Abstract
“Candidatus Endobugula sertula,” the uncultured microbial symbiont of the bryozoan Bugula neritina, produces ecologically and biomedically important polyketide metabolites called bryostatins. We isolated two gene fragments from B. neritina larvae that have high levels of similarity to polyketide synthase genes. These gene fragments are clearly associated with the symbiont and not with the host.
The marine bryozoan Bugula neritina lives in temperate habitats throughout the world. Its uncultured symbiont “Candidatus Endobugula sertula” produces polyketide secondary metabolites called bryostatins (4, 12). These compounds defend the host's larvae from predation by fish (12). The molecular structure of the bryostatins suggests that they are assembled by enzymes called polyketide synthases (PKS). These large multifunctional modular enzymes are made of repeating domains, and the nascent polyketide grows in an assembly line fashion and is modified chemically depending on the presence of certain domains as it “moves down” the enzyme complex (5). Davidson and coworkers (4) found a portion of a polyketide synthase gene cluster, a ketosynthase (KS) domain, in DNA isolated from deep California (CA) populations of B. neritina/“Ca. Endobugula sertula.” They demonstrated that there is expression of this KS gene in “Ca. Endobugula sertula” in the pallial sinus, a groove on the top anterior pole of the host larvae where an inoculum of “Ca. Endobugula sertula” is found. Hildebrand and coworkers subsequently isolated a large open reading frame (bryA) from deep and shallow CA populations of B. neritina/“Ca. Endobugula sertula” that encodes an enzyme that is thought to catalyze the loading and initial elongation and modification steps of bryostatin biosynthesis (9). To date, efforts to culture the symbiont for gene knockout and complementation studies to unequivocally demonstrate that the PKS gene cluster produces bryostatins have not been successful. The CA shallow and deep populations of B. neritina/“Ca. Endobugula sertula” have different suites of bryostatins that have the same macrolactone core but different pendant groups (3). The bryostatins in populations from North Carolina (NC) are similar to those in CA shallow populations (3, 12, 13). Here we report the isolation of two PKS gene fragments from NC populations of B. neritina/“Ca. Endobugula sertula,” one of which is almost identical to a portion of deep CA bryA and the other of which has not been reported previously.
Identification of polyketide synthase gene fragments.
Adult colonies of the bryozoan B. neritina were collected from Radio Island Jetty near Morehead City, NC. These colonies were kept in flowing seawater tables at the University of North Carolina's Institute of Marine Science. Lopanik et al. (12) described larval spawning and collection methodologies previously. Genomic DNA was extracted from B. neritina larvae using an Isoquick DNA extraction kit (Orca Research, Inc.). Degenerate PCR primers for a PKS gene were developed based on proteobacterial KS amino acid sequences deposited in the GenBank database (accession numbers AF081920 [three KS genes], AF239749 [two KS genes], and AF319998) (Table 1). These sequences were aligned using the ClustalW method, and regions where there were high levels of similarity were identified. The amino acid residues M/L/SDPQQR and LGDPIEI/L were used to design the forward and reverse primers, respectively, and the corresponding DNA sequences were aligned to determine the amount of degeneracy at each oligonucleotide position. Primers based on these sequences (Table 1) were used to amplify B. neritina larval DNA in a 25-μl PCR mixture with a “hot-start” Taq polymerase enzyme (JumpStart Taq; Sigma-Aldrich). The first two cycles consisted of melting at 94°C for 2 min, annealing at 37°C for 2 min, and extension at 72°C for 3 min, and this was followed by 39 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 45 s, and extension for 1 min and a final extension for 5 min. The products were separated by electrophoresis on a 1% agarose gel, and a band at ∼700 bp was excised from the gel. The PCR product was purified from the gel (Supelco GenElute agarose spin column) and cloned (Invitrogen TOPO TA cloning kit) into Escherichia coli. The seven resulting clones were screened by colony PCR using vector-specific primers M13F and M13R to amplify the insert. The PCR products were analyzed by restriction enzyme digestion with AluI (New England Biolabs). Five clones had unique inserts which were sequenced using an Applied Biosystems, Inc. 310 genetic analyzer. The resulting sequences were searched against the GenBank database. Internal primers based on the DNA sequences were synthesized, and the clone DNA was amplified with the corresponding M13F or M13R primer (Table 1). The products were sequenced, which resulted in the complete DNA sequence of the insert in both the forward and reverse directions. The complete sequences were aligned with similar sequences using ClustalX (19), and a phylogenetic tree was generated with Mega3.1 (11) using the minimum-evolution method and bootstrapping 10,000 times.
