Abstract
The aggressive clinical behavior of melanoma has led to the hypothesis that the developmental origins of melanocytes in the neural crest might be relevant for their metastatic propensity. We demonstrate that primary human melanocytes, transformed using a specific set of introduced genes, form melanomas that frequently metastasize to multiple secondary sites, while human fibroblasts and epithelial cells transformed using an identical set of genes generate primary tumors that rarely do so. Importantly, these melanomas exhibit a metastasis spectrum similar to that observed in human patients. These observations indicate that part of the metastatic proclivity of melanoma is attributable to lineage-specific factors expressed in melanocytes and not in other cell types analyzed. Analysis of microarray data from human nevi reveals that Slug, a master regulator of neural crest cell specification and migration, correlates in its expression pattern with other genes that are important for neural crest cell migrations during development. Moreover, Slug is required for the metastasis of the transformed melanoma cells. These findings indicate that melanocyte-specific factors present prior to neoplastic transformation can play a pivotal role in governing melanoma's progression.
Introduction
The resistance of advanced cases of melanoma to existing treatment modalities and the rapid rise in the incidence of this disease1,2 underscore the importance of acquiring a better understanding of the pathogenesis of this disease. Research in this area has led to the discovery of alterations that disrupt key tumor suppressor and oncogene pathways during melanoma formation. The end-results of these modifications include inhibition of the Rb tumor suppressor pathway 3, activation of the mitogen-activated protein kinase (MAPK) pathway 4, and activation of signaling pathways downstream of certain receptor tyrosine kinases 5. Many of these changes, originally discovered through examination of tumor biopsies, have been functionally validated using experimental models 6.
Melanoma is notorious for its tendency to rapidly progress to metastatic disease. Indeed, the observation of widespread metastatic dissemination with relatively small (or undetectable) primary tumors is not an uncommon clinical occurrence7-9. In principle, this strong metastatic propensity might be due to peculiarities in the oncogenic mutations sustained during melanoma development or to extrinsic factors, such as the ability of UV radiation to induce melanoma-specific DNA lesions. An alternative hypothesis that has been proposed, however, is that melanocytes are intrinsically predisposed as a result of their differentiation program to exhibit invasive and metastatic traits following neoplastic transformation 10. In spite of the importance of distinguishing between these two fundamentally different scenarios, the inability to fully characterize the genetic lesions present within established cancer cell lines has precluded attempts to ascribe their tumor phenotypes to either differentiation- or mutation-specific causes.
Dermal melanocytes arise from a uniquely migratory embryonic cell population termed the neural crest. Neural crest cells are specified at the dorsal side of the neural tube during development; cells of this lineage subsequently migrate throughout the embryo and give rise to numerous differentiated cell types, including dermal melanocytes 11. The derivation of cutaneous melanomas from the migratory neural crest cell population supports the intrinsic predisposition hypothesis described above. Thus, elements of the neural crest molecular circuitry responsible for migratory behavior during development might well be reactivated during melanoma pathogenesis.
To examine the effects of cell type-specific factors on melanoma phenotypes, we exploited an experimental protocol that makes possible the neoplastic transformation of primary human cells through the introduction of a defined set of genetic elements 12. This system utilizes the Simian Virus 40 early region (SV40ER), which encodes the viral large T (LT) and small T (st) oncoproteins, in conjunction with a gene encoding the catalytic subunit of the telomerase holoenzyme (hTERT) to immortalize primary human cells; the resulting immortalized cells are then transformed by introducing a ras oncogene. Since this procedure generates cancer cell lines transformed with an identical set of genes, variations in tumor phenotypes arising from genetic differences between cancer cell lines are effectively minimized. A variety of human cell types transformed in this manner generate localized tumors in the absence of metastasis 12-15. We have applied this technique to primary human melanocytes, and have uncovered distinctive behaviors of transformed melanocytes that help to explain the highly malignant phenotype associated with many human melanomas.
Results
Requirements for the Transformation and Immortalization of Melanocytes
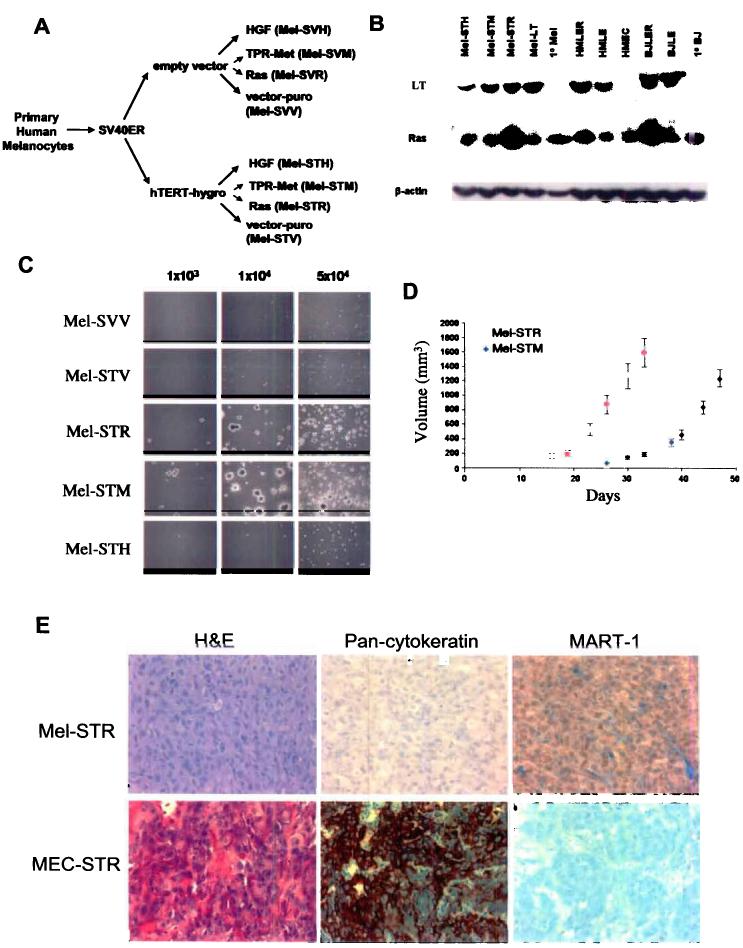
To immortalize primary human melanocytes, we used retrovirus infection to introduce the SV40ER and hTERT cDNAs into the cells. Following this genetic modification, the melanocytes proliferated indefinitely in culture (data not shown) but failed to form either anchorage-independent colonies in soft agar or tumors when injected subcutaneously into immunocompromised mice (Figure 1; Table 1). We subsequently introduced an oncogenic Ras protein (RasG12V) into the immortalized melanocytes, yielding Mel-STR cells, and observed that RasV12 efficiently transformed these cells, as demonstrated by their ability to form colonies in suspension culture and tumors in vivo (Mel-STR; Figure 1; Table 1).
