification of acyl carrier protein (ACP) and peptidyl carrier protein domains. The heterologous host strain should also provide a pool of activated short-chain carboxylic acids and/or amino acids required for secondary metabolite production and has to be resistant to the produced compound in case of its toxicity for the organism. Ideally, the native promoters should be active in the new strain, and the possibility to regulate the expression through the use of inducible promoters would be very advantageous. Taking all of these factors together, the utilization of a phylogenetically related improved strain as heterologous hosts to enable large-scale production with a high yield would be a good alternative for the biosynthesis of natural products and for combinatorial biosynthesis.
If the biosynthetic product is encoded by a single gene (e.g., the products of type III PKS) or by a relatively short coding sequences (<10 kb), then the introduction into the heterologous host is a relatively simple process. With this approach it is possible to find the products of the genes that are thought to be silent (at least under the laboratory conditions) or express genes from environmental DNA. One such example is the detection of flaviolin as the product of a type III PKS of unknown function in Sorangium cellulosum. The corresponding chalcone synthase-like gene was expressed in P. putida, giving rise to flaviolin production, whereas this compound has never been detected in any myxobacterium (12). Another example is the heterologous production of violacein using a 6.7-kbp fragment from an environmental DNA library (6).
However, the introduction of large biosynthetic gene clusters into heterologous hosts is still a challenging task which is limited not only by the biochemistry and genetics of the host organism but also by the size of the DNA that is introduced. Only a few examples for the successful expression of complete biosynthetic machineries for a natural product from myxobacteria in either phylogenetically related or distant strains are known. These examples are the production of epothilone in M. xanthus (initial yield, 0.2 mg/liter) and in Streptomyces coelicolor (yield, 0.05 to 0.1 mg/liter) (18, 39), soraphen in Streptomyces lividans (yield, <0.3 mg/liter) (49), and myxochromide in P. putida (yield, 40 mg/liter) (45). In most cases, time-consuming cloning procedures were used, and the biosynthetic gene clusters were transferred in several fragments. Whereas in streptomycetes expression from a range of vectors containing large DNA fragments (bacterial artificial chromosomes [BACs] and cosmids) is possible, the expression in M. xanthus is dependent on the integration of the DNA into the chromosome due to the lack of plasmids available for myxobacteria. Only one biosynthetic gene cluster, encoding the genes for epothilone biosynthesis has been successfully expressed in M. xanthus. The integration of the respective gene cluster has been performed stepwise through the integration of different cosmids into the chromosome (18).
A different strategy was developed to engineer the myxochromide biosynthetic gene cluster for the heterologous expression in P. putida (45). Red/ET recombineering (47) was applied for the cloning and modification of the whole, approximately 43 kbp, biosynthetic gene cluster. Recombinogenic engineering in E. coli involves the expression of the protein pair RecE/RecT or Redα/Redβ (47) and is mostly suitable for the engineering of large DNA molecules such as BACs and cosmids. Hence, it is ideal for the engineering of PKS and NRPS pathways. However, the system relies on homologous recombination, and therefore repeats within the DNA region (which are often found in PKS and NRPS genes) to be cloned can cause difficulties. Recently, this strategy was also applied for the engineering of the geldanamycin biosynthetic genes in a complementation plasmid and allowed for fast gene disruption, replacement, and generation of a new product (42).
