Abstract
Antibodies to Plasmodium falciparum are classically measured using the enzyme-linked immunosorbent assay (ELISA). Although highly sensitive, this technique is labor-intensive when large numbers of samples must be screened against multiple antigens. The suspension array technology (SAT) might be an alterative to ELISA, as it allows measurement of antibodies against multiple antigens simultaneously with a small volume of sample. This study sought to adapt the new SAT multiplex system for measuring antibodies against nine malarial vaccine candidate antigens, including recombinant proteins from two variants of merozoite surface protein 1, two variants of apical merozoite antigen 1, erythrocyte binding antigen 175, merozoite surface protein 3, and peptides from the circumsporozoite protein, ring erythrocyte surface antigen, and liver-stage antigen 1. Various concentrations of the antigens were coupled to microspheres with different spectral addresses, and plasma samples from Cameroonian adults were screened by SAT in mono- and multiplex formats and by ELISA. Optimal amounts of protein required to perform the SAT assay were 10- to 100-fold less than that needed for ELISA. Excellent agreement was found between the single and multiplex formats (R ≥ 0.96), even when two variants of the same antigen were used. The multiplex assay was rapid, reproducible, required less than 1 μl of plasma, and had a good correlation with ELISA. Thus, SAT provides an important new tool for studying the immune response to malaria rapidly and efficiently in large populations, even when the amount of plasma available is limited, e.g., in studies of neonates or finger-prick blood.
Malaria, caused by Plasmodium falciparum, is a major parasitic disease, resulting in high mortality and morbidity. Individuals living in areas where malaria is transmitted often become immune to the disease, with antibodies (Abs) being an important component of immunity (2). Although the specificity of protective Abs is unclear, several antigens (Ags) have been identified as putative targets of immunity, and these Ags are being evaluated as vaccine candidates. Accordingly, it is important to determine Ab levels (titers) in large populations of individuals living in areas where malaria is endemic. Often only small quantities of blood are available, for example, from neonates or finger-prick blood samples from adults. Therefore, an assay that measures Abs to multiple antigens with a few microliters of blood is needed.
Antibody levels to malarial Ags are classically assessed using the enzyme-linked immunosorbent assay (ELISA). This technique requires each Ag to be tested separately and involves long incubation periods, making ELISA labor-intensive and time-consuming as well as requiring significant amounts of Ag and serum or plasma. An immunoassay that measures Abs to multiple Ags simultaneously would be highly advantageous.
Flow cytometry was originally used to study whole cells, organelles, or nuclei (5), but it can also measure biological reactions on the surface of solid fluorospheres using suspension array technology (SAT) (10, 11, 15). SAT uses symmetrical microspheres internally labeled with two fluorescent dyes. The internal dyes are factory established and confer to each set of spheres an intrinsic fluorescence or spectral address. Microspheres can be coupled to specific capture reagents, including protein Ags. Since the microspheres can be distinguished by their spectral addresses, they can be combined after coupling to different Ags to produce multiplex assays, thereby allowing the rapid screening of multiple Ags using a small volume of plasma.
The technique has been used to measure Ab levels to protein Ags of several microbes (12, 14, 17) but not for more complex parasites. SAT was reported to give results comparable to those of ELISA for quantification of Abs against tetanus, diphtheria, Haemophilus influenzae type B (13), pneumococcal capsular polysaccharides (9, 14), and Neisseria meningitidis (8). Furthermore, SAT was at least as sensitive as ELISA for simultaneous detection of Abs to 10 mouse viral and microbial pathogens (7) and was reported to have better reproducibility and a greater dynamic range than ELISA (1, 4). Thus, the SAT approach is becoming an important technique in immunological studies.
The goal of this study was to develop a multiplex assay for measuring Abs to multiple malarial vaccine candidate antigens simultaneously. Nine antigens were used, including two variants (FVO and 3D7) of recombinant merozoite surface protein 1 (MSP-142) and apical merozoite antigen 1 (AMA-1), the recombinant merozoite surface protein 3 (MSP-3) C-terminal region and erythrocyte binding antigen 175 (EBA-175) region II, as well as synthetic peptides with B epitopes from circumsporozoite protein (CSP), ring erythrocyte surface antigen (RESA), and liver-stage antigen 1 (LSA-1). The optimized multiplex proved to be highly sensitive, rapid, and reproducible for the simultaneous measurement of Abs against all nine antigens using less than 1 μl of plasma. Its sensitivity was comparable to that of ELISA, it had a larger dynamic range, and cross-reactivity between Ags was not a problem. Thus, this multiplex assay is a useful new tool for immunological studies of malaria.