Figure 1.
Characterization of retrovirus-transduced primary human melanocytes. a, Schematic of the transformation protocol. b, Expression of the LT, Ras, and TPR-met proteins in primary and engineered cell lines. c, Anchorage-independent colony formation of engineered melanocyte (Mel) cell lines requires expression of the SV40ER and hTERT, together with either RasV12 or TPR-met. Assays were performed in triplicate for each cell line shown and a representative field photographed. d, Subcutaneous tumor growth of engineered melanoma cell lines in athymic nude mice in vivo (n>=12). Error bars reflect standard errors. e, Hematoxylin & eosin staining, and MART-1 and pan-cytokeratin immunostainings of histological sections prepared from engineered subcutaneous mammary epithelial cell (MEC-STR) and melanoma (Mel-STR) tumors.
Table 1.
Primary tumor formation and secondary metastasis. Incidence of tumor formation and metastasis in mice subcutaneously injected with melanocyte (Mel-STR), mammary epithelial cell (MEC-STR) and fibroblast (BJ-STR) cells engineered to express the SV40ER, hTERT and RasV12. Also shown are melanocytes expressing the SV40ER, hTERT and a vector control (Mel-STV). S,O indicate subcutaneous and orthotopic injections, respectively. NA, not applicable.
| 1o Tumor | Mel-STR | Mel-STV | MEC-STR | BJ-STR | |
|---|---|---|---|---|---|
| 93/93 | 0/12 | 21/21 (S) | 21/21 | ||
| 20/20 (O) | |||||
| Metastasis | 1/21 (S) | ||||
| Any organ | 85/93 | NA | 0/21 (O) | 0/21 | |
| Lung | 85/93 | NA | 1/21 (S) | 0/21 | |
| 0/21 (O) | |||||
| Liver | 25/93 | NA | 0/21 (S,O) | 0/21 | |
| Spleen | 21/93 | NA | 0/21 (S,O) | 0/21 | |
| Small bowel | 11/93 | NA | 0/21 (S,O) | 0/21 | |
| Lymph nodes | 27/93 | NA | 0/21 (S,O) | 0/21 |
Several tyrosine kinase receptors (RTKs) that signal through the Ras-MAPK pathway have also been implicated in melanomagenesis. In particular, melanoma cell lines frequently exhibit autocrine stimulation provoked by hepatocyte growth factor (HGF) secretion 16, and c-Met, the receptor for HGF, is frequently upregulated during melanoma progression 17,18. To test whether activation of the HGF-c-Met signaling loop would also suffice to provide the proliferative signal for transformation, we introduced a constitutively active form of the c-Met receptor (termed TPR-Met) 19 into the immortalized human melanocytes.
TPR-met was able to substitute for RasV12 in enabling the melanocytes to become transformed, as indicated by the ability of these Mel-STM cells to form suspension colonies in vitro and tumors in vivo (Figure 1c). In contrast, ectopic over-expression of HGF, which generates a c-Met autocrine growth-stimulatory loop, failed to transform the immortalized melanocytes (Figure 1c). Hence, while HGF over-expression increased the in vitro proliferation of the melanocytes (data not shown) and increased survival of minute colonies in soft agar (Figure 1c), tumor formation was not observed in vivo (Figure 1c).
Histology and Immunohistochemistry of Mel-STR Melanomas
Examination of histological sections of primary tumors generated by the Mel-STR and Mel-STM cells revealed growths exhibiting characteristics of high-grade, well-vascularized, epithelioid, amelanotic melanomas (Figure 1e). Clinical tumor samples are routinely diagnosed as melanomas by immunohistochemical staining for melanoma-specific markers as well as the absence of staining for cytokeratins. Indeed, the Mel-STR tumors stained positively for the melanoma markers MART-1 and vimentin, and lacked expression of cytokeratins (Figure 1e and Figure S1). In contrast, human mammary epithelial cells transformed through the introduction of the identical set of genes (MECSTR cells) were negative for MART-1 expression and stained positively for cytokeratins (Figure 1e).
Since the disruption of adherens junctions frequently accompanies melanoma progression, we examined the expression and localization of E-cadherin and β-catenin in the melanoma tumor sections. We observed that the Mel-STR melanomas, unlike normal melanocytes, did not express E-cadherin, and exhibited weak membranous and cytoplasmic staining for β-catenin (Supplementary Fig.2 online). Staining for the SV40 LT antigen revealed that significant regions of the tumors contained cells that did not express the LT protein and were therefore composed of recruited stromal cells of murine origin (Supplementary Fig.2 online). Histological examination revealed that these latter cells were frequently associated with regions of inflammation and necrosis and were, by several criteria, largely neutrophils (data not shown).
Transformed Melanoma Cells Form Metastatic Tumors In Vivo
We undertook an extensive anatomical analysis at necropsy of the organs of melanoma-bearing mice in order to determine whether macroscopically visible metastases developed following subcutaneous injection of Mel-STR melanoma cells. To facilitate visualization of metastases in these animals, we used Mel-STR cells into which the green fluorescent protein (GFP) gene had been introduced through use of a retroviral vector. Mice were sacrificed when they appeared moribund, which occurred in the majority of mice approximately 6-8 weeks after the initial injection of transformed cells.
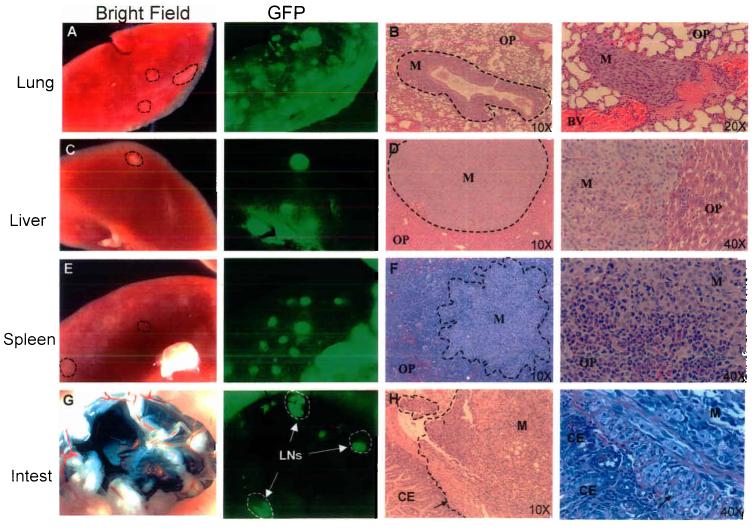
We observed numerous mice with widespread metastatic dissemination. Metastatic nodules were observed in 92% of mice bearing Mel-STR tumors, most commonly in the lungs (92%) and lymph nodes (29%), as well as in the liver (27%), spleen (22%), and small bowel (12%) (Figure 2; Table 1). In general, the histology of metastatic growths resembled that of primary tumors. In cases where mice bore metastases to the liver, spleen, or small bowel, metastatic burden was also invariably present in the lungs, raising the possibility that lung metastases may serve to further disseminate additional metastases to other organs in these animals.