In the present study we applied this method for the reassembly (“stitching”) of the whole myxothiazol gene cluster, as well as further modifications required for integration into the chromosome of the targeted heterologous hosts—either M. xanthus or P. putida. One of the most important improvements caused by this strategy is the fact that the whole secondary metabolite cluster can be cloned on one construct, modified in E. coli, and then transferred in one step into the heterologous host strain. Biosynthetic clusters are frequently isolated from cosmids or BAC DNA libraries. However, due to their sizes, many large complete biosynthetic gene clusters are not often found on a single clone. In this work, two cosmids carrying the mta genes were identified (Fig. 1a) in the cosmid library and were used for the stitching of the whole gene cluster. As shown in Fig. 1b, the gene cluster contains identical repeats 1.2 kb in size. To avoid the possibility of Red/ET-induced recombination in these regions, a combination of Red/ET recombineering and standard DNA engineering techniques was applied. Restriction sites absent from the cosmids were introduced during Red/ET cloning steps and then used to connect both parts of the complete gene cluster by ligation (Fig. 2b). To transfer the cluster into the heterologous host, further modifications of the stitched construct p15Aori-138+201 (Fig. 3) were required. The homologous region from the chromosome of M. xanthus (TA fragment), together with kanamycin resistance as a selection marker for the integration, as well as oriT for conjugation, the trpE region for integration into the chromosome of P. putida, plus a tetracycline resistance marker were introduced into the stitched construct. We intended to insert an inducible promoter so that gene transcription and metabolite production in the heterologous host could be controlled. For this purpose we introduced the Pm promoter (which is inducible in pseudomonads) upstream of the mtaB gene (Fig. 3). However, we found that this promoter is constitutively active but not inducible in M. xanthus (data not shown). Currently, no reliable inducible promoters are available for myxobacteria. Nevertheless, the same final construct was suitable for introduction into the two heterologous hosts we are exploring, and the whole construct, approximately 60 kbp in size, has been integrated into their chromosomes. To achieve this task, electroporation was used in M. xanthus, and conjugation was applied to P. putida.
While myxothiazol production in P. putida required additional metabolic engineering of the host strain (to be described elsewhere [13]), no additional modification of M. xanthus DZF1 was required. This strain has already been shown as a suitable host for epothilone production, although the yield of produced epothilone was initially much lower than in the natural producer S. cellulosum (18). Nevertheless, it was possible to improve the production to 23 mg/liter by optimizing the fermentation conditions (21).
The successful integration of the mta gene cluster in M. xanthus DZF1 was verified genetically and confirmed by the detection of the expected product myxothiazol A from the culture broth. The production reaches its maximum after 28 to 32 h. The yield of approximately 20 mg/liter in the heterologous host M. xanthus is similar to that found in M. fulvus f16 (10, 38). This strain has been optimized for the production of the compound by classical strain improvement, reaching yields of 60 mg/liter (K. Gerth, unpublished data). In addition, the yield in the heterologous strain is significantly higher than in S. aurantiaca DW4-3/1 (approximately 10 mg/liter; R. Müller, unpublished data) from which the biosynthetic genes have been isolated. One could argue that some silent or nonfunctional myxothiazol gene cluster was affected by the engineering or complemented by the genes inserted. However, according to our analysis of all of the secondary metabolic pathways in the genome of M. xanthus DK1622, there is no evidence for such a gene locus with significant similarity to the genes from S. aurantiaca DW4/3-1 (4; Müller, unpublished).
These results show that, in addition to the advantages which M. xanthus offers as a heterologous host for phylogenetically related myxobacteria (similarly high GC content, preferential codon usage, presence of a broad-spectrum P-pant transferase required for the activity of a series of PKS/NRPS megasynthetases), this strain provides suitable amounts of all necessary starter and extender units required for the biosynthesis of myxothiazol (e.g., isovaleryl-CoA, acetyl-CoA, and methylmalonyl-CoA). The use of Red/ET recombination allowed the rapid modification of the stitched gene cluster for the integration into two heterologous hosts: M. xanthus (described here) and P. putida (13). The approach used here allowed the design of a DNA construct harboring all of the required elements for the heterologous production of a natural product in a single construct. Genetic manipulation of the megasynthetase can now be achieved rapidly in E. coli, followed by gene cluster transfer into the heterologous host for the analysis of the genetic manipulation on the biosynthesis.
Heterologous expression of natural product pathways from microorganisms that are difficult to handle, slow growing, or uncultivated is a promising strategy for the utilization of their (secondary) metabolic potential. The engineering of the complete biosynthetic gene cluster by Red/ET recombination allows the cluster to be reconstituted (“stitched”) and manipulated in E. coli, followed by integration into the heterologous host strain in one step. Our work on the expression of myxothiazol in M. xanthus is an important step in the development of heterologous systems for myxobacteria that present good possibilities for natural product formation. This is the second success in using M. xanthus as a heterologous host, and no additional metabolic engineering steps were required (in contrast to E. coli, streptomycetes or pseudomonads). Further development of M. xanthus for heterologous expression will facilitate the exploration of biosynthetic gene clusters of known and unknown function.