MATERIALS AND METHODS
The antigens.
The malaria Ags used in the study included recombinant MSP142 of the FVO and 3D7 strains expressed in Escherichia coli (molecular weight, 42,000), produced by the Malaria Vaccine Development Branch (MVDB), National Institute of Allergy and Infectious Disease, National Institutes of Health, Rockville, MD; recombinant AMA-1 of the FVO and 3D7 strains expressed in yeast (molecular weight, 83,000), provided by MVDB; recombinant EBA-175 RII expressed in yeast (molecular weight, 60,000), obtained from Science Applications International Corp., Frederick, MD; and the recombinant MSP-3 C-terminal region expressed in E. coli (molecular weight, 22,000), provided by The Pasteur Institute, Paris, France. The following peptides, synthesized by AnaSpec, Inc. (San Jose, CA), were used: CSP consisting of five PNAN repeats (molecular weight, 2,100) coupled to bovine serum albumin (BSA) (CPNANPNANPNANPNANPNAN) (the cystine residue was added to enable coupling of the peptide to BSA); RESA consisting of five copies of the EENV repeat (molecular weight, 2,500) coupled to BSA through an added cystine residue (CEENVEENVEENVEENVEENV); and a 40-mer peptide from LSA-1 (molecular weight, 4,500) (LAKEKLQGQQSDLEQERLAKEKLQEQQSDLEQERLAKEKL). Since peptides with molecular weights of less than 3,000 couple poorly to microspheres (unreported data), CSP and RESA peptides were conjugated to BSA following synthesis.
Coupling of antigens to the microspheres.
Ags were coupled to the microspheres using a modification of the protocol from Luminex Corp. Aliquots of 5 million SeroMAP Microspheres (Luminex Corp., Austin, TX) with different spectral addresses (17, 18, 19, 21, 22, 44, 50, 51, 59, 64, and 69) were centrifuged at 8,000 rpm for 2 min, washed with 100 μl of distilled water, resuspended in 80 μl of activation buffer (0.1 M NaH2PO4, pH 6.2), and then activated with 10 μl of a 50-mg/ml solution of 1-ethyl 3-(3-dimethylaminopropyl) carbodiimide hydrochloride and 10 μl of a 50-mg/ml solution of sulfo-N-hyroxysulfosuccinimide. The microspheres were mixed gently and incubated at room temperature (RT) in the dark for 20 min. The activated microspheres were then washed twice with 250 μl of coupling buffer [0.05 M of 2-(N-morpholino) ethanesulfonic acid, pH 5.0] and resuspended in 100 μl of coupling buffer. Various amounts of the malarial Ags were added to the microspheres, and they were then mixed by vortexing followed by sonication at 55 kHz for 5 to 10 s (ultrasonic cleaner; Cole Parmer, Vernon Hills, IL). The total volume was adjusted to 500 μl by adding the coupling buffer. The microspheres were incubated at RT in the dark for 2 h using a rotator at 50 rpm (Bellco Glass Inc., Vineland, NJ) and then washed twice with 1,000 μl of blocking/storage buffer (phosphate-buffered saline [PBS], pH 7.2, containing 1% BSA, 0.02% Tween 20, and 0.05% sodium azide). The coupled microspheres were resuspended in 200 μl of blocking/storage buffer and stored at 4°C in the dark. To determine the optimal amount of protein for coupling, 0.2, 1, 2.5, and 5 μg/million microspheres of recombinant proteins and BSA-conjugated peptides (CSP-BSA and RESA-BSA) and 0.4, 1, 2, and 4 nmol/million microspheres of LSA-1 were assessed. Because of its smaller size, moles were used for LSA-1, as recommended by Luminex Corp.
SAT assay.