Figure 2.
Primary Mel-STR melanomas give rise to widespread metastases in vivo. Immunocompromised NOD/SCID mice were injected subcutaneously with 5×105 GFP-labeled Mel-STR cells. Organs were harvested at necropsy and were immediately visualized for GFP epifluorescence, or fixed for subsequent paraffin embedding. Images of whole-organs (a,c,e,g) and H&E-stained sections (b,d,f,h) were captured of lungs, livers, spleens, and small intestines taken from representative metastasis-bearing mice. Dashed lines demarcate tumor cell regions. M, metastasis; OP, organ parenchyma; CE, colonic epithelium; BV, blood vessel; LN, lymph node; Intest, small bowel. Arrows in h indicate a front of melanoma cells invading from the outside surface of the small bowel, through the muscularis propria, into the colonic epithelium.
Histological examination of metastasis-bearing lungs revealed that metastatic melanomas frequently encapsulated and grew along the lung vasculature (Figure 2). Metastasis-associated lung vessels invariably demonstrated vasculitis and fibrinoid necrosis. In certain cases, melanoma cells could also be observed within the lumina of blood vessels. In such cases, tumor fibrin thrombi were frequently observed within the vessel lumina. The vascular association of metastatic growths suggested a hematogenous route of dissemination.
The lymph node metastases mentioned above were located adjacent to the growing tumor mass, suggesting that the affected nodes were associated with the lymphatic drainage from the site of implantation. In a subset of mice, we also observed Mel-STR metastases in the axillary lymph nodes of the animals. Furthermore, in cases of axillary lymph node metastasis, the growths were observed exclusively (8/8 cases; Table 1) on the side of the mouse ipsilateral to the original site of injection. This pattern of growth parallels the flow of the lymphatic fluid, suggesting that the melanoma cells reach the axial lymph node through a lymphatic rather than hematogenous route.
While intestinal metastases are rarely observed in cancers that arise outside of the peritoneal cavity, a peculiar aspect of melanoma is its tendency to metastasize to the small bowel 20,21. We found that this behavior is recapitulated in our experimental model, and that 11% of injected mice had visible metastatic nodules on the surface of the small bowel (Figure 2; Table 1). Moreover, in a small number of cases, metastatic melanoma cells could be observed that were invading into the small bowel lumen (Figure 2h). Thus, with high penetrance, the metastatic spectrum of the Mel-STR cells paralleled to a remarkable extent that observed in human melanoma patients.
In stark contrast to these observations, metastatic nodules were very rarely observed in mice bearing subcutaneous fibroblast (BJ-STR) or mammary epithelial cell (MEC-STR) tumors (Table 1) 12,13. In addition, orthotopic injection of MEC-STR cells into the fourth inguinal mammary gland did not enable the resultant tumors to metastasize. Since these primary human cell types were all transformed in vitro through the introduction of an identical set of genes, these observations indicate that the unique ability of transformed melanocytes to efficiently generate metastatic nodules is in part a consequence of their pre-existing differentiation program.
Clonal and Genomic Analysis of Metastatic Nodules
The short latency with which metastases were seen after introduction of the melanoma cells into mice, together with the large number of microscopic and macroscopic metastases observed, suggested that many cells in the polyclonal population of transformed melanocytes already possessed the ability to form metastases prior to their introduction into murine hosts. Nonetheless, it remained formally possible that metastasis only occurred in vivo after rare metastasis-prone cells arose within the primary tumor cell population. To address this issue, we undertook a clonal analysis of the disseminated metastases arising in single animals in vivo.
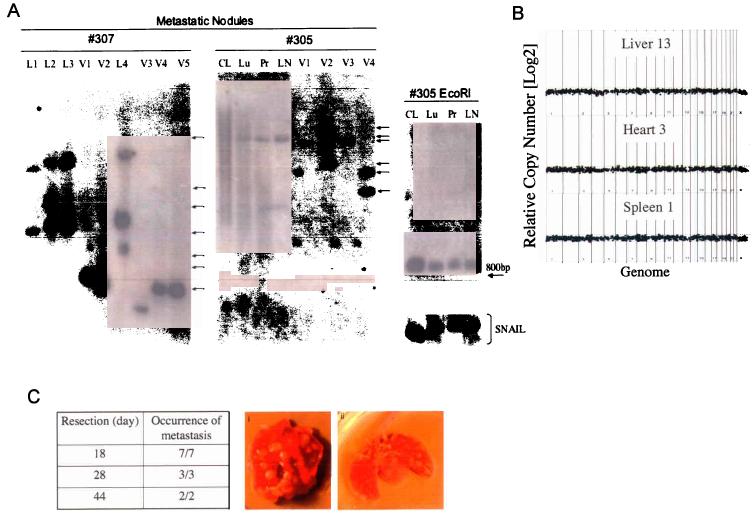
We dissected individual metastatic nodules arising in single Mel-STR-injected mice and dissociated the tissue fragments into single cells by enzyme treatment. We then plated the heterogeneous cell populations (comprised of both metastatic human tumor cells and murine stromal cells) on tissue culture dishes and selected for the melanoma cells using a drug marker that had introduced by retroviral infection into the tumor cells during the initial transformation process. This procedure allowed us to obtain pure tumor cell populations, each of which was derived from a single metastatic nodule in vivo. Because retrovirus infection leads to quasi-random insertion of proviral sequences into host cell genomes, we were able to use Southern blot analysis22 to identify the provirus integration sites in the ancestral clone(s) from which the various Mel-STR populations were derived. We therefore isolated genomic DNA from each of the metastatic nodule-derived tumor cell populations and performed Southern blot analysis, probing for GFP sequence.