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| BKS254f | RMWYGGACCCSCARCARCG |
| BKS960r | AKYTCGATBGGRTCRCCCAG |
| KSint_f | GGCAAYGGTWTKGTMCCTGGA |
| KSint_r | TCCAGGKACMAWACCRTTGGC |
| BnCOIf | TTGATACTGGGGGCTCCTGATATG |
| BnCOIr | AAGCCCGATGATAAGGGAGGGTA |
| BKS1f | GGTGCGGAGACGGGTGATTA |
| BKS1r | CAACAAGAGCGGAGGAACAGGTA |
| BKS2f | TTGCCGAGTCGTGTAATAGTCT |
| BKS2r | TTGGCACGTTGATCGAAGGTAAA |
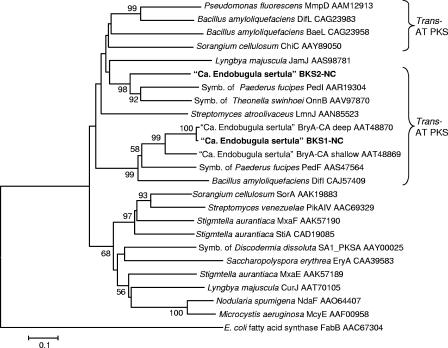
Of the five clones that had unique DNA inserts, the sequences of two of the inserts (designated BKS1 and BKS2) had high levels of similarity to other bacterial PKS genes. The other three clones contained noncoding DNA sequences. The amino acid sequences encoded by BKS1 and BKS2 were 49.5% identical to each other (Fig. 1); the DNA sequences were 54.4% identical. BKS1 was almost identical (98.7% amino acid identity, 97.2% DNA identity) to KS2 of bryA, the putative bryostatin biosynthetic gene recently isolated from deep CA populations of B. neritina/“Ca. Endobugula sertula,” and very similar (80% identical amino acids, 82.1% DNA identity) to KS2 of bryA isolated from shallow CA populations of B. neritina (Fig. 2) (9). In contrast, the BKS2-encoded amino acid sequence was 49.1% identical to the KS2-encoded amino acid sequence of deep CA BryA and 48.7% identical to the KS2-encoded amino acid sequence of shallow BryA (the DNA sequences were 52.7% and 53.7% similar, respectively). Additionally, the two inserts were fairly similar to PKS genes from the Paederus beetle bacterial symbiont, a member of the γ-Proteobacteria (Fig. 2) (16, 17), and to the onnamide cluster from a symbiont isolated from the marine sponge Theonella swinhoei (Fig. 2) (18). The two clones were closely related to PKS gene clusters that have discrete acyltransferase (AT) domains, such as those of the pederin PKS clusters (PedF and PedI) (16, 17), the onnamide cluster (OnnB) (18), the leinamycin cluster (LmnJ) (2), and the mupirocin cluster (MmpD) (6) (Fig. 2); in fact, BryA does not have integrated ATs (9). In typical type I PKS gene systems, the AT domains, which are responsible for attaching the extender unit to the enzyme, are embedded within each module of the PKS polypeptide (5); however, in several systems, there is only one AT domain, and it is found in a separate open reading frame upstream or downstream of the open reading frame encoding the PKS polypeptides (1, 2, 6, 16-18). The B. neritina/“Ca. Endobugula sertula” KS genes were also fairly similar (>48% identity at the amino acid level) to KS genes from bacteria that are not closely related according to 16S rRNA gene sequences, including Microcystis aeruginosa (cyanobacteria) and Streptomyces spp. (actinobacteria) (Fig. 2). This is not surprising as evolutionary pressures such as horizontal gene transfer and environmental factors may affect secondary metabolite biosynthetic gene cluster evolution differently than they affect 16S rRNA evolution (7, 10, 15).
FIG. 1.
Alignment of amino acid sequences of two KS domain genes isolated from B. neritina/“Ca. Endobugula sertula” with BryA KS2 from shallow and deep California populations. S, shallow; D, deep.
FIG. 2.