Acknowledgments
Research in the laboratory of R.M. was funded by the Deutsche Forschungsgemeinschaft and the Biofuture program of the BMB+F.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Aldrich, C. C., B. J. Beck, R. A. Fecik, and D. H. Sherman. 2005. Biochemical investigations of pikromycin biosynthesis employing native penta- and hexaketide chain elongation intermediates. J. Am. Chem. Soc. 127:8441-8452. [DOI] [PubMed] [Google Scholar]
- 2.Becker, W. F., G. von Jagow, T. Anke, and W. Steglich. 1981. Oudemansin, strobilurin A, strobilurin B and myxothiazol: new inhibitors of the bc1 segment of the respiratory chain with an E-beta-methoxyacrylate system as common structural element. FEBS Lett. 132:329-333. [DOI] [PubMed] [Google Scholar]
- 3.Bode, H. B., and R. Müller. 2006. Analysis of myxobacterial secondary metabolism goes molecular. J. Ind. Microbiol. Biotechnol. 33:577-588. [DOI] [PubMed] [Google Scholar]
- 4.Bode, H. B., and R. Müller. 2005. The impact of bacterial genomics on natural product research. Angew. Chem. Int. Ed. 44:6828-6846. [DOI] [PubMed] [Google Scholar]
- 5.Bollag, D. M., P. A. McQueney, J. Zhu, O. Hensens, L. Koupal, J. Liesch, M. Goetz, E. Lazarides, and M. Woods. 1995. Epothilones, a new class of microtubule-stabilizing agents with Taxol-like mechanism of action. Cancer Res. 55:2325-2333. [PubMed] [Google Scholar]
- 6.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 7.Del Vecchio, F., H. Petkovic, S. G. Kendrew, L. Low, B. Wilkinson, R. Lill, J. Cortes, B. A. Rudd, J. Staunton, and P. F. Leadlay. 2003. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 30:489-494. [DOI] [PubMed] [Google Scholar]
- 8.Gaitatzis, N., A. Hans, R. Müller, and S. Beyer. 2001. The mtaA gene of the myxothiazol biosynthetic gene cluster from Stigmatella aurantiaca DW4/3-1 encodes a phosphopantetheinyl transferase that activates polyketide synthases and polypeptide synthetases. J. Biochem. 129:119-124. [DOI] [PubMed] [Google Scholar]
- 9.Gerth, K., N. Bedorf, H. Irschik, G. Höfle, and H. Reichenbach. 1994. The soraphens: a family of novel antifungal compounds from Sorangium cellulosum (Myxobacteria). I. Soraphen A1 alpha: fermentation, isolation, biological properties. J. Antibiot. 47:23-31. [DOI] [PubMed] [Google Scholar]
- 10.Gerth, K., H. Irschik, H. Reichenbach, and W. Trowitzsch. 1980. Myxothiazol, an antibiotic from Myxococcus fulvus (myxobacterales). I. Cultivation, isolation, physico-chemical and biological properties. J. Antibiot. 33:1474-1479. [DOI] [PubMed] [Google Scholar]
- 11.Gerth, K., S. Pradella, O. Perlova, S. Beyer, and R. Müller. 2003. Myxobacteria: proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J. Biotechnol. 106:233-253. [DOI] [PubMed] [Google Scholar]
- 12.Gross, F., N. Luniak, O. Perlova, N. Gaitatzis, H. Jenke-Kodama, K. Gerth, D. Gottschalk, E. Dittmann, and R. Müller. 2006. Bacterial type III polyketide synthases: phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch. Microbiol. 185:28-38. [DOI] [PubMed] [Google Scholar]
- 13.Gross, F., M. W. Ring, O. Perlova, J. Fu, S. Schneider, K. Gerth, S. Kuhlmann, F. Stewart, Y. Zhang, and R. Müller. Red/ET-subcloning and heterologous expression of methylmalonyl-CoA biosynthesis genes of Sorangium cellulosum So ce56 in Pseudomonas putida KT2440. Chem. Biol., in press. [DOI] [PubMed]