The continuous distribution in the sizes of cleaved DNA fragments derived from the parental transformed cells demonstrated the extensive polyclonality of the Mel-STR population immediately prior to injection in vivo (Figure 3a). Upon examination of metastatic Mel-STR nodules, at least 10 distinct cell clones were present among the metastases of two animals that had been injected with the same polyclonal population of transformed melanocytes (Figure 3a, #305,7). Since these various clones were identified through examination of a small number of nodules relative to the total metastastic burden, it is apparent that a large number of additional cell clones were present among the metastases of these animals. In support of this notion, genomic DNA obtained directly from the entire lung of one metastasis-laden animal demonstrated a continuous distribution of DNA fragments, indicating the presence of numerous distinct clones of metastatic tumor cells in the lung (Figure 3a, #305Lu, Pr). This continuous distribution of integrant sizes was reminiscent of that observed in the starting population of injected cells, indicating extensive clonal heterogeneity among the metastatic Mel-STR cells.
Figure 3.
Primary Mel-STR melanomas rapidly seed distinct metastatic clones to secondary organs. a, Southern blot analysis of the metastases arising in single Mel-STR-injected mice. Lanes were loaded with BamHI-cleaved genomic DNA extracted either from organs (#305Lu, Pr, LN), or from the parental Mel-STR cell line (Parental, #305CL) immediately prior to injection, or from Mel-STR cells that were isolated from individual metastatic nodules in vivo and expanded in culture (L1-4, V1-5). Arrows indicate independent insertion sites. BamHI cleaves once within the vector provirus, upstream of the probed retroviral GFP sequence. GFP probe specificity in lanes exhibiting smears was confirmed by digesting the extracted DNAs with EcoRI, which cleaves twice within the vector provirus and collapses the smears into a single internal 880bp fragment independent of provirus integration site. Genomic integrity in samples exhibiting smears was also confirmed by probing EcoRI-digested DNA for an endogenous 1 kb region upstream of the SNAIL gene, revealing a single intact 10.7 kb band without detectable degradation. HindIII digestion of the extracted DNAs recapitulated the results observed with BamHI cleavage (data not shown). L, lung; V, liver; LN, lymph node; CL, parental Mel-STR cell line; Pr, primary tumor lysate; Lu, whole-lung lysate. b, Whole genome array CGH performed on DNA isolated from metastatic nodules isolated from the liver, spleen, and heart of a single metastasis-laden animal. No significant genome copy number alterations are observed in any of the metastatic nodules relative to the parental primary tumor obtained from the same animal. Data are displayed as log2(CY3/CY5 intensity ratios) from pter to qter with chromosome 1 on the left and chromosomes 22 and X on the right. c, Mice were injected subcutaneously with Mel-STR cells and the primary melanomas resected at the indicated times. All mice were sacrificed at 48 days post-injection. The fraction of mice with visible lung metastases upon necropsy is indicated. (i, ii) are representative melanoma-burdened lungs from mice whose primary melanomas were excised at day 18 or day 28 after injection, respectively. Arrows indicate individual metastatic nodules.
In addition to the clonal analyses, we used array CGH to directly interrogate the genomes of metastatic nodules to determine whether alterations in DNA copy number were observed relative to the primary tumors. No significant alterations in DNA copy number were observed in six independent nodules isolated from three distinct organs, relative to the primary tumor DNA. This result indicates that within the level of resolution afforded by the array CGH assay, genomic alterations sustained at the site of primary tumor growth were not responsible for enabling Mel-STR cell metastasis.
Mel-STR Tumors Rapidly Seed Metastatic Cells to Secondary Organs
To further examine the metastatic phenotype of the transformed melanoma cells, we determined the stage of tumor growth at which metastatic Mel-STR cells were seeded into secondary organs. Accordingly, we surgically resected primary Mel-STR tumors from age-matched cohorts of mice at specific times after injection (18, 28 or 44 days), and examined the mice for metastatic growths at the endpoint of the experiment (44 days). To ensure full resection of the primary melanomas, we removed a 0.5cm margin of skin during the surgery and confirmed the absence of melanoma cells by histological examination of the adjacent skin (data not shown). We found that even in mice in which the primary tumor was removed just 18 days after injection (mean wt. = 400mg), numerous metastatic nodules could be seen in the lungs of the animals (5/5) at the endpoint of the experiment (Figure 3b). Similarly, mice whose primary tumors were resected 28 days after injection (mean wt.= 600mg) also developed extensive lung metastases (4/4).
Taken together with the results of the clonal analyses, these observations enabled us to estimate a lower bound on the frequency with which metastasis-enabling alterations must be occurring in vivo if the Mel-STR cells must acquire the ability to metastasize subsequent to their introduction in vivo. This frequency, ∼1/100, is several orders of magnitude more frequent than the estimated frequencies per cell generation of gene mutation (see Supplementary Note online). Thus, taken in conjunction with the array CGH data, these data suggest that it is unlikely that additional genetic alterations beyond those initially introduced during the transformation protocol were required in vivo to enable the injected Mel-STR cells to metastasize.
The Neural Crest Cell Factor Slug is Expressed in Melanocytes Prior to Transformation
The results above indicated that the differentiation program of melanocytes could cooperate with oncogenic lesions to uniquely predispose their transformed counterparts to forming tumors that metastasize, when compared to fibroblasts and epithelial cells transformed with identical genes. Since the process of cancer cell invasion bears numerous cellular and molecular similarities to neural crest cell migration in the developing embryo, and given the known derivation of dermal melanocytes from the neural crest, we hypothesized that elements of the motility-associated molecular program mediating neural crest cell migration may contribute to melanoma's metastasis.
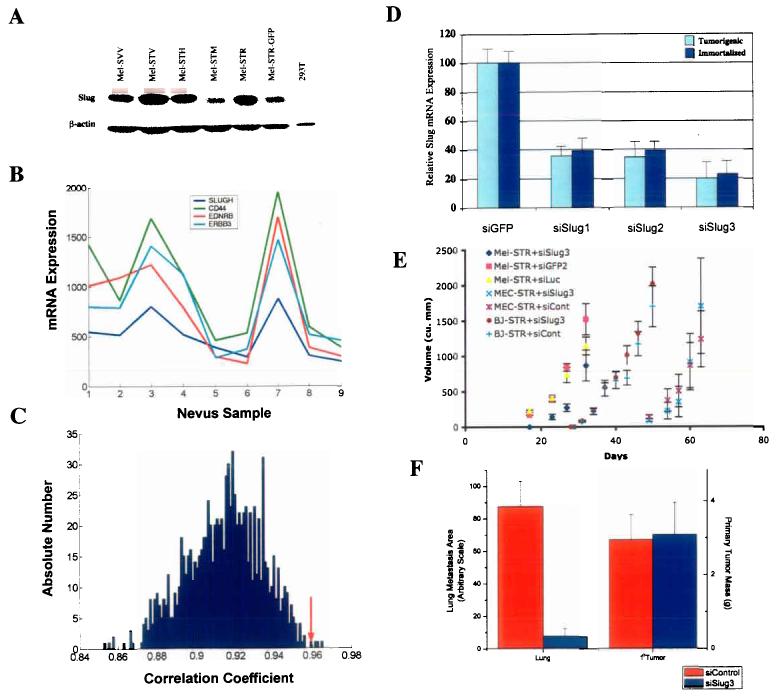
Genes of the Snail-superfamily have previously been implicated in both promoting cancer cell invasiveness 23 and governing neural crest cell migration 24. While Snail was not significantly expressed in the Mel-STR cells, the related Slug transcription factor was expressed at both the mRNA and protein levels (data not shown; Figure 4a). Interestingly, Slug expression was also observed in non-tumorigenic, immortalized melanocytes. This finding led us to use expression arrays to examine Slug mRNA levels in benign nevus samples obtained from human patients. We observed that benign nevi expressed Slug mRNA, and moreover, expression of the Slug transcript in these samples correlated strongly and significantly (p < 0.01) with genes known to be essential for neural crest cell and/or melanoblast migrations during development (Figure 4b,c). Thus, expression of the neural crest migration-associated genes Slug (SLUGH), Endothelin Receptor B (EDNRB), ErbB3, and CD44 was highly correlated in adult nevus samples 24-28. These findings indicated that components of an embryonic differentiation program involved in neural crest cell motility and migration were expressed in benign melanocytic lesions prior to neoplastic transformation.