Minimum-evolution tree based on amino acid sequences of PKS genes from B. neritina/“Ca. Endobugula sertula” and related organisms. The accession numbers of sequences obtained from GenBank are indicated. The sequences in boldface type are described in this paper. Bootstrap analysis (performed 10,000 times) percentages greater than 50% are indicated at the nodes. Scale bar = 0.1 amino acid substitution per site. The suffix NC indicates North Carolina, and the suffix CA indicates California.
Association of PKS gene fragments with “Ca. Endobugula sertula.”
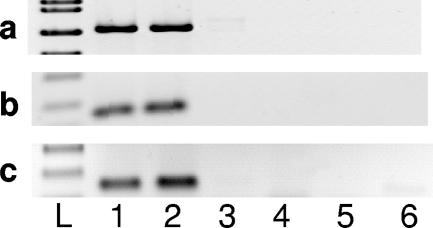
In spring 2002, three replicate groups of B. neritina larvae and, after they settled onto polystyrene petri dishes, newly settled juveniles were exposed for 10 h to the antibiotic gentamicin (75 μg ml−1) on 10 consecutive days. Individuals treated with gentamicin and control individuals (larvae and juveniles exposed to filtered seawater lacking antibiotic) were transplanted into the field and grown until they were reproductive adults. Lopanik et al. (12) provided a full description of the experimental details previously. Aliquots of the next-generation larvae from experimental and control adults were collected, and DNA was extracted for analysis. Purified DNA was amplified using (i) primers specific for the B. neritina cytochrome oxidase I gene, (ii) primers specific for the “Ca. Endobugula sertula” 16S rRNA gene (8), and (iii) primers developed from the two PKS gene DNA sequences isolated from B. neritina/“Ca. Endobugula sertula” (BKS1 and BKS2) (Table 1). For each reaction, the presence of the gene was assessed by electrophoresis of the reaction products through an agarose gel. The B. neritina cytochrome oxidase I gene was amplified from all next-generation larvae collected from the control and gentamicin-treated B. neritina colonies (data not shown). Both the BKS1 and BKS2 genes appeared to be associated with “Ca. Endobugula sertula,” as they were not detected in larvae spawned from the gentamicin-treated group, in which the symbiont loads were reduced by >99% (12), but were present in larvae with “Ca. Endobugula sertula” (Fig. 3). In the aposymbiotic larvae the bryostatin concentrations were found to be reduced by 97%, and these larvae were not chemically deterrent to fish compared to control larvae (12). Our data support the findings of Davidson et al. (4), who performed a similar symbiont knockout experiment, in which they observed a 95% reduction in “Ca. Endobugula sertula” levels in the treatment colonies and a 50% reduction in bryostatin levels in adult B. neritina colonies grown from larvae treated with antibiotics.
FIG. 3.
PCR of symbiotic and aposymbiotic B. neritina larval DNA with primers specific for 16S rRNA genes (8) (a) and the polyketide synthase gene fragments BKS1 (b) and BKS2 (c). Primers are described in Table 1. Lanes 1 and 2, control (symbiotic; n = 2); lanes 3, 4, and 5, treatment (aposymbiotic; n = 3); lane 6, no-template control; lane L, molecular weight ladder.
We isolated two KS genes from B. neritina/“Ca. Endobugula sertula” DNA that were very similar to KS genes from other bacteria, including members of the γ-Proteobacteria (e.g., Paederus symbiont and Pseudomonas fluorescens) (Fig. 1). BKS1 was almost identical to KS2 of bryA isolated from deep CA populations of B. neritina/“Ca. Endobugula sertula.” This observation is particularly interesting because the populations of B. neritina/“Ca. Endobugula sertula” used in this study (shallow NC) are genetically more similar to shallow CA populations based on the B. neritina cytochrome oxidase I gene and the “Ca. Endobugula sertula” 16S rRNA gene sequences (3, 14). Furthermore, the shallow CA and NC populations have the most similar bryostatin profiles (3, 12, 13). While the differences in the KS genes between shallow NC and shallow CA populations of B. neritina/“Ca. Endobugula sertula” are unlikely to result in differences in bryostatin composition in these populations (3) as these KS genes likely produce the polyketide core common to all of the bryostatins, the results may support the notion that secondary metabolism genes are often the result of horizontal gene transfer and recombination (7, 10, 17).