- 14.He, J., and C. Hertweck. 2005. Functional analysis of the aureothin iterative type I polyketide synthase. ChemBioChem 6:908-912. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in non-motile mutant of Myxococcus. Proc. Natl. Acad. Sci. USA 74:2932-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irschik, H., R. Jansen, K. Gerth, G. Höfle, and H. Reichenbach. 1995. Chivosazol A, a new inhibitor of eukaryotic organisms isolated from myxobacteria. J. Antibiot. 48:962-966. [DOI] [PubMed] [Google Scholar]
- 17.Jansen, R., H. Irschik, H. Reichenbach, V. Wray, and G. Höfle. 1994. Disorazoles, highly cytotoxic metabolites from the Sorangicin-producing bacterium Sorangium cellulosum, strain So ce12. Liebigs Ann. Chem. 8:759-773. [Google Scholar]
- 18.Julien, B., and S. Shah. 2002. Heterologous expression of epothilone biosynthetic genes in Myxococcus xanthus. Antimicrob. Agents Chemother. 46:2772-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunze, B., H. Reichenbach, R. Müller, and G. Höfle. 2005. Aurafuron A and B, new bioactive polyketides from Stigmatella aurantiaca and Archangium gephyra (myxobacteria). J. Antibiot. 58:244-251. [DOI] [PubMed] [Google Scholar]
- 20.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily: the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 21.Lau, J., S. Frykman, R. Regentin, S. Ou, H. Tsuruta, and P. Licari. 2002. Optimizing the heterologous production of epothilone D in Myxococcus xanthus. Biotechnol. Bioeng. 78:280-288. [DOI] [PubMed] [Google Scholar]
- 22.Mahmud, T., H. B. Bode, B. Silakowski, R. M. Kroppenstedt, M. Xu, S. Nordhoff, G. Höfle, and R. Müller. 2002. A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria. J. Biol. Chem. 277:23768-32774. [DOI] [PubMed] [Google Scholar]
- 23.Mahmud, T., S. C. Wenzel, E. Wan, K. W. Wen, H. B. Bode, N. Gaitatzis, and R. Müller. 2005. A novel biosynthetic pathway to isovaleryl-CoA in myxobacteria: the involvement of the mevalonate pathway. ChemBioChem 6:322-330. [DOI] [PubMed] [Google Scholar]
- 24.Marques, S., M. T. Gallegos, and J. L. Ramos. 1995. Role of sigma S in transcription from the positively controlled Pm promoter of the TOL plasmid of Pseudomonas putida. Mol. Microbiol. 18:851-857. [DOI] [PubMed] [Google Scholar]
- 25.Mermod, N., P. R. Lehrbach, W. Reineke, and K. N. Timmis. 1984. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 3:2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Morrison, C. E., and D. R. Zusman. 1979. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J. Bacteriol. 140:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller, I., S. Weinig, H. Steinmetz, B. Kunze, S. Veluthoor, T. Mahmud, and R. Müller. A unique mechanism for methyl ester formation via an amide intermediate found in myxobacteria. ChemBioChem 7:1197-1205. [DOI] [PubMed]
- 29.Niggemann, J., M. Herrmann, K. Gerth, H. Irschik, H. Reichenbach, and G. Höfle. 2004. Tuscolid and tuscoron A and B: isolation, structural elucidation and studies on the biosynthesis of novel Furan-3(2H)-one-containing metabolites from the myxobacterium Sorangium cellulosum. Eur. J. Org. Chem. 2004:487-492. [Google Scholar]
- 30.Peiru, S., H. G. Menzella, E. Rodriguez, J. Carney, and H. Gramajo. 2005. Production of the potent antibacterial polyketide erythromycin C in Escherichia coli. Appl. Environ. Microbiol. 71:2539-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichenbach, H. 2001. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 27:149-156. [DOI] [PubMed] [Google Scholar]
- 32.Reichenbach, H., and G. Höfle. 1999. Myxobacteria as producers of secondary metabolites, p. 149-179. In S. Grabley and R. Thiericke (ed.), Drug discovery from nature. Springer, Berlin, Germany.