Figure 4.
Suppression of Slug expression inhibits melanoma metastasis in vivo. a, Western blot analysis of Slug protein expression. Slug protein levels were examined in melanocyte cell lines engineered with LT (SVV), LT, hTERT & HGF (STH), LT, hTERT & TPR-Met (STM), LT, hTERT & RasV12 (STR), or LT, hTERT, RasV12 & GFP (STR-GFP), and in transformed kidney cells (293T). b, mRNA expression levels as determined by microarray analysis. Transcript levels of SLUGH, CD44, EDNRB and ERBB3 were correlated in 9 human nevus samples. c, Reference statistics for CD44 (35). 1000 correlation coefficients were obtained by randomly permuting the SLUGH expression profile template. Shown is a histogram plot of correlations corresponding to the rank of CD44 in the SLUGH neighborhood list. The correlation coefficient of CD44 with the non-permuted SLUGH template is indicated by a red arrow (0.958). There are only 4 permuted correlations larger than this value, resulting in strong statistical significance (p=0.004). d, Quantitative RT-PCR performed on immortalized and transformed melanocyte populations stably expressing either siSlug (siSlug1, siSlug2, siSlug3) or control (siGFP2) siRNAs. Data were normalized to GAPDH expression and plotted as a percentage relative to Slug transcript levels in siGFP2-expressing cells. e, In vivo tumor growth curves of Mel-STR, MEC-STR, and BJ-STR cells stably expressing either siSlug3 or control (siGFP2, siLuc) siRNA contructs. f, Quantification of lung metastasis burden in siSlug3- or control siRNA (siGFP2, siLuc)- Mel-STR melanoma lines. Random lung sections were stained with a human-specific vimentin antibody and the area of brown color corresponding to stained Mel-STR cells was quantified using NIH Image software (35).
Slug is Essential for Mel-STR Melanoma Metastasis in vivo
We utilized retrovirus siRNA-mediated inhibition to examine whether the expression of Slug, a master regulator of neural crest cell specification and migration, was in fact contributing to the metastatic ability of the Mel-STR melanoma cells. Accordingly, we designed three independent retrovirus siRNAs directed against SLUGH (siSlug1-3) and stably introduced them into the Mel-STR cells. As two independent controls, we also introduced siRNAs against the GFP (siGfp2) and luciferase (siLuc) proteins into the same cell population. Using quantitative RT-PCR, we determined that siSlug1 and siSlug2 reduced endogenous SLUGH mRNA levels by ∼66%, whereas siSlug3 reduced endogenous mRNA levels by 80% (Figure 4d). Essentially identical levels of relative SLUGH mRNA transcript reduction were observed when these same vectors were introduced into the Mel-STV cells (Figure 4d).
To test whether a reduction in Slug levels affected melanoma growth or progression, we introduced the Mel-STR+siSlug3 cells, as well as two control lines (Mel-STR +siGfp2 or +siLuc), subcutaneously into immunodeficient mice. SLUGH inhibition resulted in a slight decrease in primary tumor growth rates when compared to the control melanoma lines (Figure 4e). No apparent differences were observed in the histologies of the primary tumors that developed in the Mel-STR+siSlug3 line versus control lines (data not shown).
In contrast, there was a marked reduction in the incidence of metastasis when primary tumor cells experiencing Slug inhibition were seeded in host mice. Metastatic lung tumor burden in the Mel-STR+siSlug3 tumors was reduced by more than 10-fold when compared to control siGfp2 or siLuc tumors (Figure 4f). This was evaluated by staining lung tissue sections for the Mel-STR-specific marker human vimentin, and subsequently quantifying the stained area in random fields with NIH Image software (see Supplementary Fig.1 online). Importantly, to control for differences in primary tumor growth rates, mice were sacrificed at times such that the average primary tumor burden in the two experimental groups was comparable (Figure 4f). These results provide strong indication that Slug is an important component of the metastatic program in the Mel-STR melanoma cells.
Discussion
While a fundamental feature of human melanoma is its tendency to metastasize to numerous organs throughout the body, very few animal models recapitulate this essential aspect of the disease. We have demonstrated that human dermal melanocytes, transformed by the introduction of the SV40ER, hTERT, and RasG12V genes, can form primary tumors that are invasive and highly metastatic to secondary sites in the body. Moreover, the anatomical sites of metastasis exhibited by the melanoma cells created in this manner are analogous to those observed in human patients. To our knowledge, this is the first human melanoma model that exhibits multi-organ metastases from a subcutaneous site.
The introduction of the identical set of genes into human epithelial and fibroblast cell types has resulted, time and again, in localized tumor formation without apparent metastatic dissemination. Importantly, implementation of an isogenic transformation protocol controls for the possibility that oncogene-specific differences are responsible for the various cancer phenotypes observed upon experimental conversion of primary human epithelial, fibroblast, and melanocyte cell types to a neoplastic state. Our results therefore indicate that the rapid progression of human melanomas to a metastatic state can be attributed in part to lineage-specific factors associated with the melanocyte differentiation program.
The commonly observed presence of nevus cell aggregates in human lymph nodes29-31 supports the notion that normal melanocytes prior to transformation express a differentiation program that harbors factors capable of promoting migratory behaviors. These are characterized by clusters of histologically normal melanocytes detected within the capsule of lymph nodes, particularly those from the axillary, cervical, and inguinal regions. This unique clinical observation, made in the absence of any malignant histology, suggests that dermal melanocytes indeed possess latent migratory abilities.