Immediately before use, stock suspensions of antigen-coated microspheres were thoroughly resuspended by vortexing and sonication. Aliquots of the microspheres were transferred to Eppendorf tubes and diluted to 100 microspheres/μl using PBS with 1% BSA. Filter plates (Multiscreen BV; Millipore, Billerica, MA) were washed with 200 μl of PBS plus 0.05% Tween 20 (washing buffer) and then washed with 200 μl of PBS plus 1% BSA (dilution buffer) using a vacuum manifold (ZIP Plate Micro SPE plate; Millipore, Billerica, MA). Then, 50 μl of diluted microspheres was distributed to microtiter wells (5,000 microspheres/well) and 50 μl of diluted plasma was added. The plates were placed on a Microplate Shaker (Lab-line, Melrose Park, IL) and incubated at RT in the dark for 1 h at 500 rpm. After incubation, the plates were washed five times with 200 μl of washing buffer, and the microspheres were resuspended in 50 μl of dilution buffer. A total of 50 μl of 2 μg/ml of R-phycoerythrin-conjugated, F(ab′)2 fragment-specific, goat anti-human immunoglobulin G (heavy plus light chains) (Jackson Immunoresearch, West Grove, PA) was added to each well, and plates were incubated for 30 min in the dark at RT on a microplate shaker. After the incubation period, the plates were washed five times with the washing buffer and the microspheres were resuspended in 100 μl dilution buffer. A total of 50 μl of the resuspended microspheres was analyzed using a Liquichip M100 reader (QIAGEN, Valencia, CA). The reader was programmed to read a minimum of 100 microspheres per spectral address, and results are expressed as median fluorescent intensity (MFI).
To optimize the assay, samples for use as positive controls were identified by screening plasma that had been collected from 90 Cameroonian adults by our group after receiving institutional review board approval from Georgetown University and from the National Ethical Committee of Cameroon. The samples were screened using microspheres coated with 6 μg/million microspheres of each antigen, i.e., sphere coupled in the presence of excess Ag. The plasma samples with consistently high MFI were selected and used as positive controls. As negative controls, three pools of plasma collected from 40 Americans who had never travelled to areas where malaria was endemic and who tested negative by ELISA against an extract of asexual-stage malaria parasites were used.
ELISA.
To compare results obtained by the optimized multiplex SAT assay and ELISA, 50 sera from Cameroonian adults were tested for Abs against MSP-142 (FVO and 3D7), AMA-1 (FVO and 3D7), RESA, and LSA-1. In the ELISA, microtiter plates (Maxisorp, Nunc, Denmark) were coated with 100 μl of 1 μg/ml of the FVO and 3D7 strains of recombinant MSP-142 and AMA-1 and with 100 μl of 20 μg/ml of RESA-BSA, BSA alone, and LSA-1 diluted in 0.1 M carbonate bicarbonate buffer (pH 9.5) and incubated overnight at 4°C. The plates were washed three times with washing buffer consisting of PBS plus 0.05% Tween 20 using an automatic plate washer (Microplate Autowasher; Bio-Tek Instruments, Winooski, VT) and blocked with 200 μl of PBS plus 10% milk. After 1 h of incubation at 37°C, the plates were washed three times with the washing buffer. Then, 100 μl of test plasma diluted in PBS-1% milk and positive and negative control plasma was added to duplicate wells. The plates were incubated for 1 h at 37°C and washed five times with the washing buffer, and then 100 μl of a monoclonal antibody to human IgG (whole molecule, A-1543; Sigma, St. Louis, MO) conjugated to alkaline phosphatase was added, using dilutions of 1:4,000 for MSP-142 and AMA-1, 1:1,000 for RESA-BSA and BSA alone, and 1:2,000 for LSA-1. After 1 h of incubation at 37°C, the plates were washed five times and 100 μl of p-nitrophenyl phosphate substrate (1 mg/ml in alkaline phosphatase substrate buffer, pH 9.86) was added in each well. The plates were incubated for 30 min at RT. The optical density (OD) was read at 405 and 630 nm using an Ultra Microplate Reader (Bio-Tek Instruments, Winooski, VT).
Statistical analysis.
The Pearson correlation coefficient was used to evaluate the reproducibility of monoplex and multiplex assays as well as monoplex versus multiplex assays. To compare ELISA and SAT, the values were corrected for nonspecific binding, and the extent of correlation was assessed using the Pearson coefficient. The BSA background was also subtracted from RESA-BSA in both SAT and ELISA. A sample was categorized as Ab positive by ELISA or SAT if the average OD or MFI was greater than the means ± 3 standard deviations of the negative plasma controls. All the analyses were conducted with SAS software (version 9.1; SAS Institute, Cary, NC).
RESULTS
Optimal concentration of antigens.