- 33.Sasse, F., H. Steinmetz, J. Heil, G. Höfle, and H. Reichenbach. 2000. Tubulysins, new cytostatic peptides from myxobacteria acting on microtubuli: production, isolation, physico-chemical and biological properties. J. Antibiot. 53:879-885. [DOI] [PubMed] [Google Scholar]
- 34.Schulz, S., J. Fuhlendorff, and H. Reichenbach. 2004. Identification and synthesis of volatiles released by the myxobacterium Chondromyces crocatus. Tetrahedron 60:3863-3872. [Google Scholar]
- 35.Silakowski, B., B. Kunze, and R. Müller. 2001. Multiple hybrid polyketide synthase/non-ribosomal peptide synthetase gene clusters in the myxobacterium Stigmatella aurantiaca. Gene 275:233-240. [DOI] [PubMed] [Google Scholar]
- 36.Silakowski, B., H. U. Schairer, H. Ehret, B. Kunze, S. Weinig, G. Nordsiek, P. Brandt, H. Blöcker, G. Höfle, S. Beyer, and R. Müller. 1999. New lessons for combinatorial biosynthesis from myxobacteria: the myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274:37391-37399. [DOI] [PubMed] [Google Scholar]
- 37.Simunovic, V., J. Zapp, S. Rachid, D. Krug, P. Meiser, and R. Müller. Myxovirescin biosynthesis is directed by an intriguing megasynthetase consisting of hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl CoA synthases, and trans-acting acyltransferases. ChemBioChem 7:1206-1220. [DOI] [PubMed]
- 38.Steinmetz, H., E. Forche, H. Reichenbach, and G. Höfle. 2000. Biosynthesis of myxothiazol Z, the ester-analog of myxothiazol A in Myxococcus fulvus. Tetrahedron 56:1681-1684. [Google Scholar]
- 39.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 40.Trowitzsch, W., G. Reifenstahl, V. Wray, and G. Höfle. 1980. Myxothiazol, an antibiotic from Myxococcus fulfus (Myxobacterales) II. Structure elucidation. J. Antibiot. 33:1480-1490. [DOI] [PubMed] [Google Scholar]
- 41.Trowitzsch-Kienast, W., V. Wray, K. Gerth, H. Reichenbach, and G. Höfle. 1986. Antibiotika aus Gleitenden Bakterien. XXVIII. Biosynthese des Myxothiazols in Myxococcus fulvus Mx f16. Liebigs Ann. Chem. 10:93-98. [Google Scholar]
- 42.Vetcher, L., Z. Q. Tian, R. McDaniel, A. Rascher, W. P. Revill, C. R. Hutchinson, and Z. Hu. 2005. Rapid engineering of the geldanamycin biosynthesis pathway by Red/ET recombination and gene complementation. Appl. Environ. Microbiol. 71:1829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinig, S., T. Mahmud, and R. Müller. 2003. Markerless mutations in the myxothiazol biosynthetic gene cluster: a delicate megasynthetase with a superfluous nonribosomal peptide synthetase domain. Chem. Biol. 10:953-960. [DOI] [PubMed] [Google Scholar]
- 44.Wenzel, S., and R. Müller. 2005. Recent developments toward the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 16:594-606. [DOI] [PubMed] [Google Scholar]
- 45.Wenzel, S. C., F. Gross, Y. Zhang, J. Fu, F. A. Stewart, and R. Müller. 2005. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering. Chem. Biol. 12:349-356. [DOI] [PubMed] [Google Scholar]
- 46.Wenzel, S. C., B. Kunze, G. Höfle, B. Silakowski, M. Scharfe, H. Blöcker, and R. Müller. 2005. Structure and biosynthesis of myxochromides S1-3 in Stigmatella aurantiaca: evidence for an iterative bacterial type I polyketide synthase and for module skipping in nonribosomal peptide biosynthesis. ChemBioChem 6:375-385. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., F. Buchholz, J. Muyrers, and F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., J. Muyrers, G. Testa, and A. Stewart. 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18:1314-1317. [DOI] [PubMed] [Google Scholar]
- 49.Zirkle, R., J. M. Ligon, and I. Molnar. 2004. Heterologous production of the antifungal polyketide antibiotic soraphen A of Sorangium cellulosum So ce26 in Streptomyces lividans. Microbiology 150:2761-2774. [DOI] [PubMed] [Google Scholar]