Elimination of the symbiont from B. neritina adults and larvae resulted in the elimination of these KS genes (Fig. 3). We cannot unequivocally demonstrate that the KS genes described from CA (9) and NC (Fig. 1) populations of B. neritina/“Ca. Endobugula sertula” are involved in the biosynthesis of the bryostatins until the symbiont is cultivated and traditional gene knockout and complementation studies confirm this or the PKS cluster is expressed in a heterologous host and bryostatin production is established. However, because BKS1 is very closely related to bryA, the PKS gene isolated in bryostatin-producing CA populations of B. neritina/“Ca. Endobugula sertula” (Fig. 2) (9), we hypothesize that these genes may be involved in bryostatin biosynthesis. bryA is too short to produce the entire backbone of bryostatin based on conventional type I PKS biosynthetic models (5); thus, other PKS genes are probably involved. It is possible that BKS2 resides in one of these other genes.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the BKS1 and BKS2 nucleotide sequences are DQ663570 and DQ663571, respectively.
Acknowledgments
We are grateful to Barb Campbell, Kathy Coyne, and Lisa Waidner for their advice concerning the molecular portions of this project. We thank Jeremy Weisz, Channing Jones, and Glen Safrit for assistance with collection of B. neritina.
Sigma Xi grants-in-aid of research to N.B.L. and SG project R/F-9 (grant to N.M.T.) funded portions of this project.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Chen, X.-H., J. Vater, J. Piel, P. Franke, R. Scholz, K. Schneider, A. Koumoutsi, G. Hitzeroth, N. Grammel, A. W. Strittmatter, G. Gottschalk, R. D. Sussmuth, and R. Borriss. 2006. Structural and functional characterization of the three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188:4024-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, Y. Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, S. K., and M. G. Haygood. 1999. Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula.” Biol. Bull. 196:273-280. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, S. K., S. W. Allen, G. E. Lim, C. M. Anderson, and M. G. Haygood. 2001. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 67:4531-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donadio, S., M. J. Staver, J. B. McAlpine, S. J. Swanson, and L. Katz. 1991. Modular organization of genes required for complex polyketide biosynthesis. Science 252:675-679. [DOI] [PubMed] [Google Scholar]
- 6.El-Sayed, A. K., J. Hothersall, S. M. Cooper, E. Stephens, T. J. Simpson, and C. M. Thomas. 2003. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10:419-430. [DOI] [PubMed] [Google Scholar]
- 7.Ginolhac, A., C. Jarrin, P. Robe, G. Perriere, T. Vogel, P. Simonet, and R. Nalin. 2005. Type I polyketide synthases may have evolved through horizontal gene transfer. J. Mol. Evol. 60:716-725. [DOI] [PubMed] [Google Scholar]
- 8.Haygood, M. G., and S. K. Davidson. 1997. Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for the bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus Endobugula sertula.” Appl. Environ. Microbiol. 63:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrand, M., L. E. Waggoner, H. Liu, S. Sudek, S. Allen, C. Anderson, D. H. Sherman, and M. Haygood. 2004. bryA: an unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem. Biol. 11:1543-1552. [DOI] [PubMed] [Google Scholar]
- 10.Jenke-Kodama, H., A. Sandmann, R. Muller, and E. Dittmann. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22:2027-2039. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformat. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Lopanik, N., N. Lindquist, and N. Targett. 2004. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia 139:131-139. [DOI] [PubMed] [Google Scholar]
- 13.Lopanik, N., K. R. Gustafson, and N. Lindquist. 2004. Structure of bryostatin 20: a symbiont-produced chemical defense for larvae of the host bryozoan, Bugula neritina. J. Nat. Prod. (Lloydia) 67:1412-1414. [DOI] [PubMed] [Google Scholar]
- 14.McGovern, T. M., and M. E. Hellberg. 2003. Cryptic species, cryptic endosymbionts, and geographical variation in chemical defences in the bryozoan Bugula neritina. Mol. Ecol. 12:1207-1215. [DOI] [PubMed] [Google Scholar]
- 15.Metsä-Ketelä, M., L. Halo, E. Munukka, J. Hakala, P. Mantsala, and K. Ylihonko. 2002. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl. Environ. Microbiol. 68:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piel, J., I. Hofer, and D. Q. Hui. 2004. Evidence for a symbiosis island involved in horizontal acquisition of pederin biosynthetic capabilities by the bacterial symbiont of Paederus fuscipes beetles. J. Bacteriol. 186:1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piel, J., D. Q. Hui, G. P. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]