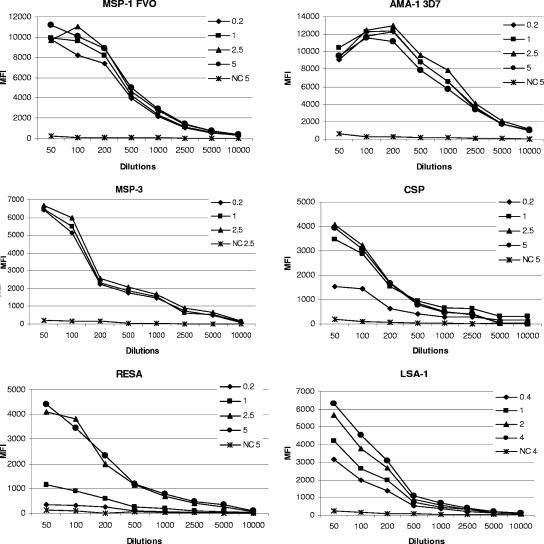
To determine the optimal concentration of Ag to use for coupling, one million microspheres were coupled using 0.2, 1, 2.5, and 5 μg of recombinant proteins and BSA-conjugated peptides, as well as 0.4, 1, 2, and 4 nmol/million microspheres of the synthetic 40-mer LSA-1. The Ag-coated microspheres were then incubated with plasma (diluted 1:50 to 1:10,000) from Ab-positive Cameroonian adults (n = 5) and a pool of Ab-negative plasma. Results showed that microspheres coupled with 0.2 to 1 μg/million microspheres of each of the six recombinant proteins gave maximal binding (representative examples are shown in Fig. 1). The two highest concentrations (2.5 and 5 μg/million microspheres) of BSA-coupled peptides from CSP and RESA produced the highest MFI (Fig. 1). Concentrations of 2 to 4 nmol/million microspheres of the synthetic peptide LSA-1 gave maximal MFI (Fig. 1). Low background levels (less than 500 MFI) were found with the pool of negative control plasma for all Ags. A high-dose prozone effect was found in a few samples at dilutions of 1:50 and 1:100 for MSP-1 (FVO and 3D7), AMA-1 (FVO and 3D7), and EBA-175 regardless of the amount of Ag coated onto the microspheres. As expected, MFI values were generally higher for the recombinant proteins that have multiple B-cell epitopes (i.e., MFI up to 19,000 for MSP142, AMA-1, EBA-175, and MSP-3) compared to the peptides that have a limited number of epitopes (i.e., MFI up to 7,000 for CSP and RESA or 10,000 for LSA-1).
FIG. 1.
Determining the optimal amount of antigen to couple the microspheres. Different amounts of Ag were coupled to microspheres, and Ab binding was determined following incubation with several dilutions of plasma from Cameroonian adults and a pool of negative control (NC) plasma. Microspheres coupled with 0.2 to 1 μg/million microspheres of recombinant proteins gave maximal binding (representative examples from MSP-142 FVO, AMA-1 3D7, and MSP-3 are shown). For peptide-BSA antigens, 2.5 μg/million microspheres worked best, but 1 μg/million microspheres also worked well for CSP. For LSA-1, 2 to 4 nmol/million microspheres gave optimal results. Low levels of background (less than 500 MFI) were found with the pool of NC plasma for all antigens (data shown for spheres coupled with the highest amount of each Ag). Similar results were found for MSP-142 3D7, AMA-1 FVO, and EBA-175.
Comparison of results from monoplex versus multiplex assays.
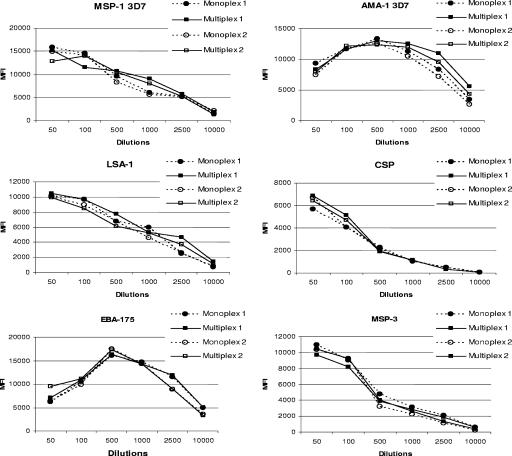
To determine if MFI were equivalent when Ag-coated microspheres were used in a monoplex (i.e., a single Ag) or in a multiplex (i.e., equal amounts of nine Ag-coated microspheres used simultaneously), plasma from nine Cameroonian adults and a pool of negative control plasma were screened (Fig. 2). Results were essentially identical between the two formats, with correlation coefficients for all nine Ags of ≥0.96, as follows (correlation coefficients are in parentheses): MSP1-FVO (0.99), MSP-3D7 (0.99), AMA1-FVO (0.96), AMA1-3D7 (0.99), EBA-175 (0.99), MSP-3 (0.99), CSP (0.99), RESA (0.99), and LSA-1 (0.99). Similar results were obtained regardless of the dilution of plasma use. Thus, multiplexing the nine malarial antigens did not change the results from those obtained when Ags were tested individually.