The present results validate a previously untested speculation made some years ago (9) that the particularly aggressive nature of melanomas is ascribable to the derivation of dermal melanocytes from the embryonic neural crest. By controlling for oncogenespecific differences, our experiments demonstrate that the differentiation program of normal melanocytes can collaborate with oncogenic lesions to predispose their transformed derivatives to forming invasive and metastatic tumors. This notion is supported by analysis of microarray data from benign human nevus tissues, which reveal co-regulated expression of genes known to be important for neural crest cell migrations during development, prior to neoplastic transformation of melanocytes. Moreover, one of these genes, encoding the neural crest cell transcription factor Slug, plays an important functional role in the metastatic spread of the Mel-STR melanoma cells.
The present results do not address whether Slug expression is sufficient to enable melanoma metastasis. The multi-step nature of metastasis makes it unlikely that a single factor can orchestrate this complex process. Moreover, the fact that Slug is expressed in benign melanocytic lesions implies that acquisition of its expression following transformation is not the single pathogenetic event that enables metastasis. Nevertheless, the preexisting expression of Slug in benign precursor lesions may reduce the number of additional rate-limiting alterations required to enable metastasis following melanocyte transformation, when compared to cancers arising from other cell types. These experiments indicate that lineage-specific factors associated with melanocyte differentiation can significantly impact the tumorigenic and metastatic phenotype observed following neoplastic transformation.
Materials and Methods
Tissue culture and Cells
Primary human melanocytes, derived from neonate foreskin, were obtained from Clonetics and the Yale Cell Culture Core Facility (New Haven, CT). Primary melanocytes were maintained in MGM-3 medium (Clonetics). Immortalized and transformed melanocyte cell lines were grown in DMEM, with 5% FBS (hereafter referred to as Mel-STR medium). Foreskin fibroblast (BJ)-derived cell lines were previously described 12 and maintained in DMEM:M199 (4:1), supplemented with 10% heat-inactivated fetal calf serum. Human mammary epithelial (HMEC)-derived cell lines were generated as previously described 13 and grown in DMEM:F12 (1:1) supplemented with EGF (10 ng/ml), insulin (10 μg/ml), hydrocortisone (1 μg/ml), and 5% calf serum.
To generate the immortalized melanocyte cell line Mel-ST, primary melanocytes were infected with pBABE-LTg-zeo, and pBABE-hTERT-hygro. These immortalized melanocytes were subsequently infected with pBABE-RasV12-puro, pBABE-TPR-metpuro, pBABE-HGF-puro, or pWZL-RasV12-blast retroviruses, to generate Mel-STR, -STM, and –STH cells, respectively. GFP-labeled cells were infected with pWZL-GFP-blast. For stable siRNA cell lines, Mel-ST and Mel-STR (pWZL-RasV12-blast) were infected with pLKO-puro shRNA vectors.
Retrovirus vectors and infections
Retroviral constructs for human HGF and mutant c-Met receptor (TPR-met) were generated by cloning the full-length cDNA of human HGF or TPR-met (kindly provided by G. Vande Woude) into the pBabe-puro vector system 32. Other pBABE-based vectors included LTg-zeo 13, hTERT-hygro 33, and RasV12-puro (S. Lowe). PWZL-based vectors 32 used were RasV12-blast (subcloned from pBABE-RasV12-puro using BamHI/SalI sites) and GFP-blast (gift from A. Orimo). For siRNA-mediated inhibition, three different siRNA sequences against human Slug (5′-AACTGGACACACATACAGTG-3′, 5′-GAGGAAAGACTACAGTCCAAG-3′, 5′-CAGACCCATTCTGATGTAAAG-3′) were cloned into the pLKO lentiviral vector system (kindly provided by S.A.Stewart and W.C.Hahn). SiRNA sequences against GFP and luciferase were described 34.
Amphotropic retroviruses were created by transient cotransfection of DNA into 293T cells. 6cm plates were cotransfected with 1 μg of appropriate packaging plasmid (pCL-10A1 (Imgenex) for moloney-based infections and DHR8.1:VSVG (9:1) for lentiviral infections) and 1 μg of carrier vector using FuGene 6 (Roche). Viral supernatants were harvested at 48 hrs post-transfection, passed through a 0.4μm filter, and supplemented with 8 μg/ml polybrene (Sigma) prior to infection.
RT-PCR
Total RNA was isolated from tumor tissues or cell lines using TRIZOL reagent (Invitrogen), following manufacturer's protocols. Quantitative real time RT-PCR analysis was performed using the iCycler apparatus (Bio-Rad) and SYBR Green PCR Core Reagents System (Perkin-Elmer Applied Biosystems). Results were evaluated using the iCycler IQ Real Time Detection System Software (Bio-Rad). Data were normalized relative to GAPDH expression. Primers used were 5′-ATTCGGGAAAGGTGAAGAGG-3′ and 5′-CAGCTGTTCTGCTGTGAAGG-3′ (Pax3), 5′-ATTCGGACCCACACATTACC-3′ and 5′-GGTTTTGGAGCAGTTTTTGC-3′ (Slugh), 5′-CTCTTTCCTCGTCAGGAAGC-3′ and 5′-GGACAGAGTCCCAGATGAGC-3′ (Snail), 5′-GCTTCCAGACTGTGGACTCG-3′ and 5′-CCGTACTGTCCGGAAGACAT-3′ (ErbB3), 5′-GACCCCTTCATTGACCTCAAC-3' and 5'-CTTCTCCATGGTGGTGAAGA-3' (GAPDH).
Western blotting
Western blotting was performed using standard protocols. Primary antibodies used were against Large T antigen (Pab101, Santa Cruz), Ras (C-20; Santa Cruz), Slug (G-18; Santa Cruz), and ß-actin (Abcam). Goat anti-mouse (115-035-146) and goat antirabbit (111-035-144) HRP-conjugated secondary antibodies were purchased from Jackson Immunoresearch.
Soft agar assays
Soft agar assays were performed as described 35.
Animals and surgery
Athymic nude mice were purchased from Taconic Laboratories (NCR nude, nu/nu). NOD/SCID mice were bred and maintained in-house. All mice were housed in a specific pathogen-free facility and were administered autoclaved food and water ad libum. For in vivo tumor experiments, 106 cancer cells were resuspended in 200ul of medium and injected subcutaneously into 10-12wk old male NOD/SCID mice or 8wk old irradiated athymic nude mice. Nude mice received 400 rad of γ-radiation using a dual 137Cesium source one day prior to injection. Mice were monitored bi-weekly and tumor diameters were measured using precision calipers. For in vivo metastasis experiments, primary tumors were permitted to grow for ∼7 weeks or until they reached a diameter of 2cm. For tumor resection experiments, primary tumors were permitted to grow for 18 or 28 days, at which point mice were anesthetized using avertin (0.4-0.7 mg/g body wgt) and the tumors excised under a dissection microscope using DeWecker microdissecting spring scissors and a hand held cautery unit. Mice were monitored daily thereafter until the endpoint of the experiment.