FIG. 2.
Comparison of MFI between monoplex and multiplex assays and assay reproducibility. Microspheres, coupled with optimal amounts of Ag (1 μg/million microspheres of MSP-142 or AMA-1; 0.2 μg/million microspheres of EBA-175 or MSP-3; 2.5 μg/million microspheres of CSP or RESA; and 4 nmol/million microspheres of LSA-1), were screened against nine malaria-positive plasma samples in two monoplex and two multiplex assays on two different days using the same batch of microspheres. The graphs show the results from one representative plasma sample for each antigen. The plasma samples and antigens not shown gave similar results.
No differences were found when the plasma samples were screened for one allelic form of MSP-1 or AMA-1 alone (monoplex assay) and when the samples were screened using the multiplex assay which contains both allelic forms (Fig. 2). Thus, these antigenic variants can be combined in the same multiplex assay.
Reproducibility of the assay.
The reproducibility of the assay was evaluated by comparing the MFI obtained in two independent monoplex and two multiplex assays conducted on two different days using the same batch of antigen-coated microspheres and six plasma dilutions from nine Ab-positive Cameroonian adults. No significant variation in MFI was observed for any of the Ags between the two assays (Fig. 2). Therefore, plate-to-plate variation using the assay was minimal, and the assay was reproducible on different days.
The interwell variability was also examined by comparing the results from duplicate wells in a multiplex assay. Based on 90 samples, the correlation coefficients between the MFI obtained in the two wells were ≥0.91 (correlation coefficients were 0.98, 0.99, 0.97, 0.97, 0.97, 0.91, 0.92, and 0.98 for MSP-142 FVO, MSP-142 3D7, AMA-1 FVO, AMA-1 3D7, MSP-3, CSP-BSA, RESA-BSA, and LSA-1, respectively). Thus, as shown by others (3), good reproducibility within the assay was observed.
Comparison of SAT and ELISA results.
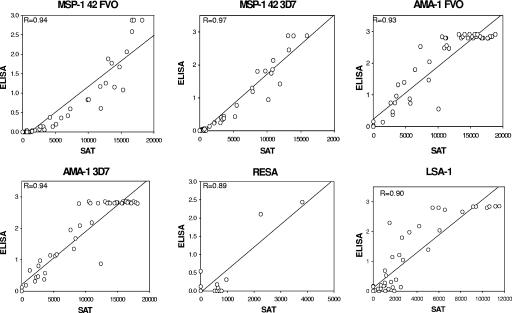
The results obtained using the optimized multiplex assay were compared with those obtained by the standardized ELISA routinely used in our laboratory. A total of 50 samples from Cameroonian adults and three pools of negative plasma were screened at a 1:1,000 dilution in the multiplex SAT and ELISA against MSP-142 FVO, MSP-142 3D7, AMA-1 FVO, and AMA-1 3D7 and at a 1:100 dilution against RESA-BSA, BSA alone, and LSA-1. The two dilutions were selected because, at these dilutions, MFI (Fig. 1) and OD values (data not shown) routinely fall on the linear part of titration curves for most samples. A high level of correlation between MFI in the SAT and OD in the ELISA was observed (Fig. 3). Correlation coefficients (as R values, in parentheses) were the following: MSP-142 FVO (0.94), MSP-142 3D7 (0.97), AMA-1 FVO, (0.93), AMA-1 3D7 (0.94), RESA-BSA (0.89), and LSA-1 (0.90). Overall, the numbers of positive and negative samples were also in good agreement between SAT and ELISA, with concordances between 84 and 98% for each of the six antigens (Table 1). Fewer than 4% of the samples that were positive by ELISA were negative by SAT, whereas up to 12% of samples positive by SAT were negative by ELISA. Thus, the sensitivity of the SAT assay is comparable to, or better than, that of the ELISA.
FIG. 3.