Southern blot analysis
Fresh tumor tissue or metastatic nodules were microdissected from organs of individual mice at necropsy under a dissection microscope (see Supplementary Fig.3 online). Tissue fragments were minced and digested for 3 hours with agitation at 37°C, in culture medium (DME+10%FBS) containing collagenase (1 mg/ml; Boehringer Mannheim), hyaluronidase (125 U/ml; Sigma) and HEPES buffer (10 mM; Gibco). Dissociated cells were plated in Mel-STR medium and subjected to puromycin drug selection (1 ug/ml) for one week. Genomic DNA was isolated from the nodule-derived tumor cell populations and purified by phenol/chloroform extraction. 15μg of DNA was digested with BamHI, and Southern blotting performed as described 36. In the case of the lung of mouse #305, gDNA from the entire metastasis-burdened organ was isolated as described above (in the absence of microdissection or culture), and 30ug loaded for blotting. As a probe template, an 800bp EcoRI fragment from pWZL-GFP was used to synthesize 32P-labeled probe using the Rediprime II kit (Amersham).
Array CGH
Whole genome CGH array comprised of 1860 PAC and BAC clones were prepared as described37. Labeling of melanoma genomic DNA, array CGH hybridization and image processing were performed as previously described38 with slight modification. Briefly, 500 ng each of cell line and normal female genomic DNA were fragmented by DPNII digestion, labeled by random priming (Bioprime Labeling System, Invitrogen, Carlsbad, CA) with CY3- and CY5-dUTP (Amersham, Piscataway, NJ) respectively, coprecipitated with 50 □g of human cot-1 DNA (Life Technologies) and resuspended in 60 □l hybridization buffer (50% formamide, 10% dextran sulfate, 2X SSC, 4% SDS, 200□g yeast tRNA). This mixture was denatured at 75°C for 10 min followed by incubations at 37°C for 60 min. Just prior to hybridization, array slides were UV cross-linked. A rubber cement dam was placed around each array, hybridization mix was added and the slide was placed in a plastic slide holder, prewarmed to 37°C, containing 200 □l of wash buffer (50% formamide, 2X SSC) to prevent evaporation. Hybridization was carried out at 37°C for 48-72 hours on a gently rocking platform. Following hybridization, slides were immersed for 15 min at 48°C in wash buffer, followed by washes at 48°C in 2X SSC, 0.1% SDS for 30 min, and PN buffer (0.1 M sodium phosphate buffer, 0.1% NP40, pH 8.0) at room temperature for 10 min. Slides were then rinsed in 2X SSC and mounted with 4', 6-diamidino-2-pheylindole (DAPI, 0.5 □M) in 90% glycerol, 10% PBS for imaging using a 16-bit CCD camera. The CY3, CY5 and DAPI images were segmented and analyzed to determine CY3/CY5 ratios for each array element using custom software as described39. Data are displayed as log2(CY3/CY5 intensity ratios) from pter to qter with chromosome 1 on the left and chromosomes 22 and X on the right based on the position of the BAC on the July, 2003 freeze of the UC Santa Cruz genome sequence assembly (http://genome.ucsc.edu).
Visualization of GFP-labeled metastatic nodules
Whole-organ bright-field and fluorescence images were captured using a Leica MZ 9.5 Dissection Microscope.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissues. Five micron sections were de-paraffinized, rehydrated through a graded alcohol series and subjected to antigen retrieval procedures for immunohistochemistry, as described previously 40. Sections were incubated in mouse monoclonal antibodies against MART-1 (Ventana), pan-cytokeratin (Ventana), human E-cadherin (1:50 Santa Cruz), human β-catenin (1:50, Santa Cruz) or Large T antigen (Pab101, Santa Cruz 1:50). Immunocomplexes were visualized using the ABC peroxidase method (Vector Laboratories, Burlingame, CA). Sections were counterstained with hematoxylin.
Quantification of lung metastasis
Entire lungs were removed from mice bearing subcutaneous melanomas and fixed in 10% neutral-buffered formalin for at least 12 hours. Immunohistochemistry was performed on lung tissues with a mouse monoclonal human-specific vimentin antibody, which specifically stains the human melanoma cells (see Supplementary Fig.1 online). The area of melanoma burden was quantified using NIH Image software in at least 2 separate hotspot fields per lung, with at least 10 lungs per cell line. Software procedures were detailed previously 41.
RNA extraction and microarray hybridization
Benign nevi were surgically isolated from normal human patients and flash frozen. mRNA was isolated using CsCl gradient centrifugation, fluorescently labeled and subsequently hybridized on an Affymetrix U133A array (22283 genes), following the manufacturer's protocol.
Neighborhood analysis
Neighborhood analysis of SLUGH was performed in a dataset that consists of 9 nevi samples. Genes significantly correlated in their expression pattern with SLUGH were selected by non-parametric permutation testing, using a previously described procedure 42. First, all 22283 genes were ranked according to their correlation coefficients with respect to the SLUGH expression profile. 1000 random permutations of the SLUGH profile were then generated, and for each permutation the 22283 genes were re-ranked according to correlation with the shuffled SLUGH profile. A histogram of the distribution of correlation coefficients among 1000 permutations for the CD44 gene is shown in Figure 4c. For any gene with a given rank in the original SLUGH correlation list, one determines how many of the 1000 permuted correlation coefficients corresponding to the same rank (but not necessarily the same gene) are greater than or equal to the original, non-permuted correlation coefficient. A p-value can then be assigned by dividing this quantity by the total number of permutations. This procedure for SLUGH in the 9 nevus samples dataset yielded 235 genes with p<0.05, of which 31 had p<0.01. Supplementary Table 1 online lists all 235 genes with their correlations and corresponding p-values.
Supplementary Material
Acknowledgements
We are grateful to Drs. Scott Dessain, Homayoun Vaziri, Gaorav Gupta, Sheila Stewart, William Hahn, Akira Orimo, Samo Godar, Ittai Ben-porath, Jing Yang, and Tamer Onder for helpful discussions and suggestions during the course of this work. We thank Dr. Todd Golub for helpful discussions regarding microarray experiments. We also thank Tony Chavarria for assistance with animal husbandry, and Donna LaCivita for primary melanocyte isolation. P.B.G. is supported by a US Army Pre-doctoral Breast Cancer Fellowship (DAMD17-02-1-0468). This work was supported by NIH/NCI grant RO1CA78461-05 (R.A.W.). R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Foundation Cancer Research Professor.