Correlation between the results from ELISA and multiplex SAT. A total of 50 samples from Cameroonian adults were screened in the multiplex assay and by ELISA against six malarial Ags at a dilution of 1:1,000 (AMA-1 and MSP-142) or 1:100 (RESA and LSA-1). Results were compared using a linear regression curve, and the correlation coefficients were determined. Each data point represents a sample that was Ab positive in one or both assays. Ab-negative samples are not shown. A high level of correlation was observed for all the antigens tested. Correlation coefficients are based on n = 50 samples. ELISA is measured in OD, and SAT is measured in MFI.
TABLE 1.
Correlation between SAT and ELISA resultsa
| Ag | % of samples that were:
|
|||
|---|---|---|---|---|
| Positive by SAT and ELISA | Positive by SAT but negative by ELISA | Negative by SAT but positive by ELISA | Negative by SAT and ELISA | |
| MSP-1 FVO | 66 | 2 | 6 | 26 |
| MSP-1 3D7 | 62 | 0 | 12 | 26 |
| AMA-1 FVO | 86 | 4 | 0 | 10 |
| AMA-1 3D7 | 88 | 2 | 0 | 10 |
| RESA | 10 | 4 | 12 | 74 |
| LSA-1 | 66 | 4 | 6 | 24 |
A total of 50 plasma samples from Cameroonian adults were used.
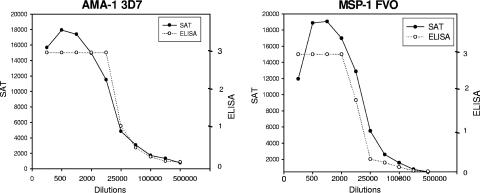
Previous studies have reported differences in the dynamic range (i.e., range of dilutions on the linear part of the curve) for the SAT assay and ELISA (1, 4). Therefore, multiple dilutions of plasma from individuals who have very high titers of Ab for MSP-142 and for AMA-1 were screened in parallel by SAT and ELISA (Fig. 4). Using these recombinant proteins, the titration curve was relatively linear from 1:500 to 1:25,000 (i.e., 50- to 100-fold range of dilutions) in the multiplex assay but were only from 1:2,000 to 1:25,000 (i.e., 10- to 12.5-fold range of dilutions) by ELISA. These results demonstrate a wider dynamic range with multiplex SAT compared to ELISA for Ags that induce high-level Ab responses.
FIG. 4.
Titration curves by ELISA and SAT for MSP-1 and AMA-1. Pools of plasma from individuals with very high Ab titers to MSP-142 3D7 and AMA-1 FVO were screened at 10 dilutions (1:50 to 1:500,000) by ELISA and multiplex SAT. The linear range with SAT was consistently greater than that for ELISA.
DISCUSSION
Multiplex assays have been developed for detecting Abs to combinations of viral and bacterial pathogens (7, 13) and to different bacterial serotypes (8, 9). The multiplex assay described herein is to the first to measure Ab responses to P. falciparum malarial proteins, including sporozoite (CSP), liver-stage Ag (LSA-1), and asexual blood-stage Ags (MSP-142, AMA-1, EBA-175, MSP-3, and RESA) simultaneously. The assay for malaria proved to be rapid, allowing us to screen over 250 samples against the nine Ags in an afternoon. The assay also requires small amounts of Ag and minimal amounts of plasma. It has a wider dynamic range and is as sensitive as ELISA. Thus, this multiplex immunoassay is a useful new tool for research on malaria.
A major concern with multiplexing microspheres coated with different Ags is that combining Ags might result in Ab competition or blocking. In this study, no significant difference was found when antigen-coated spheres were used alone or in combination (Fig. 2). Measurement of Abs against the two variants of AMA-1 and MSP-142 was not affected by multiplexing the microspheres. The two variants for AMA-1 differ by 25 out of 533 amino acids, i.e., they share 95.3% homology. Very similar results were obtained by ELISA and multiplex SAT for the two variants, suggesting that most of the B-cell epitopes are common to the two variants. One might have predicted that by mixing the two highly similar variants the MFI would decrease in the multiplex compared to the monoplex at high plasma dilutions. However, even when the plasma samples were diluted 1:100,000, no appreciable differences were seen between the mono- and multiplex assays, suggesting that equivalent amounts of Ab bound to each of the variants (Fig. 2). In the case of MSP-1, the amino acid composition of the two variants differs by about 50%. Ab differences between the two variants both by ELISA and SAT were seen. Only two samples were positive for 3D7 but not FVO by ELISA, whereas by SAT three samples were positive for FVO but not 3D7 and three other samples were positive for 3D7 but not FVO. These results suggest that the two variants have some variant-specific epitopes and that the multiplex SAT may be better than ELISA in detecting these differences. The simultaneous measurement of Abs in the multiplex assay may represent a more “natural” method than measuring Abs in separate assays. In vivo, Abs against multiple Ags and variants interact. Therefore, multiplexing may allow one to gain a better understanding of the competition of Abs in vivo.