References
- 1.Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179–85. doi: 10.1111/j.1365-2133.2004.05708.x. [DOI] [PubMed] [Google Scholar]
- 2.Beddingfield FC., 3rd The melanoma epidemic: res ipsa loquitur. Oncologist. 2003;8:459–65. doi: 10.1634/theoncologist.8-5-459. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova J, et al. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res. 1996;56:5475–83. [PubMed] [Google Scholar]
- 4.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9:6483–8. [PubMed] [Google Scholar]
- 5.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3:559–70. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 6.Tietze MK, Chin L. Murine models of malignant melanoma. Mol Med Today. 2000;6:408–10. doi: 10.1016/s1357-4310(00)01781-0. [DOI] [PubMed] [Google Scholar]
- 7.Shaw HM, McCarthy WH, McCarthy SW, Milton GW. Thin malignant melanomas and recurrence potential. Arch Surg. 1987;122:1147–50. doi: 10.1001/archsurg.1987.01400220057011. [DOI] [PubMed] [Google Scholar]
- 8.Corsetti RL, Allen HM, Wanebo HJ. Thin < or = 1 mm level III and IV melanomas are higher risk lesions for regional failure and warrant sentinel lymph node biopsy. Ann Surg Oncol. 2000;7:456–60. doi: 10.1007/s10434-000-0456-4. [DOI] [PubMed] [Google Scholar]
- 9.Bedrosian I, et al. Incidence of sentinel node metastasis in patients with thin primary melanoma (< or = 1 mm) with vertical growth phase. Ann Surg Oncol. 2000;7:262–7. doi: 10.1007/s10434-000-0262-z. [DOI] [PubMed] [Google Scholar]
- 10.Nesbit M, S. V, Herlyn M. Biology of melanocytes and melanoma. In: Balch C, A. H, Sober A, et al., editors. Cutaneous Melanoma. Quality Medical, St. Louis; 1998. p. 463. [Google Scholar]
- 11.Bronner-Fraser M. Neural crest cell migration in the developing embryo. Trends Cell Biol. 1993;3:392–7. doi: 10.1016/0962-8924(93)90089-j. [DOI] [PubMed] [Google Scholar]
- 12.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 13.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundberg AS, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–86. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 15.Rich JN, et al. A genetically tractable model of human glioma formation. Cancer Res. 2001;61:3556–60. [PubMed] [Google Scholar]
- 16.Li G, et al. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene. 2001;20:8125–35. doi: 10.1038/sj.onc.1205034. [DOI] [PubMed] [Google Scholar]
- 17.Cruz J, Reis-Filho JS, Silva P, Lopes JM. Expression of c-met tyrosine kinase receptor is biologically and prognostically relevant for primary cutaneous malignant melanomas. Oncology. 2003;65:72–82. doi: 10.1159/000071207. [DOI] [PubMed] [Google Scholar]
- 18.Natali PG, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. Br J Cancer. 1993;68:746–50. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano S, Ponzetto C, Di Renzo MF, Cooper CS, Comoglio PM. Tyrosine kinase receptor indistinguishable from the c-met protein. Nature. 1989;339:155–6. doi: 10.1038/339155a0. [DOI] [PubMed] [Google Scholar]
- 20.Phillips DL, Benner KG, Keeffe EB, Traweek ST. Isolated metastasis to small bowel from anaplastic thyroid carcinoma. With a review of extra-abdominal malignancies that spread to the bowel. J Clin Gastroenterol. 1987;9:563–7. doi: 10.1097/00004836-198710000-00017. [DOI] [PubMed] [Google Scholar]
- 21.Elsayed AM, Albahra M, Nzeako UC, Sobin LH. Malignant melanomas in the small intestine: a study of 103 patients. Am J Gastroenterol. 1996;91:1001–6. [PubMed] [Google Scholar]
- 22.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–17. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 23.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–8. [PubMed] [Google Scholar]
- 24.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 25.Lee HO, Levorse JM, Shin MK. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259:162–75. doi: 10.1016/s0012-1606(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda K, et al. Expression of CD44H in the cells of neural crest origin in peripheral nervous system. Neuroreport. 1996;7:1713–6. doi: 10.1097/00001756-199607290-00003. [DOI] [PubMed] [Google Scholar]
- 27.Britsch S, et al. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998;12:1825–36. doi: 10.1101/gad.12.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense slug RNA. Dev Biol. 1999;213:101–15. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- 29.Subramony C, Lewin JR. Nevus cells within lymph nodes. Possible metastases from a benign intradermal nevus. Am J Clin Pathol. 1985;84:220–3. doi: 10.1093/ajcp/84.2.220. [DOI] [PubMed] [Google Scholar]
- 30.Bortolani A, Barisoni D, Scomazzoni G. Benign “metastatic” cellular blue nevus. Ann Plast Surg. 1994;33:426–31. doi: 10.1097/00000637-199410000-00013. [DOI] [PubMed] [Google Scholar]
- 31.Johnson WT, Helwig EB. Benign nevus cells in the capsule of lymph nodes. Cancer. 1969;23:747–53. doi: 10.1002/1097-0142(196903)23:3<747::aid-cncr2820230331>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Counter CM, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cifone MA, Fidler IJ. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci U S A. 1980;77:1039–43. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuperwasser C, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–9. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgson G, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–64. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 38.Snijders AM, et al. Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene. 2003;22:4370–9. doi: 10.1038/sj.onc.1206482. [DOI] [PubMed] [Google Scholar]
- 39.Jain AN, et al. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–32. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuperwasser C, Pinkas J, Hurlbut GD, Naber SP, Jerry DJ. Cytoplasmic sequestration and functional repression of p53 in the mammary epithelium is reversed by hormonal treatment. Cancer Res. 2000;60:2723–9. [PubMed] [Google Scholar]
- 41.Petersen SL, Gardner E, Adelman J, McCrone S. Examination of steroid-induced changes in LHRH gene transcription using 33P-and 35S-labeled probes specific for intron 2. Endocrinology. 1996;137:234–9. doi: 10.1210/endo.137.1.8536618. [DOI] [PubMed] [Google Scholar]
- 42.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 43.Chang YS, et al. Mosaic blood vessels in tumors: frequency of cancer cells in contact with flowing blood. Proc Natl Acad Sci U S A. 2000;97:14608–13. doi: 10.1073/pnas.97.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Materials and methods are available as supporting material on Nature Genetics Online.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.