Very good intra- and interassay reproducibility was observed in the SAT assay (Fig. 2). Correlation coefficients greater than 0.90 were found for variation between duplicate wells within the same plate as well as between multiplex assays conducted on different days. High reproducibility may be directly related to the high dynamic range of the SAT assay and expressing the results as medians instead of means, since extreme values do not influence the results. The use of samples in duplicates has been recommended by Dasso et al. (4), whereas Carson and Vignali (3) did not find it necessary. Our results suggest that a single dilution of plasma is probably adequate for semiquantitative analysis of antimalarial Ab levels.
The reaction kinetics of SAT is faster than that of ELISA, because the SAT is particle based (13). In this study, the total incubation time for ELISA was more than twice that for SAT (Table 2). The shorter incubation and, thus, shorter total assay time of the SAT assay has been described previously (13). In addition, no overnight incubation of Ag with microtiter wells is required. Since each Ag is directly coupled to spheres with a unique spectral address, different combinations of Ags can be multiplexed, providing flexibility with the assay in different experiments. Using the SAT, one can screen more than 250 samples in a single day against the nine malarial antigens, using three filter plates; by ELISA, this would represent about 100 plates if the samples are run in duplicate, requiring several weeks. Thus, the multiplex assay is both reproducible and rapid.
TABLE 2.
Technical comparison of ELISA and SAT
| Assay | Incubation time (min) | Amt of Ag required (μg/plate) | Plasma or serum vol |
|---|---|---|---|
| ELISA | 210 | 10-200 | ∼1 μl/antigen |
| SAT | 90 | 0.2-2.5 | ∼1 μg for all antigens |
The SAT has a sensitivity equal to, or better than, ELISA for measuring cytokines (6) and has been reported to have a higher dynamic range (3). The use of a filter plate is believed to play a role in the increase in sensitivity (16). In Ab studies, SAT has been reported to be more (1) or slightly less (13) sensitive than ELISA. We observed a good correlation in sensitivity of ELISA and SAT for the six antigens tested (Fig. 3). The samples used were screened at a dilution of 1:1,000 for the recombinant protein and 1:100 for the peptides. A larger number of samples was positive by SAT than by ELISA for MSP-1, RESA, and LSA-1, but Abs to AMA-1 were detected in several samples by ELISA but not SAT (Table 1). Most of the discrepancy resulted from samples slightly positive by SAT and negative by ELISA. Other studies have also reported some discrepancies between SAT and ELISA (7). In some cases, results from SAT were confirmed using a third technique, such as indirect fluorescent-antibody assay.
The simultaneous measurement of Abs to multiple malarial Ags has numerous advantages, especially when only small amounts of plasma are available. The use of SAT has practical applications for characterizing the immune response to malaria in infants and young children. Infants are protected against malaria during their first months of life, and maternal antibodies are thought to play an important role. However, the exact specificity of the protective Abs has not been identified. Research in this field has been limited by the small amount of blood one can draw from an infant. The SAT assay will allow scientists to follow changes in Ab levels in infants and eventually to gain a better understanding of the immunology of malaria in young children. The SAT can also be used for the rapid screening of blood samples from hundreds of persons living in areas where malaria is endemic and, thus, allow a rapid identification of putative protective Abs. The multiplex assay can also help to identify new malarial vaccine candidates and shorten the laboratory phase of vaccine trials, therefore allowing for more rapid determination of the protection conferred by the vaccine.
Acknowledgments
This study was supported by grant UO1 AI43888 and grant R21AI53798 from NIAID.
We thank Sheryl Dumbar and Allan Ward from Luminex Corp. for their support in the development of the assay and W. Bancroft for providing EBA-175 recombinant protein. We also thank F. F. Mandy and colleagues at the Public Health Agency of Canada for their support in assay development.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Biagini, R. E., D. L. Sammons, J. P. Smith, B. A. MacKenzie, C. A. Striley, V. Semenova, E. Steward-Clark, K. Stamey, A. E. Freeman, C. P. Quinn, and J. E. Snawder. 2004. Comparison of a multiplexed fluorescent covalent microsphere immunoassay and an enzyme-linked immunosorbent assay for measurement of human immunoglobulin G antibodies to anthrax toxins. Clin. Diagn. Lab. Immunol. 11:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouharoun-Tayoum, H., P. Attanath, A. Sabchareon, T. Chongsuphajaisiddhi, and P. Druilhe. 1990. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro but act in cooperation with monocytes. J. Exp. Med. 172:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carson, R. T., and D. A. Vignali. 1999. Simultaneous quantification of 15 cytokines using a multiplex flow cytometric assay. J. Immunol. Methods 227:41-52. [DOI] [PubMed] [Google Scholar]
- 4.Dasso, J., J. Lee, H. Bach, and R. G. Mage. 2002. A comparison of ELISA and flow microsphere-based assays for quantification of immunoglobulins. J. Immunol. Methods 263:23-33. [DOI] [PubMed] [Google Scholar]
- 5.Jaroszeski, M. J., and G. Radcliff. 1999. Fundamentals of flow cytometry. Mol. Biotechnol. 11:37-53. [DOI] [PubMed] [Google Scholar]
- 6.Kellar, K. L., R. R. Kalwar, K. A. Dubois, D. Crouse, W. D. Chafin, and B. E. Kane. 2001. Multiplex fluorescent bead-based immunoassays for quantification of human cytokines in serum and in culture supernatants. Cytometry 45:27-36. [DOI] [PubMed] [Google Scholar]
- 7.Khan, I. H., L. V. Kendall, M. Ziman, S. Wong, S. Mendoza, J. Fahey, S. M. Griffey, S. W. Barthold, and P. A. Luciw. 2005. Simultaneous serodetection of 10 highly prevalent mouse infectious pathogens in a single reaction by multiplex analysis. Clin. Diagn. Lab. Immunol. 12:513-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lal, G., P. Balmer, H. Joseph, M. Dawson, and R. Borow. 2004. Development and evaluation of a tetraplex flow cytometric assay for quantitation of serum antibodies to Neisseria meningitides serogroups A, C, Y, and W-135. Clin. Diagn. Lab. Immunol. 11:272-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lal, G., P. Balmer, E. Stanford, S. Martin, R. Warrington, and R. Borow. 2005. Development and validation of a nonaplex assay for simultaneous quantitation of antibodies to nine Streptococcus pneumoniae serotypes. J. Immunol. Methods 296:135-147. [DOI] [PubMed] [Google Scholar]
- 10.Mandy, F. F., T. Nakmura, M. Bergeron, and K. Sekiguchi. 2001. Overview and application of suspension array technology. Clin. Lab. Med. 21:713-729. [PubMed] [Google Scholar]
- 11.Nolan, J. P., and L. A. Sklar. 2002. Suspension array technology: evolution of the flat array paradigm. Trends Biotechnol. 20:9-12. [DOI] [PubMed] [Google Scholar]
- 12.Opalka, D., C. E. Lachman, S. A. MacMullen, K. U. Jansen, J. F. Smith, N. Chirmule, and M. T. Esser. 2003. Simultaneous quantification of antibodies to neutralizing epitopes on virus-like particle for human papillomavirus type 6, 11, 16, and 18 by a multiplex Luminex assay. Clin. Diagn. Lab. Immunol. 10:108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickering, J. W., T. B. Martins, M. C. Schroder, and H. R. Hill. 2002. Comparison of a multiplex flow cytometric assay with enzyme-linked immunosorbant assay for quantification of antibodies to tetanus, diphtheria, and Haemophilus influenzae type b. Clin. Diagn. Lab. Immunol. 9:872-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickering, J. W., T. B. Martins, and R. W. Greer. 2002. A multiplex fluorescent microsphere immunoassay for antibodies to pneumococcal capsular polysaccharides. Am. J. Clin. Pathol. 117:589-596. [DOI] [PubMed] [Google Scholar]
- 15.Seideman, J., and D. Peritt. 2002. A novel monoclonal antibody screening method using the Luminex-100 microsphere system. J. Immunol. Methods 267:165-171. [DOI] [PubMed] [Google Scholar]
- 16.Vignali, D. A. 2000. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 243:243-255. [DOI] [PubMed] [Google Scholar]
- 17.Willman, J. H., H. R. Hill, T. B. Martins, T. D. Jaskowski, E. R. Ashwood, and C. M. Litwin. 2001. Multiplex analysis of heterophil antibodies in patients with indeterminate HIV immunoassay results. Am. J. Clin. Pathol. 115:764-769. [DOI] [PubMed] [Google Scholar]